Abstract
AMPA receptors mediate fast excitatory synaptic transmission and are essential for synaptic plasticity. ANQX, a photoreactive AMPA receptor antagonist, is an important biological probe used to irreversibly inactivate AMPA receptors. Here, using X-ray crystallography and mass spectroscopy, we report that ANQX forms two major products in the presence of the GluR2 AMPAR ligand-binding core (S1S2J). Upon photostimulation, ANQX reacts intramolecularly to form FQX or intermolecularly to form a covalent adduct with Glu705.
Introduction
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (AMPARsa) are a subtype of glutamate-gated ion channels that mediate most fast excitatory synaptic transmission by ensuring rapid responses to synaptically released glutamate.1–3 In addition, activity-dependent changes in the number of AMPARs at synapses modulates the strength of synaptic transmission, an essential component of the mechanism underlying various forms of synaptic plasticity including learning and memory.4–8
AMPARs are composed of four modular subunits (GluR1–4 or GluRA–D), each consisting of an amino-terminal domain (NTD) that modulates receptor assembly, a ligand-binding domain (LBD) that gates the pore of the receptor, three transmembrane segments (M1, M3, M4), a reentrant loop (M2) that lines the pore of the channel, and a cytoplasmic C-terminal domain that influences receptor trafficking (Figure S1, Supporting Information).1–3
High-resolution crystal structures of an engineered ligand-binding core (S1S2J) with several bound ligands have provided insight into the structure and function of full-length receptors.9,10 Gouaux and co-workers provided the first high-resolution structures of the GluR2 AMPAR ligand-binding core (Figure S1, Supporting Information).11–13 These structures revealed that the ligand-binding core, formed from two discontinuous polypeptide segments (Figures S1 and S2, Supporting Information), adopts a clamshell-like shape that is open in two states, unliganded (apo) and with a competitive antagonist bound. The clamshell is closed with agonist bound. Notably, structurally related ligands within a given class produce distinct degrees of clamshell closure.14–17 Coupled with electrophysiological experiments carried out on full-length receptors, these studies suggested that the degree of closure affects the conductance (ion permeation) of the channel, providing a model for channel gating. In addition to modulating channel biophysics, ligand binding also appears to influence the trafficking of AMPARs. For example, both agonists and antagonists have been shown to induce the internalization of AMPARs from neuronal plasma membranes.18 Although the mechanistic basis for this effect is not understood, it is likely that conformational changes within the ligand-binding domain are translated to the intracellular C-terminal domains, which play a critical role in receptor trafficking.
Quinoxaline-2,3-diones are a major class of competitive AMPAR antagonists, frequently used in studies focused on characterizing the activity of AMPARs.19 Key members of this family of antagonists are 6,7-dinitroquinoxaline-2,3-dione (DNQX) and 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX). Recently we reported the development of ANQX, a new member of the family of quinoxaline-2,3-diones containing an ortho-nitrophenylazide, which irreversibly inhibits AMPARs in the presence of ultraviolet light and provides a means of rapidly inactivating receptors expressed on the surfaces of neurons.20–22 In the absence of ultraviolet light, ANQX reversibly inhibits AMPARs. The mechanism by which ANQX irreversibly antagonizes AMPARs has not been previously described. Here we report that ANQX forms two products upon photolysis in the presence of the GluR2 ligand-binding core (S1S2J). Following irradiation with ultraviolet light, ANQX loses dinitrogen to form a highly reactive nitrene that either reacts intramolecularly to form [1,2,5]oxadiazolo[3,4-G]quinoxaline-6,7(5H,8H)-dione 1-oxide (FQX) or intermolecularly to form a covalent adduct with Glu705 located in the binding pocket. The high-resolution crystal structure of FQX bound to the S1S2J revealed that FQX binds in the same orientation and promotes the same degree of closure of the S1S2J around the ligand as the previously reported structures complexed with DNQX and CNQX.12,23 Together, these data indicate a common orientation of quinoxaline-2,3-diones within the AMPAR LBD and reveal the mechanism of receptor photoinactivation with ANQX.
Results
ANQX is a photoreactive quinoxaline-2,3-dione that, upon irradiation with ultraviolet light, loses dinitrogen to form a highly reactive nitrene that undergoes either intra- or intermolecular reactions. We identified the products of both reactions using X-ray crystallography and mass spectrometry. Irradiation of S1S2J-ANQX cocrystals provided a high-resolution structure of the intramolecular reaction product, FQX, reversibly bound to S1S2J. Irradiation of S1S2J in solution in the presence of ANQX yielded sufficient quantities of the intermolecular reaction product to identify GluR705 as the site of cross-linking.
Crystal Structure of the S1S2J–FQX Complex
The high-resolution X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with FQX was solved at 1.87 Å resolution (Figure 1, Table 1 of Supporting Information). The complex crystallized in the orthorhombic P212121 space group with four protein monomers in the asymmetric unit (Table 1 of Supporting Information). Only two of the monomers are related by noncrystallographic 2-fold symmetry axis. A monomer of the S1S2J dimer complexed with DNQX was used as the search model for molecular replacement. Pairwise superposition of the Cα-atoms of the four molecules yielded pairwise rmsd values ranging between 0.21–0.41 Å.
Figure 1.
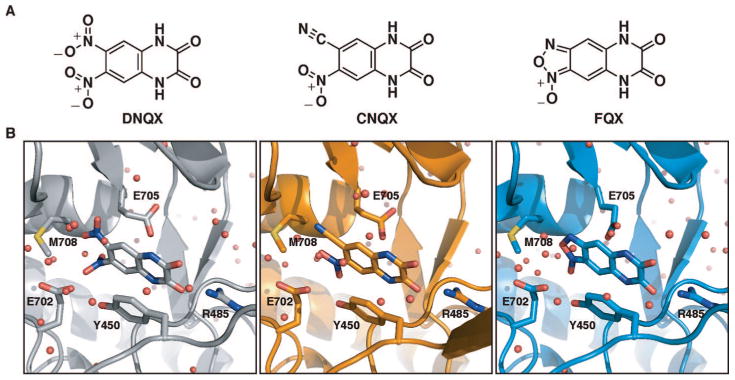
Structural formulas and orientation of the quinoxaline-2,3-diones discussed in the present study within the GluR2 ligand-binding core (S1S2J). (A) The competitive antagonists 6,7-dinitroquinoxaline-2,3-dione (DNQX), 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), and [1,2,5]oxadiazolo[3,4-G]quinoxaline-6,7(5H,8H)-dione 1-oxide (FQX). (B) Close-up views of DNQX, CNQX and FQX bound to the S1S2J.
Superposition of the S1S2J–FQX monomers with the previously reported S1S2J-DNQX monomer revealed that FQX binds analogously to DNQX within the S1S2J, with an average rmsd of 0.40 Å (Figure 2). The degree of clamshell closure in the S1S2J–FQX structure is identical to that observed in the previously reported S1S2J–DNQX and S1S2J–CNQX structures.12,23 In all three structures, the clamshell is opened by 15° compared to the glutamate-bound structure. In addition, key residues positioning FQX in the binding site are identical to those orienting DNQX (Figure 2) and CNQX in the ligand-binding core. The quinoxaline ring of FQX is positioned directly below Tyr450, forming a favorable π-stacking interaction. The FQX carbonyl oxygens form hydrogen bonds with Arg485, and the amide nitrogens form hydrogen bonds with the backbone carbonyl of Pro478 and a well-defined water molecule, respectively. The two oyxgens of the FQX furoxan ring form a hydrogen bond with a water molecule that is held in position through hydrogen bonds with Tyr405 and Met708. The only binding site residue that differs between the S1S2J–FQX structure and the DNQX and CNQX structures is Glu705, which adopts different conformations in each of the S1S2J–FQX monomer structures due to rotation about the Cβ-Cγ bond.
Figure 2.
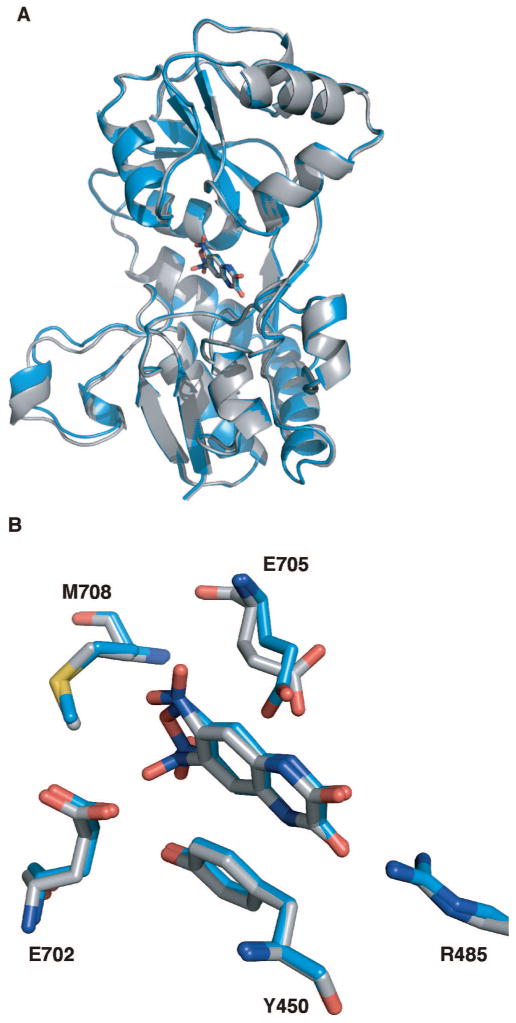
Aligned structures of FQX and DNQX bound to the GluR2 ligand-binding core (S1S2J). (A) FQX (blue ribbon), DNQX (gray ribbon), and CNQX (not shown) produce the same degree of closure of the ligand-binding core. (B) Close-up view of the ligand-binding core showing the residues in close proximity to the ligands FQX and DNQX. Glutamate (E) 705 adopts a different conformation in the FQX structure (blue sticks) than the DNQX structure (gray sticks) and CNQX structure (not shown).
Mass Spectral Analysis of the S1S2J -ANQX Solution Complex
The photocross-linked S1S2J–ANQX adduct was obtained through continuous irradiation and perfusion with “fresh” (unphotolyzed) ANQX over the histidine (6×) tagged S1S2J protein immobilized on Ni-NTA beads. Treatment of S1S2J with ANQX and ultraviolet light, but not ultraviolet light alone, resulted in the formation of covalently modified S1S2J bearing a 220 Da mass increase as determined by electrospray ionization orthogonal-time-of-flight (ESI-o-TOF) (Figure 3A). This is the expected mass increase for the formation of a covalent adduct between photolyzed ANQX (−N2, MW 220) and S1S2J (MW 32218). To determine the location of this adduct, the covalently modified S1S2J was digested with trypsin and the resulting peptides were analyzed by nanoscale liquid chromatography and tandem mass spectrometry (MS/MS). A peptide fragment containing the active site residues Glu705 and Met708 was found bearing a 220 Da adduct in the ANQX-treated sample, which corresponds to the expected mass increase for the formation of a peptide–ANQX fragment (Figure 3B, top panel). In control samples treated with UV light in the absence of ANQX, the 220 Da adduct was not observed (Figure 3B, bottom panel). Comparison of the fragment analysis for the ANQX treated versus untreated samples showed that an adduct formed with Glu705 (y11, E*, top panel, Figure 3B, top panel) and not Met708 (y8, M, Figure 3B, top panel). That is, there is a 220 Da mass difference between the Glu705 fragment in the ANQX treated sample (y11, E*, m/z 1602.72 + NH4, Figure 3B, top panel) compared to that of the Glu705 fragment in the untreated sample (y11, E, m/z 1399.38, Figure 3B, bottom panel). There is no mass difference for the Met708 fragment between the ANQX treated and untreated samples (y8, M, Figure 3B). This peptide fragmentation data provides sufficient coverage to assign the site of covalent modification to Glu705 (E*) and not Met708 (M). We conclude that Glu705 is the preferred site of reaction with photoactivated ANQX.
Figure 3.
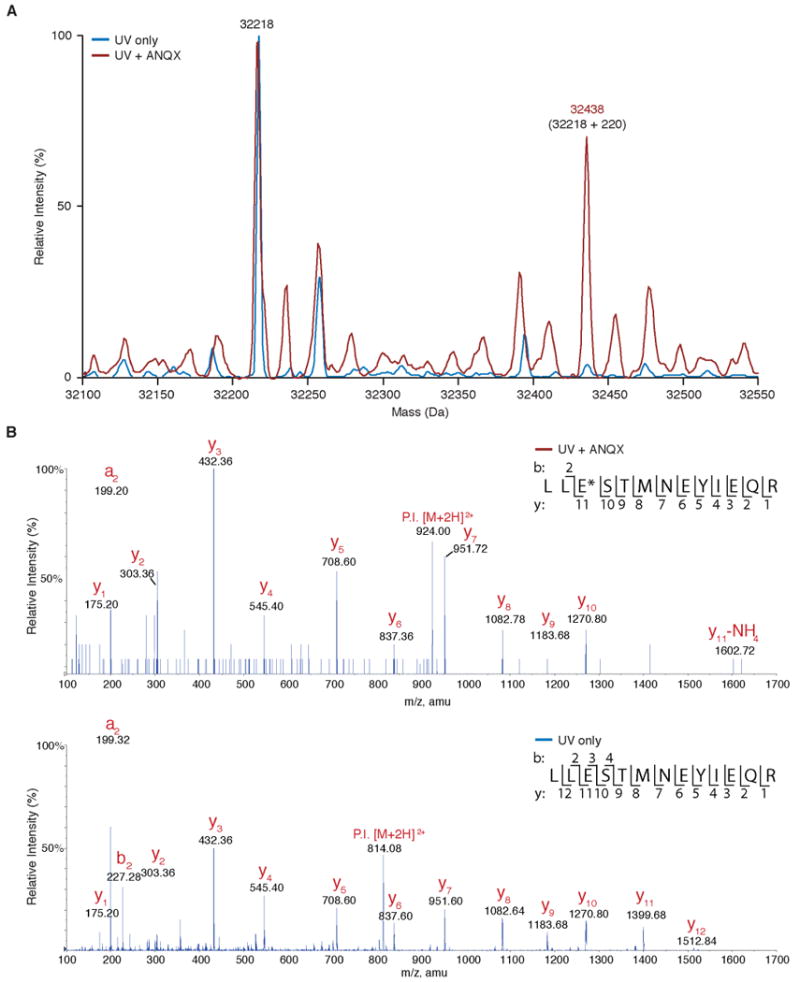
Photocross-linking of ANQX to the GluR2 ligand-binding core (S1S2J) analyzed using mass spectrometry. (A) ESI-o-TOF mass spectrum of the S1S2J (32218 Da) following irradiation (90 s) in the absence (blue trace) and presence (red trace) of ANQX (100 μM). Irradiation in the presence of ANQX produces a new major peak (32438 Da) that corresponds to the expected increase in mass (+220 Da) for the ANQX–protein adduct. (B) Mass spectra (MS/MS) of the peptide fragment spanning Glu705 following irradiation (90s) in presence (top panel) and absence (bottom panel) of ANQX. Irradiation in presence of ANQX produced a new peak (m/z 1602.72, E*, top panel), corresponding to the expected mass increase for the formation of an ANQX adduct, and the loss of unlabeled Glu705 (m/z 1399.68, E, bottom panel) peak.
Discussion
In the present study, we used X-ray crystallography and mass spectroscopy to identify the products of photolyzing ANQX in the presence of the GluR2 ligand-binding core (S1S2J). Photolysis of ANQX–S1S2J cocrystals yielded a 1.87 Å crystal structure of the intermolecular reaction product, FQX reversibly bound to the GluR2 ligand-binding core (S1S2J). FQX is formed via intramolecular reaction of the nitrene formed following the loss of dinitrogen from ANQX (Scheme 1).24
Scheme 1.
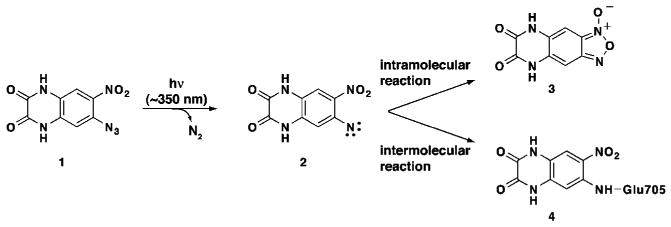
Photochemistry of ANQXa
a Upon irradiation with UV light, ANQX (1) forms a highly reactive nitrene (2), which forms two products within the S1S2J binding pocket. The intramolecular reaction results in the formation of FQX (3). The intermolecular reaction results in the formation of a covalent bond with Glu705 in the ligand-binding core (4).
In a previous electrophysiological study, ANQX was used to irreversibly photoinactivate AMPARs on neurons.21 In these experiments, we noted an initial increase in the AMPAR current following the first pulse of ultraviolet light, which suggested that a major product of photolyzed ANQX is the formation of a lower affinity antagonist. Consistent with this observation, the IC50 of FQX (7.6 μM25) is nearly 8-fold higher than the IC50 for ANQX (1.0 μM20). The cross-linking efficiency of nitrenes is notoriously poor, with ortho-nitrophenylazides tending to react intramolecularly upon photolysis.24 Therefore, it is not surprising that we only observed the major reaction product (FQX) reversibly bound to the S1S2J, following irradiation of ANQX–S1S2J cocrystals. An intense effort was made to obtain crystals of the intermolecular reaction product, covalently modified S1S2J–ANQX. However, irradiation of S1S2J–ANQX cocrystallization drops resulted in protein that did not form crystals even after several months and irradiation of the S1S2J immobilized on Ni-NTA beads in the presence of ANQX resulted in protein that was too impure for protein crystallography even after FPLC purification.
Comparison of the FQX structure with the previously reported DNQX and CNQX structures revealed that all three antagonists bind in the same orientation and produce the same degree of clamshell closure within the ligand-binding core. Mass spectral analysis revealed that Glu705 is the site covalently modified by ANQX. On the basis of the position of FQX in the binding pocket in one of the monomers, the nitrene nitrogen on ANQX is positioned 2.5 Å from Glu705. This residue can adopt a range of conformations with distances of 1.7–6.2 Å from the nitrene nitrogen on ANQX. Thus, the cross-link can occur within the range of a single bond without distortion. Together, these data suggest that the formation of the covalent adduct with photolyzed ANQX does not produce a change in the degree of clamshell closure around the ligand. Because Glu705 is a flexible residue in the S1S2J–FQX crystal structure, it may be the case that only certain rotamers of Glu705 react with the activated ANQX nitrene.
Taken together, these data demonstrate that ANQX is a useful biological probe for inactivating AMPARs because it is not a promiscuous cross-linker. We have shown that ANQX forms a covalent adduct with Glu705 and not Met 708. If this reaction does not occur, ANQX reacts intramolecularly to form FQX, which is a low-affinity antagonist of AMPARs.
Experimental Section
ANQX was synthesized as previously reported.20 The GluR2 ligand-binding core was expressed in Escherichia coli and purified to homogeneity as previously reported by Gouaux and co-workers. The purified S1S2J protein was cocrystallized with ANQX at 4 °C using vapor-diffusion (hanging-drop) with a 1:1 ratio of protein to well solution. While submerged in liquid nitrogen, S1S2J–ANQX cocrystals were exposed to ultraviolet light. Synchrotron data for the resulting S1S2J–FQX complex were collected at 100 K on the Advanced Light Source beamline 8.3.1 at Lawrence Berkeley National Laboratory. Data processing was performed using Elves and HKL2000. A complete data set from an S1S2J–FQX crystal diffracted to 1.87 Å, exhibited the space group P212121 and contained four molecules per asymmetric unit (Table 1 of Supporting Information). Molecular replacement solutions for S1S2J–ANQX crystal structure were obtained using as a search model one of the monomers of S1S2J–DNQX dimer structure. Photocross-linking of ANQX to the S1S2J was accomplished by irradiating a solution of the 6-HIS tagged S1S2J immobilized on Ni-NTA beads in the presence of ANQX. The resulting sample was denatured and digested with trypsin/chymotrypsin and then analyzed by nanoscale LC/MS2 using a Q TRAP mass spectrometer coupled to a liquid chromatography system. Tandem mass spectra were acquired automatically in IDA mode, and the resulting data were analyzed with MASCOT.
Supplementary Material
Acknowledgments
We thank James Holton at the Advance Light Source (ALS) 8.3.1 beamline, Pascal Egea (UCSF) for assistance collecting and processing data, and Eric Gouaux for the S1S2J construct. This work was supported by grants from the Sandler Foundation and the Herbert Boyer Foundation.
Footnotes
The structure S1S2J-FQX has been deposited in the PDB and has been assigned the following code: 3BKI.
Abbreviations: AMPAR, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; ANQX, 6-azido-7-nitro-1,4-dihydroquinoxaline-2,3-dione; DNQX, 6,7-dinitroquinoxaline-2,3-dione; CNQX, 6-cyano-7-nitroquinoxaline-2,3-dione; FQX, [1,2,5]oxadiazolo[3,4-G]quinoxaline-6,7(5H,8H)-dione 1-oxide; S1S2J, GluR2 ligand-binding core; PDB ID: 3BKI.
Supporting Information Available: Experimental methods, a cartoon depicting the portion of the GluR2 subunit corresponding to the S1S2J domain, 2Fo – Fc omit electron density for FQX and surrounding ligands, and statistics for data collection and refinement. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 2.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 3.Palmer CL, Cotton L, Henley JM. The molecular pharmacology and cell biology of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Pharmacol Rev. 2005;57:253–277. doi: 10.1124/pr.57.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bliss TVP, Collingridge GL. A Synaptic Model of Memory: Long-Term Potentiation in the Hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 5.Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 6.Song I, Huganir RL. Regulation of AMPA receptors during synaptic plasticity. Trends Neurosci. 2002;25:578–588. doi: 10.1016/s0166-2236(02)02270-1. [DOI] [PubMed] [Google Scholar]
- 7.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 8.Collingridge GL, Isaac JR, Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci. 2004;5:952–961. doi: 10.1038/nrn1556. [DOI] [PubMed] [Google Scholar]
- 9.Gouaux E. Structure and function of AMPA receptors. J Physiol. 2004;554:249–253. doi: 10.1113/jphysiol.2003.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer ML. Glutamate receptor ion channels. Curr Opin Neurobiol. 2005;15:282–288. doi: 10.1016/j.conb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Olson L, Horning M, Armstrong N, Mayer M. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 14.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 15.Hogner A, Kastrup JS, Jin R, Liljefors T, Mayer ML. Structural basis for AMPA receptor activation and ligand selectivity: crystal structures of five agonist complexes with the GluR2 ligand-binding core. J Mol Biol. 2002;322:93–109. doi: 10.1016/s0022-2836(02)00650-2. [DOI] [PubMed] [Google Scholar]
- 16.Jin R, Gouaux E. Probing the function, conformational plasticity, and dimer–dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal. Biochemistry. 2003;42:5201–5213. doi: 10.1021/bi020632t. [DOI] [PubMed] [Google Scholar]
- 17.Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005;25:9027–9036. doi: 10.1523/JNEUROSCI.2567-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin JW, Ju W, Foster K, Lee SH, Ahmadian G. Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptors internalization. Nat Neurosci. 2000;3:1282–1290. doi: 10.1038/81814. [DOI] [PubMed] [Google Scholar]
- 19.Nikam SS, Kornberg BE. AMPA receptor antagonists. Curr Med Chem. 2001;8:155–170. doi: 10.2174/0929867013373877. [DOI] [PubMed] [Google Scholar]
- 20.Chambers JJ, Gouda H, Young DM, Kuntz ID, England PM. Photochemically knocking out glutamate receptors in vivo. J Am Chem Soc. 2004;126:13886–13887. doi: 10.1021/ja048331p. [DOI] [PubMed] [Google Scholar]
- 21.Adesnik H, Nicoll RA, England PM. Photoinactivation of native AMPA receptors reveals their real-time trafficking. Neuron. 2005;48:977–985. doi: 10.1016/j.neuron.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 22.England PM. Rapid photoinactivation of native AMPA receptors using ANQX. Sci STKE. 2006;331:11. doi: 10.1126/stke.3312006pl1. [DOI] [PubMed] [Google Scholar]
- 23.Menuz K, Stroud RM, Nicoll RA, Hays FA. Subunits switch AMPA receptor antagonists to partial agonists. Science. 2007;318:815–817. doi: 10.1126/science.1146317. [DOI] [PubMed] [Google Scholar]
- 24.Murata S, Tomioka H. Photochemistry of o-Nitrophenylazide in Matrices. The First Direct Spectroscopic Observation of o-Dinitrosobenzene. Chem Lett. 1992:57–60. [Google Scholar]
- 25.Baudy RB, Sulkowski TS. 5h,8h-2-oxa-1,3,5,8-tetraaza-cyclopenta[b]-naphthalene-6,7-diones neuroprotective agents U S. USXXAM US 5719153 A 19980217 CAN 128:176169 AN 1998:146566 1998
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


