Abstract
The intranuclear trafficking of human telomerase involves a dynamic interplay between multiple nuclear sites, most notably Cajal bodies and telomeres. Cajal bodies are proposed to serve as sites of telomerase maturation, storage, and assembly, as well as to function in the cell cycle-regulated delivery of telomerase to telomeres in human cells. Here, we find that telomerase RNA does not localize to Cajal bodies in mouse cells, and instead resides in separate nuclear foci throughout much of the cell cycle. However, as in humans, mouse telomerase RNA (mTR) localizes to subsets of telomeres specifically during S phase. The localization of mTR to telomeres in mouse cells does not require coilin-containing Cajal bodies, as mTR is found at telomeres at similar frequencies in cells from wild-type and coilin knockout mice. At the same time, we find that human TR localizes to Cajal bodies (as well as telomeres) in mouse cells, indicating that the distinct trafficking of mTR is attributable to an intrinsic property of the RNA (rather than a difference in the mouse cell environment such as the properties of mouse Cajal bodies). We also find that during S phase, mTR foci coalesce into short chains, with at least one of the conjoined mTR foci co-localizing with a telomere. These findings point to a novel, Cajal body-independent pathway for telomerase biogenesis and trafficking in mice.
Keywords: telomerase, telomeres, Cajal bodies, RNA localization, mouse telomerase RNA
Introduction
Telomeres are capping structures that comprise the physical ends of eukaryotic chromosomes. In vertebrates, they consist of tandemly repeated arrays of TTAGGG and a number of associated proteins [1, 2]. Telomeres serve protective functions, preventing the ends of chromosomes from being recognized as double stranded DNA breaks [3] and serving as barriers to the loss of genetic information that results from the inability of DNA polymerases to fully replicate the ends of linear DNA. However, a portion of the telomere is lost with each cell division in most adult human somatic cells, and eventually telomere attrition triggers the cell to stop dividing and enter a state of proliferative senescence or undergo apoptosis. Increasing numbers of human telomere shortening diseases are being recognized and investigated [4–6].
Telomerase is the specialized reverse transcriptase that synthesizes telomeres and combats telomere erosion [7]. The telomerase enzyme is minimally comprised of two essential components: telomerase RNA (TR), which provides the template for repeat addition, and telomerase reverse transcriptase (TERT), which synthesizes the repeats. In humans, telomeres are synthesized early in development [8, 9]. Telomerase activity is not detected in most adult somatic cells, but notably, the enzyme is active in over 90% of human cancers and is responsible for the prolonged proliferative capacity of these cells [10–12]. (However, telomerase activity is readily detected in murine somatic cells [13–16].) Insights into the biogenesis and regulation of this enzyme therefore have the potential to lead to the development of novel anti-cancer therapeutics [17, 18].
A recent series of studies has revealed important aspects of the trafficking of the telomerase ribonucleoprotein complex (RNP) critical to the biogenesis and function of the enzyme in human cells [19–25]. Throughout interphase, human TR is found in Cajal bodies [21, 23], dynamic nuclear structures that have been implicated in the biogenesis of various cellular RNPs (reviewed in [26–28]). Localization of hTR to Cajal bodies is important for function of the telomerase enzyme [19, 24]. Accumulation of hTR in Cajal bodies occurs in cancer cells where telomerase activity is present, but not telomerase-negative normal cells [22, 23, 25], consistent with the hypothesis that Cajal bodies serve as sites of telomerase assembly. Human TERT is detected in distinct nucleoplasmic foci, separate from Cajal bodies by immunofluorescence (IF) [23], though exogenously expressed GFP-tagged, hTERT can be detected within Cajal bodies [25] as well as nucleoli [29–31]. During S phase, both human TR and TERT move to foci found immediately adjacent to Cajal bodies, and then, peaking in mid-S phase when human telomeres are replicated and synthesized [32–34], both hTR and hTERT associate with subsets of telomeres [19, 21–24]. These findings indicate that Cajal bodies are important for telomerase biogenesis and may act to deliver telomerase to the telomere.
The trafficking of telomerase components has emerged as a key process in the biogenesis and function of the enzyme, and the factors important for trafficking are being defined. The localization of human TR to both Cajal bodies and telomeres depends on TERT [22], suggesting that assembly and trafficking are tightly linked processes. The trafficking of telomerase to both Cajal bodies and telomeres depends on a newly identified telomerase component, called TCAB1 (Telomerase Cajal-body protein 1; also called WDR79) [24, 35]. There is also evidence that the core telomere binding protein, TPP1 is critical for telomerase-telomere associations [36–40]. However, much remains to be learned regarding the factors and mechanisms that influence the biogenesis and trafficking of telomerase.
To gain a better understanding of the regulation of telomerase trafficking, we initiated studies in the mouse model system where genetic approaches have contributed to our understanding of basic telomere and telomerase biology, particularly with regard to cancer and aging [41–45]. Here, we report a fluorescence in situ hybridization (FISH) procedure specific for detection of mouse telomerase RNA (mTR) and the characterization of mTR localization patterns in cultured mouse cell lines using this approach. Unexpectedly, we found that mTR does not co-localize with coilin or other Cajal body markers, and instead, is found in distinct nuclear foci. However, a fraction of the mTR localizes to telomeres selectively during S phase of the cell cycle. In many cases, we observe chains of connected mTR foci during S phase, and frequently find that at least one of the foci in a chain co-localizes with a telomere. Our findings suggest an alternative mechanism for recruitment of telomerase RNA to telomeres during S phase in murine cells, which involves the convergence of non-Cajal body mTR foci at telomeres.
Materials and Methods
Cell culture and transfection
MEF-26 (WT), MEF-42 (coilin −/−), MEF-14 (mTR −/−) (a gift of Carol Greider, Johns Hopkins University, Baltimore, MD), 3T3 (ATCC, Manassas, VA), n2a (a gift of Brian Condie, University of Georgia, Athens, GA), c2c12 (ATCC), and A9 cells were grown on coverslips in DMEM (Mediatech, Herndon, VA) supplemented with 10% fetal calf serum (FCS) (Mediatech) at 37°C with 5% CO2. For hTR overexpression, cells were transfected with pBS-U1-hTR [19] (gift of J. Lingner, EPF of Lausanne, Switzerland) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
FISH probes
All probes were synthesized by Qiagen (Valencia, CA) as follows: mTR 117–169 (probe 1),
CT*CCCGGCGAACCT*GGAGCTCCTGCGCT*GACGTTTGTTTTT*GAGGCTCGGGT*A; mTR 296–342 (probe 2),
CT*CGGGGACCAGT*TCCATTCCTGT*CCTTGCGGCGCT*CGCCCGGCCT*G; mTR 224–282 (probe 3),
GT*GCCCCGCGGCT*GACAGAGGCGAGCT*CTTCGCGGCGGCAGCGGAGT*CCTAAGACCCT*A; mTR 57–104 (probe 4),
CT*CTGCAGGTCT*GGACTTTCCT*GGCCCGCTGGAAGT*CAGCGAGAAAT*A; U3 (probe 5),
TT*CAGAGAAACTTCTCT*AGTAACACACTAT*AGAACTGATCCCT*GAAAGTATAGT*C; mU85 (probe 6),
AT*TACCAAAGATCT*GTGTGTCATCT*CTCAGTGGCCAT*GACACAGCTAAGT*C; telomere (probe 7),
CT*AACCCTAACCCT*AACCCTAACCCT*AACCCTAACCCT*AACCCTAACCCT*A; hTR 128–183 (probe 8),
GCT*GACATTTTT*TGTTTGCTCT*AGAATGAACGGT*GGAAGGCGGCA; hTR 331–393 (probe 9),
CT*CCGTTCCTCTTCCT*GCGGCCTGAAAGGCCT*GAACCTCGCCCT*CGCCC CCGAGT*G; hTR 393–449 (probe10),
AT*GTGTGAGCCGAGT*CCTGGGTGCACGT*CCCACAGCTCAGGGAAT*CGCGCCGCGCT*C. T* indicates aminoallyl-modified thymidines. All mTR and hTR probes (probes 1–4 and 8–10 above) were conjugated with cy3 mono-functional reactive dye according to the manufacturers protocol (GE Healthcare, Little Chalfront, Buckinghamshire, United Kingdom). The remaining probes were labeled with Alexa Fluor 647 or Oregon green dye according to the manufacturers protocol (Molecular Probes/Invitrogen, Carlsbad, CA).
mTR and hTR FISH
FISH was performed essentially as described [23, 25]. Hybridizations were carried out overnight at 37°C. 200 ng of each of two of the cy3-labeled mTR probes (most often probes 1 and 2) or 25 ng of each of the three cy3-labeled hTR probes (probes 8, 9 and 10) above were used per coverslip. 5–10 ng Oregon green or Alexa Fluor 647 U3 (probe 5), 100 ng Oregon green mU85 (probe 6), or 10 ng Oregon green telomere (probe 7) were also included in the hybridization when indicated. Cells were rinsed twice in 50% formamide, 2× SSC for 30 minutes at 37°C. Cells were subsequently washed once in 50% formamide, 2× SSC, 0.1% NP-40 for 30 minutes at 37°C. The coverslips were mounted in Prolong gold mounting media (Molecular Probes/Invitrogen), cured at room temperature for 1 hour and stored at −20°C until microscope analysis.
RNAse treatment
After fixation and permeabilization, cells were rehydrated in 1× PBS containing 1.5 mM MgCl2 at room temperature for 5 min. Then the cells were incubated with RNAse A (0.2 mg/ml in 1× PBS containing 1.5 mM MgCl2) at 37 C for 2 h. mTR FISH was performed after the cells were washed 3 times with 1× PBS and once with 50% formamide, 2× SSC.
Indirect Immunofluorescence (IF)
Following FISH, cells were washed three times with 1× PBS. Next, cells were incubated with one of the following primary antibodies at the indicated dilution for 1 hour at room temperature: rabbit anti-Nopp140 (1:1,000, a gift from U. Thomas Meier, Albert Einstein College of Medicine, New York, NY), rabbit anti-TRF1 (1:500), rabbit anti-TPP1 (1:500), or rabbit anti-Tin2 (1:500) (all 3 a gift from Susan Smith, Skirball Institute, New York, NY). Cells were washed three times in 1× PBS and then incubated with secondary antibody (1:100 Cy2 conjugated goat anti-rabbit IgG (H+L), Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour at room temperature. All antibodies were diluted in 0.05% Tween-20 in PBS (PBST). Cells were then subjected to three final 1× PBS washes and mounted as above.
For the following antibodies, IF was performed prior to FISH analysis: mouse anti-p80 coilin (1:1,000), mouse anti-fibrillarin (17C12, 1:1,000), mouse anti-SMN (2B1, 1:100, Novus Biologicals, Littleton, CO). Briefly, cells were grown on coverslips overnight and then washed one time in 1× PBS. Cells were fixed in 4% formaldehyde in 1× PBS for 10 minutes at room temperature. Cells were rinsed twice in 1× PBS and blocked in 1% BSA (Sigma-Aldrich, St. Louis, MO) in 1× PBS for 1 hour at room temperature. Cells were then incubated with the antibodies at the indicated dilutions for 1 hour at room temperature followed by incubation with a cy2-conjugated goat anti-mouse secondary antibody (1:100, Jackson Immunoresearch Laboratories) for 1 hour at room temperature. These antibodies were diluted in 1% BSA in 1× PBS. Following three 1× PBS washes, cells were again fixed in 4% formaldehyde in 1× PBS for 10 minutes at room temperature. Cells were rinsed twice in 1× PBS and once in 50% formamide, 2× SSC prior to FISH analysis.
S phase synchronization
Synchronous populations of MEF and 3T3 cells were obtained by double thymidine block. Cells were treated with 2 or 5 mM thymidine (Sigma-Aldrich) for 16 hours. Cells were released by rinsing twice with 1× PBS and replacing the normal growth media for 8 hours. Cells were retreated with 2 or 5 mM thymidine for another 16 hours. At various time points after release, cells were fixed and analyzed by PCNA staining or FACS analysis to verify cell cycle phase.
PCNA (proliferating cell nuclear antigen) staining
To distinguish S phase cells, cells on coverslips were rinsed once in 1× PBS and fixed for 10 minutes at room temperature in 4% formaldehyde, 10% acetic acid in 1× PBS. Cells were rinsed twice in 1× PBS and permeablized in 70% ethanol at 4°C overnight. The cells were incubated with mouse anti-PCNA antibody (PC10, 1:2500, Abcam, Cambridge, MA) for 1 hour at room temperature. Cells were rinsed 3 times in 1× PBS and then incubated with secondary antibody (1:100, AMCA-conjugated goat anti-mouse IgG (H+L), Jackson Immunoresearch Laboratories) for 1 hour at room temperature. Cells were rinsed 3 times in 1× PBS and fixed in 4% formaldehyde in 1× PBS for 10 minutes at room temperature. Cells were rinsed twice in 1× PBS and once in 50% formamide, 2× SSC prior to FISH analysis. Both antibodies were diluted in PBST.
FACS (fluorescence activated cell sorting) analysis
Cells were collected and resuspended to a single cell suspension in 0.5 mL 1× PBS. The cell suspension was transferred to a tube containing 4.5 mL of 70% ethanol and stored at −20°C until analysis (at least overnight). On the day of FACS analysis, the ethanol suspended cells were centrifuged for 5 minutes at 200 × g. The pellet was washed once in 5 mL of 1× PBS, centrifuged again for 5 minutes at 200 × g and resuspended in a solution of 0.02 mg/mL propidium iodide (Sigma-Aldrich), 0.1% Triton X-100, and 2 mg DNase-free RNAse A in 1× PBS. The cells were stained in this solution for 30 minutes at 37°C. FACS analysis was performed using a FACSCALIBUR scanner (Becton Dickinson Biosciences, San Jose, CA) and data was analyzed using FlowJo software (Tree Star, Inc., Stanford, CA).
Microscopy
Slides were analyzed using a Zeiss Axioskop 2 Mot Plus fluorescence microscope (Carl Zeiss Microimaging, Thornwood, NY). Images were acquired at 63× (Plan Apochromat objective, numerical aperture 1.4) using a cooled charge-coupled device Orca-ER camera (Hamamatsu, Bridgewater, NJ) and IPLab Spectrum software.
Results
Specific detection of mouse telomerase RNA in cultured mouse cells
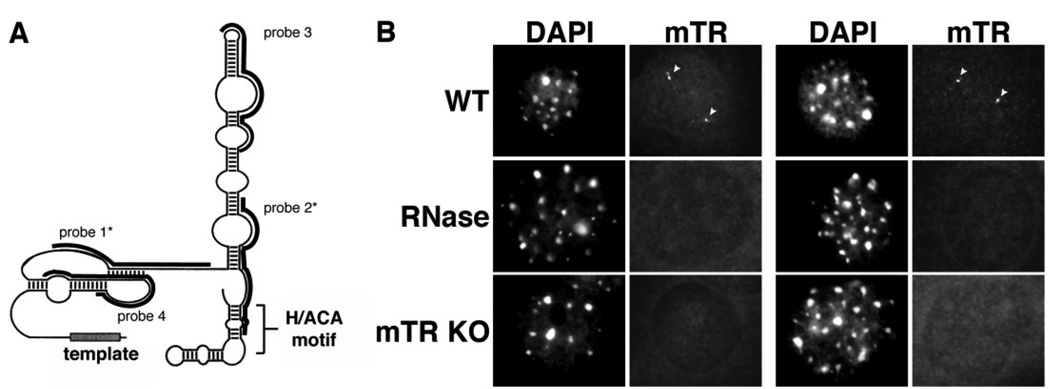
We developed a FISH procedure for the detection of mouse telomerase RNA (mTR) in order to examine its subcellular localization in mouse cells (where telomerase is active). We designed four probes against different regions of mTR; the regions encompassed by the probes are indicated in Figure 1A. (See Materials and Methods for probe sequences.) Figure 1B shows the results of hybridization with a combination of probe 1 and probe 2 in cultured mouse embryonic fibroblast (MEF) cells. Approximately 2/3 of the MEF cells displayed 1–3 small, clearly defined, spherical nuclear foci (Figure 1B). Hybridization with each of the four individual mTR probes gave similar localization patterns, and maximal signal was obtained with combinations of two probes (data not shown). The combination of probes 1 and 2 was used throughout the remainder of this work.
Figure 1. Mouse telomerase RNA is found in small spherical foci within the nuclei of cultured mouse cell lines.
A. Schematic structure of mTR. The predicted secondary structure of mTR is shown [70]. Black bars indicate the regions encompassed by each oligonucleotide probe. Asterisks denote the two probes (probes 1 and 2) used throughout this manuscript. B. FISH procedure specifically detects the presence of mTR. mTR FISH was performed on wild type MEF cells (WT, RNAse panels) or MEF cells derived from mTR −/− mice (mTR KO panels). Arrowheads denote intranuclear mTR foci present in the WT cells (WT panels), which are lost upon treatment of cells with RNAse A prior to FISH (RNAse panels). DAPI was used as a nuclear stain. Scale bar, 10 microns.
The results of control experiments indicate that the procedure is specific for detection of mTR. First, mTR signal was lost upon treatment of the cells with RNAse A prior to hybridization with the mTR probes (Figure 1B). As controls, we also tested the RNAse-treated cells with probes against U3 snoRNA (positive control for treatment) and the telomere repeat (negative control for treatment). As expected, the U3 signals were eliminated or severely reduced in the RNAse treated cells, while telomere signals were virtually unaffected (data not shown). In addition, mTR foci were not observed in MEF cells derived from mTR knockout mice (Figure 1B). Finally, the foci were not observed in HeLa (human cervical carcinoma) cells using the mTR FISH probes (data not shown), further indicating the specificity of the procedure for detection of mTR.
mTR does not localize to Cajal bodies in mouse cells
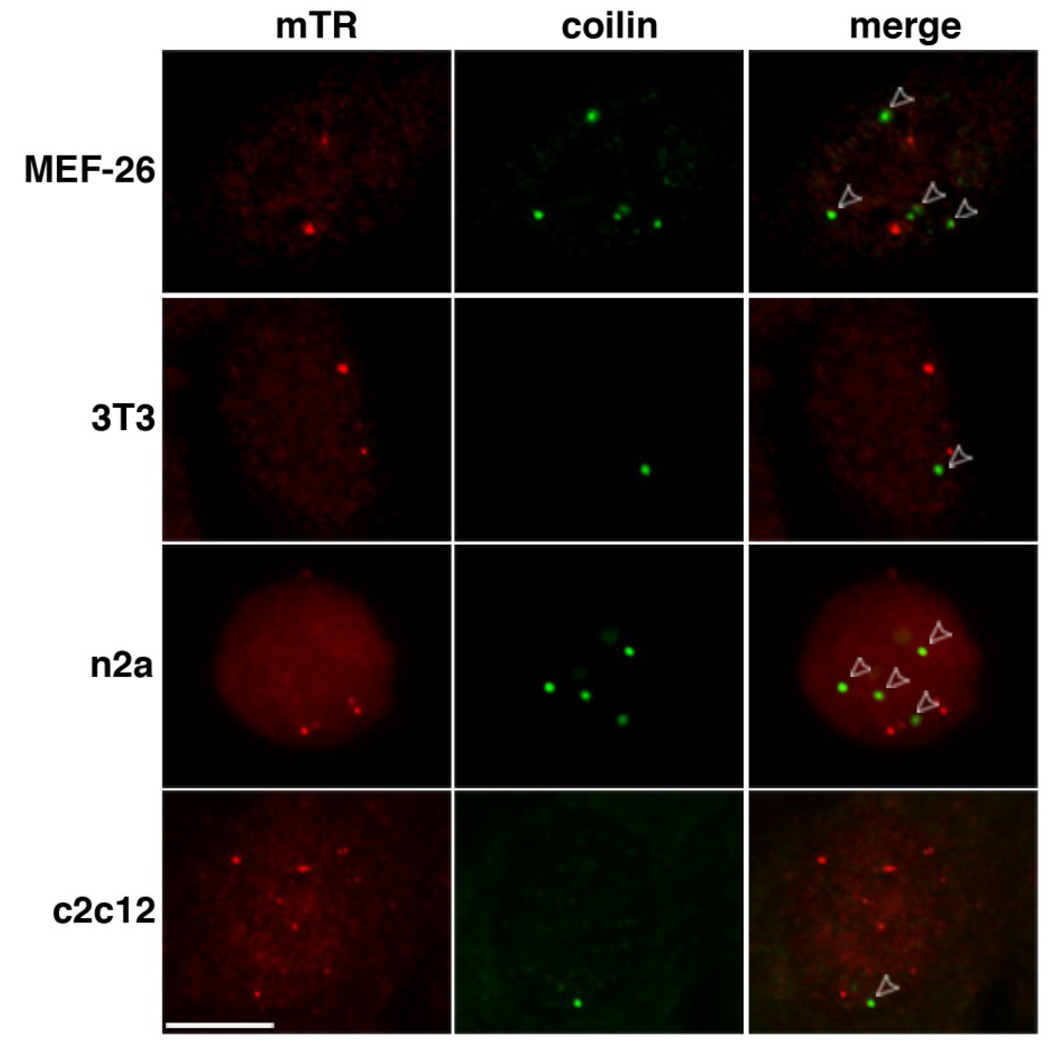
Telomerase RNA localizes to Cajal bodies throughout interphase in human cell lines [20, 25]. To determine if the mTR foci observed in the MEF cells correspond to Cajal bodies, we combined mTR FISH with immunofluorescence (IF) using antibodies against coilin, a marker protein of Cajal bodies [46]. Surprisingly, the mTR foci present in the MEF cells did not co-localize with coilin (Figure 2, MEF-26). We also found that mTR is localized in foci distinct from Cajal bodies in other mouse cell lines representing various tissue sources (Figure 2).
Figure 2. mTR foci do not colocalize with coilin in a variety of mouse cell lines.
Mouse embryonic fibroblast (MEF-26, 3T3), n2a neuroblastoma, and c2c12 myoblast cell lines were co-analyzed for mTR (detected by FISH, red) and coilin (marker protein for Cajal bodies, detected by IF, green). Merge panels indicate an overlay of mTR and coilin panels. Open arrowheads point to Cajal bodies that do not overlap with mTR foci. Scale bar, 10 microns.
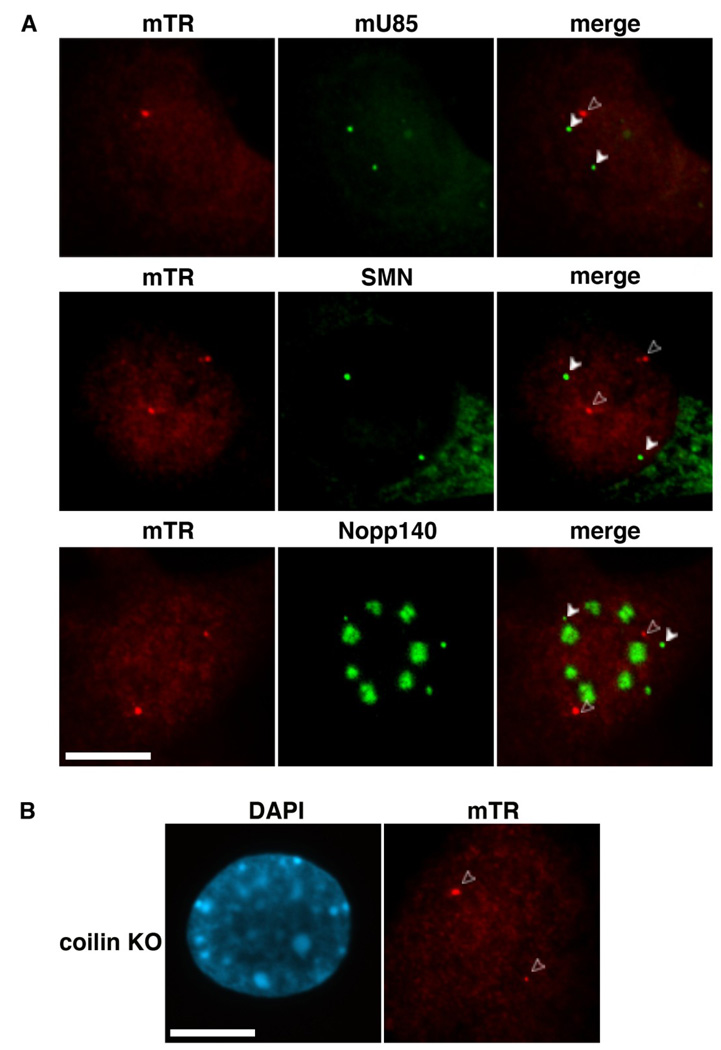
We further tested for co-localization of mTR with a series of additional molecules characteristically found in Cajal bodies. U85, a small Cajal body (sca)RNA, and SMN, the survival of motor neurons protein are prototypical markers for Cajal bodies, whereas Nopp140 localizes to both Cajal bodies and nucleoli in mammalian cells [47–50]. mTR did not co-localize with these additional Cajal body markers in the MEF cells (Figure 3). We also tested whether mTR co-localized with the snoRNP protein fibrillarin or U3 snoRNA (markers with distributions similar to Nopp140), and found no significant overlap (data not shown).
Figure 3. mTR resides in foci separate from Cajal bodies.
A. mTR foci do not correspond to known markers for Cajal bodies. mTR FISH (red, mTR panels) was performed in tandem with one of three markers for Cajal bodies: U85 scaRNA (top row, detected by FISH), SMN (middle row, detected by IF, signal present in Cajal bodies and cytoplasm), or Nopp140 (bottom row, detected by IF, signal present in Cajal bodies and nucleoli). Arrowheads denote Cajal bodies; open arrowheads point to mTR foci that do not localize to Cajal bodies. B. mTR localizes to intranuclear foci in MEF cells derived from coilin knockout (KO) mice. mTR FISH was performed on coilin KO MEF cells. Open arrowheads point to mTR foci. DAPI was used to stain the DNA. Scale bars, 10 microns.
In addition, we examined mTR localization in MEF cells derived from coilin knockout mice [51] and found no reduction in mTR foci (Figure 3B). In these MEFs where coilin is not expressed, standard components of Cajal bodies such as U85, SMN and Nopp140 can be found in distinct nucleoplasmic foci termed residual Cajal bodies [51, 52]. mTR also did not co-localize with any of these residual Cajal body markers in the coilin knockout MEFs (data not shown). Taken together, our data unexpectedly indicate that mTR accumulates in nuclear structures distinct from Cajal bodies in cultured mouse cells.
mTR is found at a subset of telomeres in S phase cells
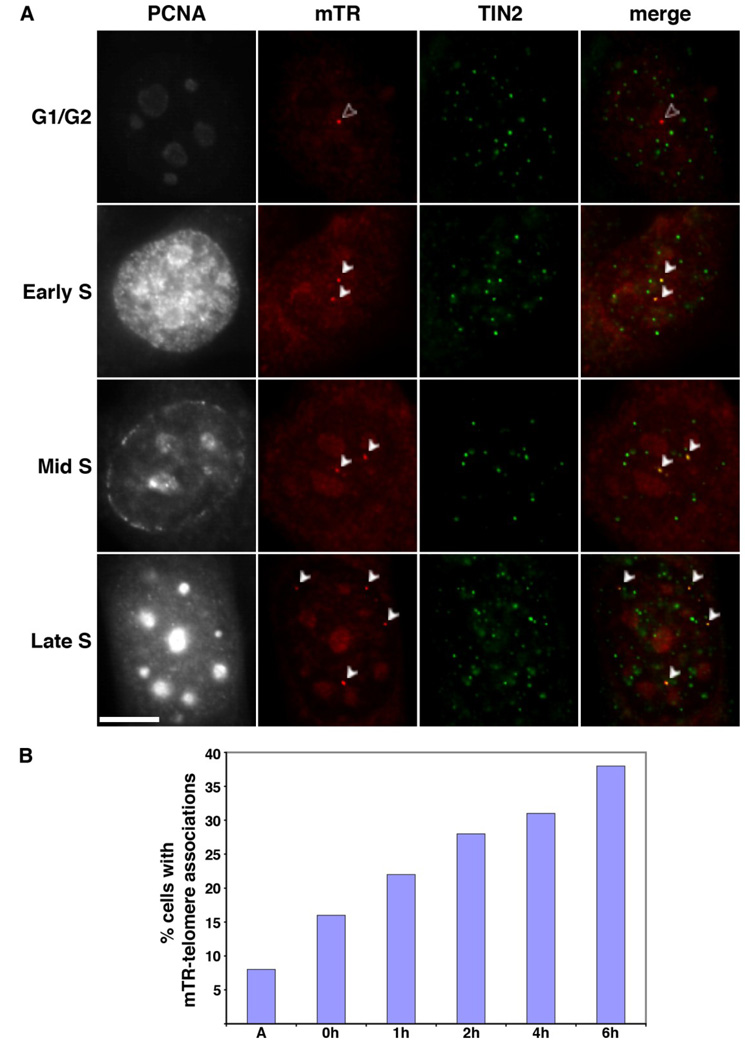
Having found that mTR does not localize to Cajal bodies, we investigated whether any of the mTR foci correspond to telomeres. When mTR FISH was combined with IF using antibodies against the telomere binding proteins TRF1, TIN2, or TPP1, or with FISH using a probe directed against the telomeric repeats, we found that mTR co-localized with a subset of telomeres in approximately 10% of the MEF (and 3T3) cells with distinct mTR foci (Figure 4 and data not shown). This is similar to the percentage of cells in which TR-telomere associations are observed in unsynchronized human cancer cell lines, where TR localizes to telomeres specifically in S phase [21–23]. In order to see if telomere localization was an S phase-specific event in the mouse cells, we stained the MEF and 3T3 cells with antibodies against PCNA (proliferating cell nuclear antigen). PCNA is expressed by interphase cells, but specifically associates with chromatin in cells undergoing DNA replication [53]. Based on established PCNA staining patterns, we were able to distinguish which cells were in S phase and to discern the S sub-phase as early, mid, or late (Figure 4A). We found that mTR-telomere associations were restricted to S phase cells and occurred throughout S phase (Figure 4A).
Figure 4. mTR localizes to subsets of telomeres in S phase.
A. Localization of mTR to telomeres occurs throughout S phase. mTR FISH (red) and TIN2 IF (green, marker for telomeres) were performed on 3T3 cells at various stages of the cell cycle. PCNA staining was performed to identify S phase cells, as well as S sub-phase (i.e. early, mid, or, late, as indicated). Merge panels show a superimposition of mTR and TIN2 panels, yellow indicates an overlap of signal. The foci where mTR and telomeres colocalize are indicated by white arrowheads. Open arrowheads (in G1/G2 panel) denotes mTR foci that do not overlap with a telomere. Scale bar, 10 microns. B. The frequency of mTR-telomere associations increases as cells progress through S phase. The percentage of 3T3 cells with mTR foci that display mTR at the telomere (assessed via TIN2) is plotted relative to time in hours (h) after release from a double thymidine block. A, asynchronous cells. Data was collected from multiple slides prepared as part of at least two separate experiments.
To further investigate the S phase-specific localization of mTR to telomeres, we synchronized 3T3 and MEF cells using a double thymidine treatment and analyzed mTR localization at various points in the cell cycle (Figure 4, Supplemental Figure 1A, and MEF data not shown). We found that mTR-telomere associations peaked in late S phase, when co-localization was observed in 38% of the 3T3 cells with mTR foci (Figure 4B, 6h post release from the double thymidine block, 60 of 157 cells). As can be seen in Figure 4B, the frequency of telomere association gradually increased as cells progressed through S phase. At G1/S (0h post release), mTR was found at at least one telomere in 16% of cells (18 of 113 cells with mTR foci). In early S (1h and 2h post release) and mid S (4h post release), the percentage increased steadily from 22% (1h post release, 38 of 174 cells with mTR foci) to 28% (2h post release, 55 of 196 cells with mTR foci) to 31% (4h post release, 50 of 162 cells with mTR foci). The frequencies of mTR-telomere associations observed were similar when determined relative to two different telomere markers (TIN2 and TPP1). Synchronized MEF cells behaved similarly to the 3T3 cells (data not shown). Our data indicate that mouse TR localizes to telomeres specifically during S phase and that co-localization of mTR and telomeres increases over the course of S phase.
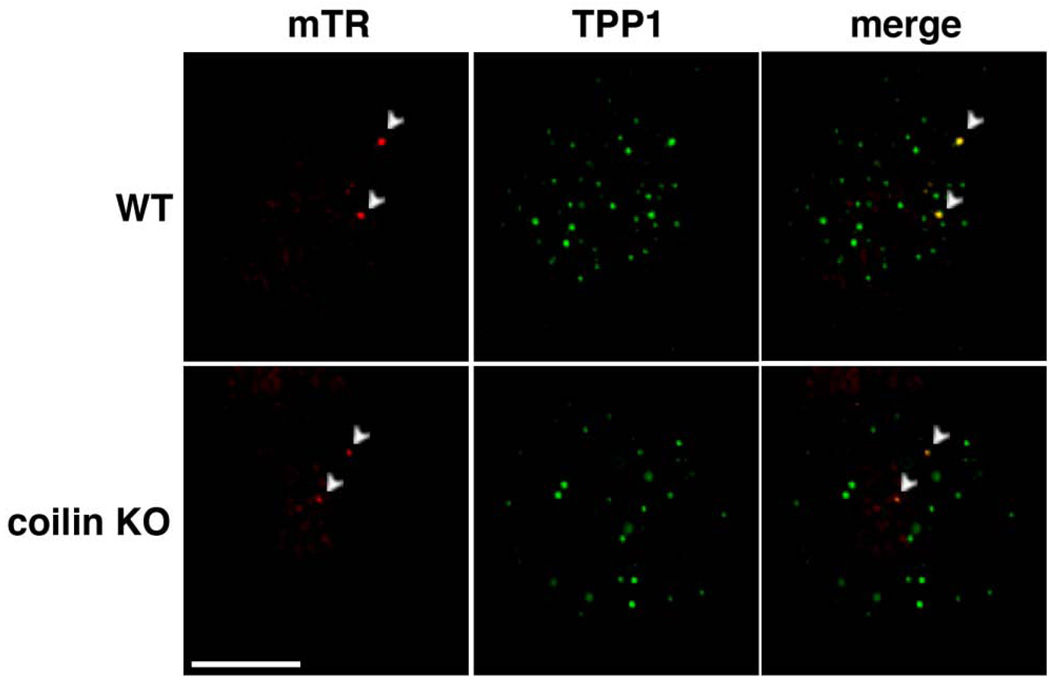
Recent reports have implicated Cajal bodies in the delivery of telomerase to telomeres in human cancer cells [19, 21, 23, 54]. However, we did not observe associations of mTR with Cajal bodies in cultured mouse cells (Figures 2 and 3), suggesting that Cajal bodies may not be necessary for delivery of mTR to the telomere. To further address a potential requirement for Cajal bodies in telomerase recruitment to telomeres in mouse cells, we performed mTR FISH and telomere IF on the coilin KO MEF cells (Figure 5). We found that mTR localized to the telomere in the absence of coilin and normal coilin-containing Cajal bodies (Figure 5). The frequency of mTR-telomere associations was the same (approximately 12% of cells) in the wild-type and coilin KO MEFs.
Figure 5. mTR localizes to the telomere in coilin KO MEFs.
mTR FISH (red) and TPP1 IF (green, marker for telomeres) were performed on MEF cells derived from wild type (WT) and coilin −/− (coilin KO) MEFs. Merge panels display an overlay of mTR and TPP1 signals. Arrowheads denote foci where both mTR and TPP1 signals overlap. Scale bar, 10 microns.
Human telomerase RNA localizes to Cajal bodies (and telomeres) in mouse cells
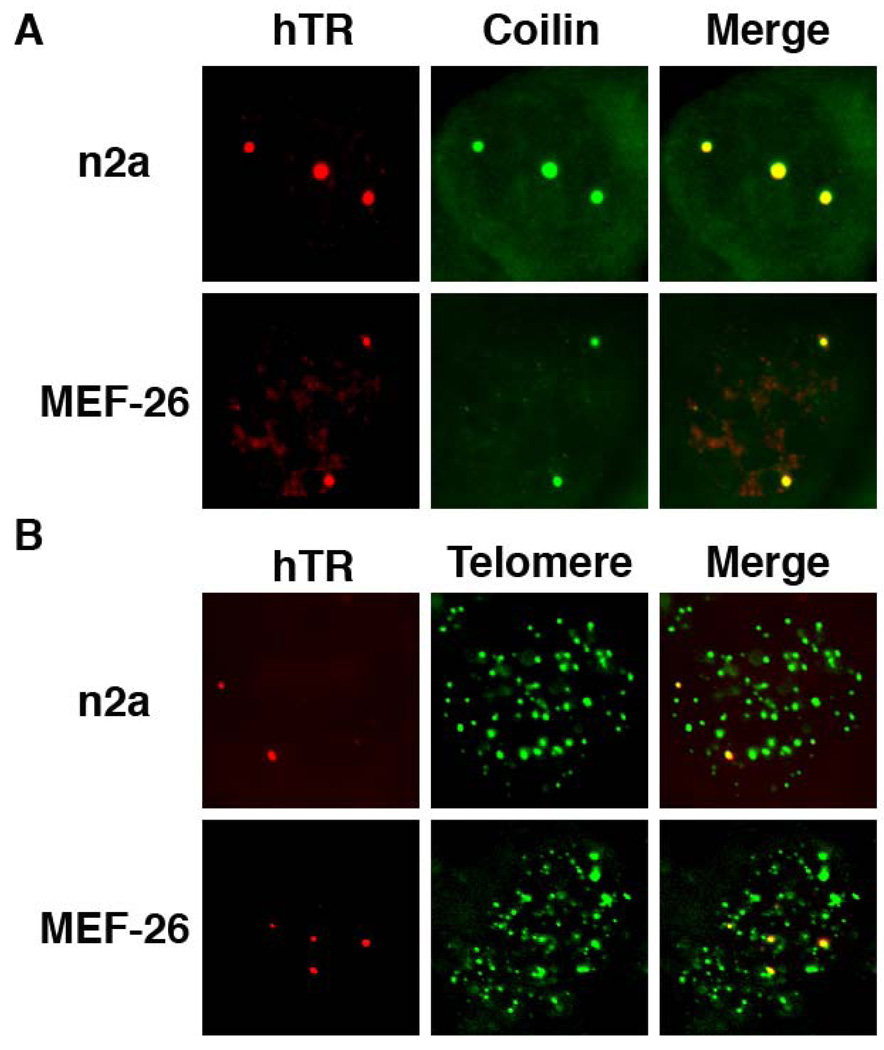
The lack of localization of mTR to Cajal bodies in mouse cells could be attributable to a difference in the mouse TR or in some aspect of the mouse cell environment (such as the nature of the Cajal bodies). To investigate the molecular basis for the difference in the localization patterns of mouse and human telomerase RNA, we examined the localization of hTR expressed in mouse cells. hTR and telomere localization were visualized by FISH, and Cajal bodies by coilin IF in both n2a and MEF26 cells (Figure 6). Unlike mTR, human telomerase RNA localized to Cajal bodies in mouse cells (Figure 6). hTR was also found at telomeres in synchronized S phase cells (Figure 6); co-analysis of PCNA staining patterns in the n2a mouse cells indicated that hTR-telomere co-localization was restricted to S phase. We did not observe any significant occurrence of hTR at foci outside of Cajal bodies and telomeres in the mouse cells. The results indicate that inherent features of mouse telomerase RNA dictate the distinct trafficking observed for TR in mouse.
Figure 6. Human telomerase RNA localizes to Cajal bodies and telomeres in mouse cells.
A. hTR co-localizes with Cajal bodies in mouse cells. Human telomerase RNA (hTR) was expressed in n2a and MEF-26 mouse cell lines and co-analyzed for hTR (detected by FISH, red panels) and coilin (detected by IF, green panels). Merge panels display an overlay of hTR and coilin signals. B. hTR co-localizes with telomeres in mouse cells. hTR-transfected n2a and MEF-26 mouse cells were examined for hTR (detected by FISH, red panels) and telomere (detected by FISH, green panels) signals. Merge panels display an overlay of hTR and telomere signals.
A novel localization pattern is associated with TR-telomere association in mouse cells
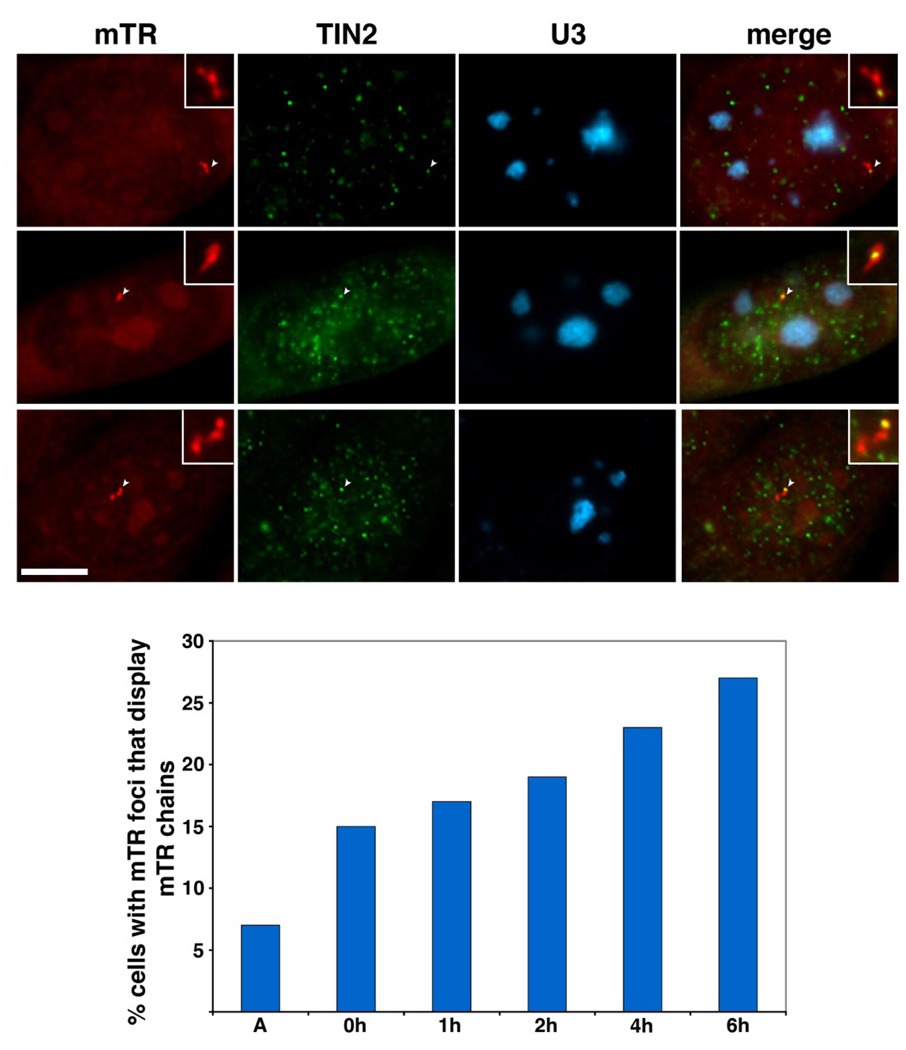
Given that telomerase does not require Cajal bodies for delivery to telomeres in mouse cells, we wondered how mTR arrives at its functional site. During the course of our analysis with synchronized cells we noticed an interesting pattern of mTR localization: mTR foci linked together in chains of 2–5 foci. We analyzed the localization of the chains of mTR foci relative to U3 snoRNA (FISH) and telomere (TIN2 or TPP1 IF) localization patterns, and found that in 60% of the chains, at least one of the mTR foci directly co-localized with a telomere (Figure 7A). Less than 10% of the chains overlapped nucleoli (visualized via U3 snoRNA) (Figure 7A). The appearance of the mTR chains coincides with the timing of mTR trafficking to telomeres; both MEF and 3T3 cells displayed a gradual increase in mTR chains as cells progressed through S phase (Figure 7B, Supplemental Figure 1B and data not shown). Taken together, our data suggest that the localization of mTR to telomeres during S phase is associated with the formation of distinct chains of mTR foci in mouse.
Figure 7. mTR foci coalesce during S phase of the cell cycle.
A. mTR foci link together to form chains that are frequently associated with telomeres but not nucleoli. 3T3 cells were stained for mTR (detected by FISH, red panels), TIN2 (telomere marker detected by IF, green panels), and U3 snoRNA (nucleolar marker detected by FISH, blue panels). Merge panels display a superimposition of all 3 panels. Insets show an enhanced magnification of the mTR chains. Arrowheads point to mTR foci within the chain that overlap with a telomere. Scale bar, 10 microns. B. The frequency of mTR chains gradually increases over S phase of the cell cycle. The percentage of 3T3 cells with mTR foci that display a mTR chain are plotted relative to time after release from a double thymidine block. A, asynchronous cells. Data compiled from two separate experiments.
Discussion
Studies in human cells have established a remarkable mechanism for the regulation of telomerase activity: regulated trafficking of the enzyme [19, 21–24]. In this work, we have delineated key steps in telomerase RNA trafficking in the genetically tractable mouse system, where we found fundamental similarities as well as interesting differences that expand our understanding of telomerase biology and open up new avenues of investigation.
Targeting telomerase to telomeres in mouse
Our results indicate that, in mouse as in human cancer cells, telomerase RNA localizes to subsets of telomeres during S phase (Figure 4A). Retinoblastoma (Rb) proteins that regulate S phase transitions and progression have been shown to play a role in telomere elongation in mouse [55], providing a link between telomerase regulation (perhaps trafficking) and the cell cycle that merits further investigation. But why telomerase is found at subsets of telomeres at any given time during S phase in both human and mouse remains a question of great interest.
One possibility is that telomerase visits all of the telomeres during the course of S phase, but is only found at a subset at any given time. We observe telomerase RNA at telomeres throughout most of S phase in both mouse (Figure 4) and human [23], and recent work provides evidence that human telomerase extends the majority of (or all) telomeres each cell cycle [34].
On the other hand, the observed localization pattern may reflect selective recruitment of telomerase to a fraction of telomeres during each cell cycle. Indeed, considerable evidence indicates that telomerase selectively extends the shortest telomeres in mouse cells [56, 57]. When mice with short telomeres are crossed with those with long telomeres, the shorter telomeres are extended first in the offspring [56]. Likewise, when telomerase levels are limiting (in a mouse heterozygous for TERT expression), the shortest telomeres in the population are maintained (while the longer ones grow shorter over time) [57]. Preferential elongation of short telomeres in human cells was also recently observed [58]. In yeast, it has been shown that only a small fraction of telomeres are extended by telomerase within any given cell cycle and that telomerase exhibits a preference to act on the shortest telomeres [59]. However, a direct correlation between telomere length and the frequency of telomerase recruitment has not yet been established for mammalian cells.
In mouse, we found that telomerase recruitment to telomeres reaches a maximum in late S phase (Figure 4B), whereas in human cells, the association peaks during mid S phase [23]. The distinct timing of telomerase recruitment in the two species may point to factors important in the recruitment process that differ between the organisms. For example, telomerase recruitment may be coordinated with replication of telomeric DNA, which may occur with distinct kinetics in various mammalian cells. Differences in average telomere length (20–150 kb in mouse cells versus 5–15 kb in human cells [60]) or in core telomerase-or telomere-associated components that regulate recruitment [1, 2, 36, 37] may also be important. Differences in the epigenetic modification of telomeric DNA or telomere-bound proteins, or in expression of telomeric repeat-containing (TERRA) non-coding RNAs could also influence telomerase recruitment dynamics in the two species [61–64]. However, while the precise kinetics may vary, our results suggest that S phase-specific recruitment of telomerase to telomeres is a conserved process in mammals.
Telomerase RNAs possess signals for distinct trafficking pathways
While in human cells it appears that localization of telomerase RNA to telomeres depends on trafficking through (and perhaps delivery by) Cajal bodies, in mouse cells we found that telomerase RNA trafficking does not appear to involve Cajal bodies (Figures 2,3,5,7). This divergence could reflect a difference in the mouse cell environment (such as the nature of Cajal bodies in mouse cells), however we found that hTR expressed in the mouse cell environment localizes to mouse Cajal bodies and telomeres just as is observed in human cells (Figure 6). The finding that hTR retains its distinct trafficking pattern in mouse cells indicates that telomerase RNAs possess signals that dictate subcellular transport pathways independent of other cellular components.
At the same time, other evidence indicates that TERT and other cellular factors also play important roles in telomerase trafficking that could affect differences observed among species. In human, localization of hTR to both Cajal bodies and telomeres depends on association with hTERT [22, 23]. A number of previous studies have demonstrated that hTR assembles with mTERT to form catalytically active telomerase complexes both in vitro and in mouse cells [14, 15, 65–67]. Interestingly however, while hTR assembles with mTERT [14, 15, 65–67] and is trafficked to telomeres in mouse cells (Figure 6), when hTR and mTERT are co-expressed in human cells (VA13 cells that are naturally devoid of hTR and hTERT) localization of hTR to telomeres is not detected [66]. These findings suggest that interactions between TERT and cellular components also contribute to localization.
Mouse telomerase RNA resides in novel nuclear foci distinct from Cajal bodies
In contrast to human cancer cells [20, 25] and Xenopus (frog) oocytes [68, 69], telomerase RNA does not accumulate in Cajal bodies in cultured mouse cells and instead is found in distinct, nucleoplasmic foci during most of the cell cycle (Figures 2 and 3). Our results further suggest that the distinct localization patterns of hTR and mTR derive from inherent properties of the RNAs (Figure 6). It is interesting that mTR is not found in Cajal bodies despite the presence of an intact CAB box motif with a sequence (UGAG) identical to that shown to be required to target hTR and small Cajal body RNAs (scaRNAs) to Cajal bodies [19, 20, 50, 70]. Furthermore, the protein that recognizes the CAB box motif (TCAB1 or WDR79) and is required for localization of TR to both Cajal bodies and telomeres (as well as for telomerase activity) in human cells is highly conserved in mouse [24, 35]. The most obvious difference between hTR and mTR is in the 5’ terminal sequences [70, 71]. hTR includes 45 nts upstream of the template region, and a portion of this sequence participates in intramolecular basepairing within the 5’ pseudoknot domain of the RNA to form the P1 stem [70]. In contrast, mTR contains just 2 nts upstream of the template and no P1 stem is formed. Additional work will be required to determine if differences in the 5’ regions or other more subtle variations in sequence and structure account for the ability or inability of the RNAs to associate with Cajal bodies. We certainly do not exclude the possibility of limited association of mTR with Cajal bodies in the mouse cells, however telomerase RNA clearly accumulates in distinct foci in mouse cells.
The relationship between mTR, the novel mTR foci identified here, and Cajal bodies may warrant further investigation. Interesting associations have been found between Cajal bodies and other related foci, and molecules found in Cajal bodies under some conditions can be found in distinct foci under other cellular conditions. During S phase in human cancer cells, hTR is found in distinct foci attached to Cajal bodies prior to localization to telomeres [23]. In addition, the Cajal body constituent SMN can be found in foci known as gems (gemini or twins of Cajal bodies [47]) in cells deficient in coilin methylation [72] and at certain times in development [73, 74]. The cell lines examined in this work (Figure 2) are derived from embryonic tissue (MEF, 3T3) or reflect an undifferentiated state (n2a, c2c12), and thus it is possible, for example, that mTR accumulates in Cajal bodies in mouse cells in other developmental states.
The novel mouse telomerase RNA foci identified in this work appear to play a role (akin to that of Cajal bodies in human cells [21, 23]) in the delivery of TR to telomeres. In mouse cells we found that TR localizes to telomeres in the absence of obvious accumulation within Cajal bodies (Figure 4A, and see also Figures 2 and 3) and in the absence of coilin (Figure 5). Instead, chains of mTR foci form and co-localize with telomeres specifically during S phase (Figure 7). The significance of the formation of groups of attached TR foci in both human (Cajal body-associated foci) and mouse (chains of mTR foci) cells at the time of delivery to telomeres is intriguing but currently unclear.
While there may be differences in the pathways in mouse versus man, the paradigm that has emerged from examining telomerase RNA localization in mammalian cells is that the RNA subunit of telomerase resides in subnuclear structures physically separate from telomeres and is mobilized to telomeres specifically during S phase. Moreover, the structures appear to play roles in the delivery of TR to the telomeres. The development of an effective mouse telomerase RNA detection procedure opens the door to a wealth of genetic approaches that should lead to a deeper understanding of how telomerase trafficking is regulated.
Supplementary Material
The percentage of 3T3 cells with PCNA staining (S phase marker detected by IF) is plotted relative to time in hours (h) after release from a double thymidine block. A, asynchronous cells. The plots shown in A and B correspond to the data presented in Figures 4 and 7, respectively.
Acknowledgements
We would like to thank Tammy Morrish and Carol Greider (Johns Hopkins University, Baltimore, MD) and Greg Matera (University of North Carolina, Chapel Hill, NC) for providing the mTR knockout and coilin knockout MEF cell lines, respectively. We would also like to thank U. Thomas Meier (Albert Einstein College of Medicine, New York, NY) and Susan Smith (New York University, New York, NY) for providing the Nopp140 and TPP1 and TIN2 antibodies, respectively. Joachim Lingner (EPF Lausanne, Switzerland) is thanked for providing the hTR expression construct. This work was supported by a National Cancer Institute (NCI) grant (R01 CA104676) to MPT and RMT. RLT was supported by an NIH training grant to the Department of Genetics at University of Georgia (GM07103) and a University of Georgia Dissertation Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Xin H, Liu D, Songyang Z. The telosome/shelterin complex and its functions. Genome Biol. 2008;9:232. doi: 10.1186/gb-2008-9-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.Armanios M. Syndromes of Telomere Shortening. Annu Rev Genomics Hum Genet. 2009 doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artandi SE. Telomeres, telomerase, and human disease. N Engl J Med. 2006;355:1195–1197. doi: 10.1056/NEJMp068187. [DOI] [PubMed] [Google Scholar]
- 6.Vulliamy TJ, Dokal I. Dyskeratosis congenita: the diverse clinical presentation of mutations in the telomerase complex. Biochimie. 2008;90:122–130. doi: 10.1016/j.biochi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 8.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 9.Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 11.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 12.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 13.Artandi SE, Alson S, Tietze MK, Sharpless NE, Ye S, Greenberg RA, Castrillon DH, Horner JW, Weiler SR, Carrasco RD, DePinho RA. Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc Natl Acad Sci U S A. 2002;99:8191–8196. doi: 10.1073/pnas.112515399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Rivera L, Herrera E, Albar JP, Blasco MA. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc Natl Acad Sci U S A. 1998;95:10471–10476. doi: 10.1073/pnas.95.18.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prowse KR, Avilion AA, Greider CW. Identification of a nonprocessive telomerase activity from mouse cells. Proc Natl Acad Sci U S A. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 18.Shay JW, Keith WN. Targeting telomerase for cancer therapeutics. Br J Cancer. 2008;98:677–683. doi: 10.1038/sj.bjc.6604209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 20.Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jady BE, Richard P, Bertrand E, Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP. Telomerase reverse transcriptase is required for the localization of telomerase RNA to cajal bodies and telomeres in human cancer cells. Mol Biol Cell. 2008;19:3793–3800. doi: 10.1091/mbc.E08-02-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 27.Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- 28.Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 29.Wong JM, Kusdra L, Collins K. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat Cell Biol. 2002;4:731–736. doi: 10.1038/ncb846. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Chen Y, Zhang C, Huang H, Weissman SM. Nucleolar localization of hTERT protein is associated with telomerase function. Exp Cell Res. 2002;277:201–209. doi: 10.1006/excr.2002.5541. [DOI] [PubMed] [Google Scholar]
- 31.Etheridge KT, Banik SS, Armbruster BN, Zhu Y, Terns RM, Terns MP, Counter CM. The nucleolar localization domain of the catalytic subunit of human telomerase. J Biol Chem. 2002;277:24764–24770. doi: 10.1074/jbc.M201227200. [DOI] [PubMed] [Google Scholar]
- 32.Ten Hagen KG, Gilbert DM, Willard HF, Cohen SN. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol Cell Biol. 1990;10:6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright WE, Tesmer VM, Liao ML, Shay JW. Normal human telomeres are not late replicating. Exp Cell Res. 1999;251:492–499. doi: 10.1006/excr.1999.4602. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tejera AM, Stagno d’Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010;18:775–789. doi: 10.1016/j.devcel.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 39.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 40.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blasco MA. Mice with bad ends: mouse models for the study of telomeres and telomerase in cancer and aging. Embo J. 2005;24:1095–1103. doi: 10.1038/sj.emboj.7600598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blasco MA. Telomere length, stem cells and aging. Nat Chem Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- 43.Chang S. Modeling aging and cancer in the telomerase knockout mouse. Mutat Res. 2005;576:39–53. doi: 10.1016/j.mrfmmm.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 44.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hackett JA, Greider CW. Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene. 2002;21:619–626. doi: 10.1038/sj.onc.1205061. [DOI] [PubMed] [Google Scholar]
- 46.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. Embo J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 48.Matera AG, Frey MR. Coiled bodies and gems: Janus or gemini? Am J Hum Genet. 1998;63:317–321. doi: 10.1086/301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. Embo J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;54:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. Embo J. 2003;22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Cao M, Gonzalo S, Dean D, Blasco MA. A role for the Rb family of proteins in controlling telomere length. Nat Genet. 2002;32:415–419. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 56.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 57.Liu Y, Kha H, Ungrin M, Robinson MO, Harrington L. Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc Natl Acad Sci U S A. 2002;99:3597–3602. doi: 10.1073/pnas.062549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Britt-Compton B, Capper R, Rowson J, Baird DM. Short telomeres are preferentially elongated by telomerase in human cells. FEBS Lett. 2009;583:3076–3080. doi: 10.1016/j.febslet.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 59.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 60.Kipling D, Cooke HJ. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 61.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. Embo J. 2009 doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoeftner S, Blasco MA. A 'higher order' of telomere regulation: telomere heterochromatin and telomeric RNAs. Embo J. 2009;28:2323–2336. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27:6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- 64.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 65.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. Embo J. 2003;22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fakhoury J, Marie-Egyptienne DT, Londono-Vallejo JA, Autexier C. Telomeric function of mammalian telomerases at short telomeres. J Cell Sci. 2010;123:1693–1704. doi: 10.1242/jcs.063636. [DOI] [PubMed] [Google Scholar]
- 67.Middleman EJ, Choi J, Venteicher AS, Cheung P, Artandi SE. Regulation of cellular immortalization and steady-state levels of the telomerase reverse transcriptase through its carboxy-terminal domain. Mol Cell Biol. 2006;26:2146–2159. doi: 10.1128/MCB.26.6.2146-2159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lukowiak AA, Narayanan A, Li ZH, Terns RM, Terns MP. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. Rna. 2001;7:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z, Tomlinson RL, Terns RM, Terns MP. Telomerase trafficking and assembly in Xenopus oocytes. J. Cell Science. 2010 doi: 10.1242/jcs.063750. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 71.Garforth SJ, Wu YY, Prasad VR. Structural features of mouse telomerase RNA are responsible for the lower activity of mouse telomerase versus human telomerase. Biochem J. 2006;397:399–406. doi: 10.1042/BJ20060456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell. 2002;3:329–337. doi: 10.1016/s1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- 73.Young PJ, Le TT, Dunckley M, Nguyen TM, Burghes AH, Morris GE. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res. 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]
- 74.Young PJ, Le TT, thi Man N, Burghes AH, Morris GE. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The percentage of 3T3 cells with PCNA staining (S phase marker detected by IF) is plotted relative to time in hours (h) after release from a double thymidine block. A, asynchronous cells. The plots shown in A and B correspond to the data presented in Figures 4 and 7, respectively.