Abstract
The temporospatial regulation of genes encoding transcription factors is important during development. The hlh-8 gene encodes the C. elegans mesodermal transcription factor CeTwist. Elements in the hlh-8 promoter restrict gene expression to predominantly undifferentiated cells of the M lineage. We have discovered that hlh-8 expression in differentiated mesodermal cells is controlled by two well-conserved E box elements in the large first intron. Additionally, we found these elements are bound in vitro by CeTwist and its transcription factor partner, CeE/DA. The E box driven expression is eliminated or diminished in an hlh-8 null allele or in hlh-2 (CeE/DA) RNAi, respectively. Expression of hlh-8 is also diminished in animals harboring an hlh-8 intron deletion allele. Altogether, our results support a model in which hlh-8 is initially expressed in the undifferentiated M lineage cells via promoter elements and then CeTwist activates its own expression further (autoregulation) in differentiated cells derived from the M lineage via the intron elements. This model provides a mechanism for how a transcription factor may regulate distinct target genes in cells both before and after initiating the differentiation program. The findings could also be relevant to understanding human Twist gene regulation, which is currently not well understood.
Keywords: Twist, mesoderm, C. elegans, intron elements, E box, hlh-8, bHLH transcription factor, autoregulation, hlh-8 (tm726)
INTRODUCTION
The regulation of key transcription factors is central to proper development. Twist is a transcription factor that is essential in mesoderm development and misregulation of Twist leads to several human diseases. Twist loss-of-function mutations result in an autosomal dominant craniosynostotic disorder called Saethre-Chotzen syndrome characterized by premature closure of the cranial sutures (Wilkie, 1997). On the other hand, up-regulation of Twist has been implicated in cancer metastasis (Yang et al., 2004). Thus, insights into Twist gene regulation are critical for understanding disease progression of both cancer and craniosynostotic disorders.
Twist is a basic helix-loop-helix (bHLH) transcription factor. bHLH proteins regulate their target genes by binding to DNA with the basic domain at a canonical E box site CANNTG. The helix-loop-helix domain is important for dimerization. The Twist pathway is conserved from Caenorhabditis elegans to humans (Wang et al., 2006) with one homolog of Twist in C. elegans, CeTwist. The bHLH domain in CeTwist is 59%–63% identical to Twist in other species (Harfe et al., 1998b). Therefore, the target sequences and dimer partners are likely to be conserved between humans and C. elegans. There is currently limited information regarding the human gene regulation. Both hypoxia-inducible factors (HIFs) (Gort et al., 2007) and a member of the Hox family, MSX2, have been suggested as direct regulators of human Twist (Satoh et al., 2008). However, in humans, Twist is expressed under non-hypoxic conditions and MSX2 is unlikely to be entirely responsible for Twist expression. Therefore, understanding the regulation of CeTwist may provide information about the control of human Twist gene expression.
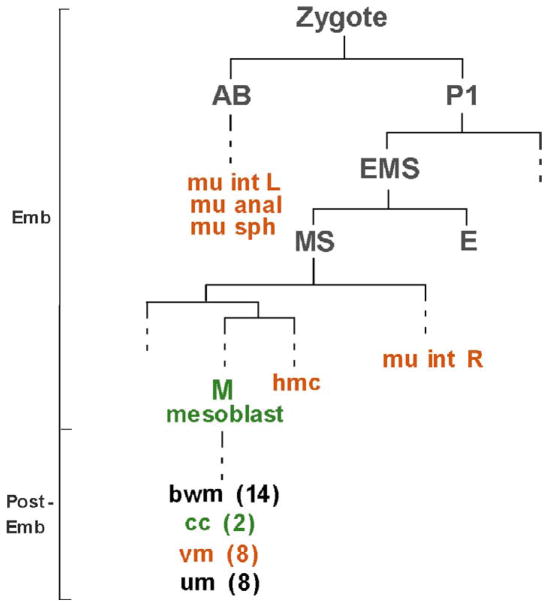
CeTwist is expressed in a subset of non-gonadal mesodermal derived tissues, including muscles (Fig. 1). It is expressed in the four enteric muscles that are required for defecation, and in the sex muscles, which are required for egg laying (Harfe et al., 1998b). The sex muscles arise from the M mesoblast cell and Twist is expressed in the undifferentiated cells of this lineage (Harfe et al., 1998b). CeTwist is also found in the coelomocytes, which are derived from the M lineage (Harfe et al., 1998b) and is predicted to be expressed in the head mesodermal cell (hmc) based on target gene activation (Zhao et al., 2007). Differentiation of the tissues where CeTwist is found occurs both embryonically and post-embryonically and in non-lineally related cells (Sulston and Horvitz, 1977; Fig. 1). Therefore, the precise temporospatial regulation of the gene is expected to be complex.
Figure 1. CeTwist is expressed in four different cell lineages.
An abbreviated lineage of C. elegans during embryogenesis (Emb) and post-embryogenesis (Post-Emb). Founder cells are grey. Cells that express CeTwist are shown in color, green for CeTwist expression controlled by elements in the promoter region and orange for CeTwist expression controlled by intron 1 elements. Dashed lines correspond to cell divisions not shown. Abbreviations: hmc, head mesodermal cell; mu int L and mu int R, left and right intestinal muscles, respectively; mu anal, anal depressor; mu sph, anal sphincter; bwm, body wall muscles; cc, coelomocytes, vm, vulval muscles; um, uterine muscles. (Sulston and Horvitz, 1977; adapted from Zhao et al., 2007).
The promoter of the hlh-8 gene, which codes for CeTwist, has been examined and elements controlling coelomocyte and undifferentiated M lineage expression have been identified (Harfe et al., 1998b). The M lineage expression is regulated by two Hox factors, LIN-39 and MAB-5, and a PBC homology cofactor, CEH-20 (Liu and Fire, 2000). Hox genes are important in the patterning and formation of the anterior/posterior axis in many organisms (Hueber and Lohmann, 2008). Due to the anterior, middle, and posterior location of the hmc, sex, and enteric muscles, respectively, it is unlikely that the same Hox genes control hlh-8 expression in all of these tissues. Furthermore, as in humans, there is a link between hypoxia and CeTwist. RNAi experiments that knockdown CeTwist suppress the phenotype of a constitutively active HIF mutant (Gort et al., 2007). In addition, there are potential hypoxia-response element sites in the promoter and introns of hlh-8. However, it is unlikely that HIFs are the sole regulators of hlh-8 since target gene expressionis observed under non-hypoxia situations (Wang et al., 2006). Therefore, further investigation of hlh-8 gene regulation is warranted.
This study focuses on additional cis-acting elements involved in hlh-8 regulation. We identify two hlh-8 intron E boxes, E1 and E2, which are necessary for expression of CeTwist in the vulval muscles, enteric muscles and the hmc. We show through the use of an hlh-8 presumptive null mutant and in vitrobinding studies that CeTwist directly regulates its own expression (autoregulation) through the E2 site. We also utilize a CeTwist mutant that contains an hlh-8 intron deletion to further explore the gene regulation. Our current studies support a model where the promoter and Hox factors provide a basal level of CeTwist in the undifferentiated cells of the M lineage. Autoregulation then provides a higher level of CeTwist in the vulval muscles, which differentiate from the M lineage, and potentially in other differentiated mesodermal cells. Autoregulation has been shown for other bHLH factors in mammals; however this is the first time it has been demonstrated for any Twist family gene. In addition, the altered level of CeTwist protein that is expressed from the E2 could be required for distinct target gene regulation in differentiated cells.
MATERIALS AND METHODS
C. elegans strains and maintenance
Animals were maintained according to standard conditions and techniques (Brenner, 1974). Investigations were done at 20°C. Three C. elegans strains were used in this study: N2 wild type, hlh-8(nr2061) referred to as hlh-8(−) (Corsi et al., 2000), and hlh-8(tm726) denoted here as hlh-8(iΔ). The hlh-8(iΔ) allele was isolated by the National Bioresource Project of Japan. The strain was backcrossed eight times, and the deletion was confirmed through sequencing.
Previously integrated gfp reporter constructs were introduced into hlh-8(iΔ) animals by standard genetic mating and confirmed through PCR and outcrossing to N2 animals. The following gfp reporter lines were used in this study: arg-1::gfp ccIs4443(II) (Corsi et al., 2002); egl-15::gfp ayIs2(IV) (Harfe et al., 1998a); NdEbox::gfp ccIs4656(IV) which contains the regulatory DNA of ceh-24(Harfe and Fire, 1998); hlh-8::gfp ayIs7(IV) which contains sequences upstream of hlh-8 in plasmid pBH47.70 (Harfe et al., 1998b); and ccIs4438 [hlh-8::gfp] (IV) (Yanowitz et al., 2004) which is expressed in all 6 coelomocytes and contains the coelomocyte enhancer from the hlh-8 promoter (Harfe et al., 1998b).
To gain an accurate expression period of the egl-15::gfp reporter, individual L4 animals were scored every hour to two hours for GFP expression. Concurrently, the stage of development for each hermaphrodite was determined by observing the morphology of the developing vulva. This allowed for accurate assessment of the initiation of egl-15::gfp expression. Once the animals reached adulthood, they were scored several times a day to determine when the egl-15::gfp was no longer expressed. Animals were scored for the M lineage division defects using the hlh-8::gfp ayIs7(IV) reporter and individual larvae starting from the L1 stage were scored several times a day until the adult stage was reached.
Construction of hlh-8 intron gfp transgenic lines
Reporter constructs were made from the hlh-8 first intron regions that were amplified via PCR and inserted into the multiple cloning site of the egl-18::gfp minimal promoter vector pKKMCS (gift from J. Wagmaister and D. Eisenmann; Wagmaister, et al., 2006) and the cloning junctions were sequenced. In order to test E1 and E2 contribution in the intron, the construct pSM7(E1E2) was used for Site-Directed Mutagenesis with mutant primers and the Quick Change Site-Directed Mutagenesis Kit (Stratagene Cat# 200516). The mutant constructs were confirmed through sequencing prior to injection into N2 animals (see below).
To examine the intron and promoter regions together, reporter constructs pSM27 and pSM28 were made from the 3.7 kb hlh-8 promoter, first exon, and first intron by Sequence Overlapping Extension (SOE) PCR (Hobert, 2002). Due to the length of the construct, a modified SOE PCR process was used to change E1 and E2 sites from CATCTG to AATCAG in pSM28. Briefly, mutagenic primers were used to change the E2 site in two standard PCR reactions, and then SOE PCR was used to fuse the two products together. These steps were repeated with mutagenic primers to change the E1 site. The resulting product was then cloned into a vector containing the hlh-8 promoter and entire genomic region and sequenced to confirm the E1 and E2 E boxes were mutated. The confirmed construct was used as template for SOE PCR to fuse gfp cDNA to the hlh-8 DNA.
N2 animals were transformed with the plasmid gfp reporter constructs (100 μg/ml) or SOE gfp reporter constructs (60 μg/ml) and the transformation marker pRF4 (50 μg/ml) by standard microinjection techniques (Mello et al., 1991). At least two independent lines were isolated and 30 animals per line were scored for gfp expression.
Homologous alignments of distantly related nematodes
Sequences were obtained and BLASTs were performed on WormBase (www.WormBase.org). ClustalW alignment of homologous regions was generated from http://www.ebi.ac.uk/Tools/clustalw2/index.html. Shading of the alignment was produced from BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
Analyzing the binding affinity of CeTwist dimers to E boxes through EMSA
Electrophoretic Mobility Shift Assays (EMSA) used recombinant CeTwist and CeE/DA that were purified from E. coli strains as described in Zhao et al., 2007. A wild-type and E box mutated set of four pairs of 20mers were designed (Fig. 5B). The probes were radiolabeled with γ-AT32P followed by incubation with the purified CeTwist and/or CeE/DA protein according to Harfe et al., 1998b. The input concentrations of proteins were determined by SDS-PAGE examination. The protein-probe mixture was separated with a 6% native polyacrylamide gel (Invitrogen Cat# EC63655BOX) followed by autoradiography and phosphorimage analysis.
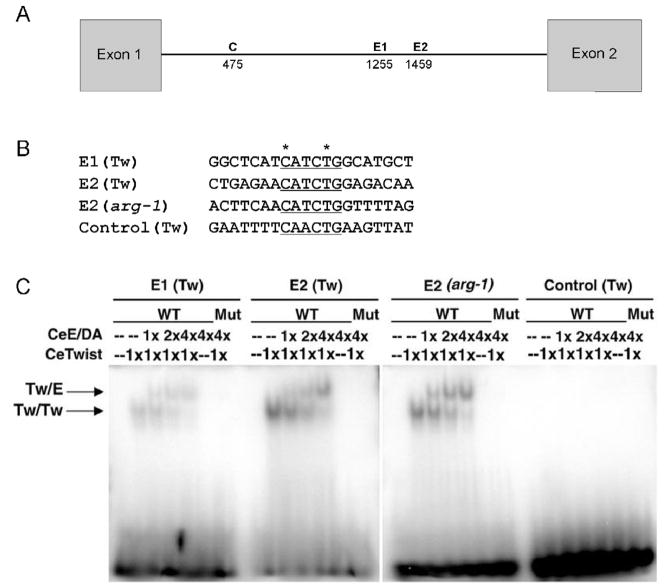
Figure 5. CeTwist dimers can bind to E1 and E2 in vitro.
An EMSA was done with CeTwist and its known binding partner CeE/DA. (A) Line drawing indicating the position of the 3 intron E boxes used in the EMSA: E1 (Tw), E2 (Tw), Control (C) (Tw). (B) 20mer probes used in gel shift assays contain the E box (underlined) and 14 flanking nucleotides. The asterisks indicate nucleotides that were mutated as a negative control. (C) Native gel containing the radiolabeled probes plus varying amounts of purified protein. 2x indicates twice as much CeTwist protein was added to the reaction than in 1x and half as much as in 4x. Arrows point to the bands corresponding to CeTwist/CeE/DA heterodimers (Tw/E-upper) or CeTwist/CeTwist homodimers (Tw/Tw-lower). WT corresponds to using the probes in (B). Mut corresponds to the mutated E box probes changed from CANNTG to AANNAG. Twist dimers bound with higher affinity to E2 (Tw) than E1 (Tw) and did not bind to the Control (Tw) E box.
Knockdown of CeE/DA by RNA interference (RNAi)
Animals containing the pSM10(E2a) plasmid were subjected to RNAi feeding treatment (Kamath et al., 2000). Nematode Growth Media agar plates containing 100 μg/ml ampicillin and 0.35 mM IPTG were used to culture HT115(DE3) E. coli. The hlh-2 dsRNA expression in HT115(DE3) was induced by a 24-hour room temperature incubation. L1 larvae were fed either HT115(DE3) expressing hlh-2 dsRNA or containing an empty L4440 vector. Animals were moved every 24 hours to a new RNAi feeding plate and adult animals were scored.
Reverse Transcription PCR (RT-PCR) and splice product cloning
Glass beads (Sigma) and Trizol Reagent (Invitrogen Cat# 15596-018) were used to extract total RNA from hlh-8(iΔ) and N2 animals (Wang et al., 2006). M-MuLV Reverse Transcriptase (New England Biolabs Cat# M0253S) was used with a poly-A primer to make cDNA-mRNA hybrids, which were subjected to PCR with primers from hlh-8 exon 1 and exon 5. Actin primers were used for total mRNA quantity control. Spliced products were individually extracted from an agarose gel and subjected to TA-cloning using vector pCR®2.1 (Invitrogen Cat# K2020). The cDNA clones were sequenced to identify the splice-site locations.
RESULTS
hlh-8 intron 1 sequences control expression in a subset of differentiated mesodermal tissues
Elements that control the expression of hlh-8 in undifferentiated M lineage cells and in coelomocytes were previously identified in the upstream 8.3 kb promoter region (Harfe et al., 1998b). However, translational constructs containing the entire hlh-8 genomic sequence have a broader expression pattern (Harfe et al., 1998b). We investigated the large first intron for additional elements that control hlh-8 expression. We used a plasmid, pKKMCS, containing an egl-18::gfp minimal promoter that can be activated by juxtaposition to a tissue-specific enhancer element to express gfp in a temporospatial orientation reflecting the activity of the element (Wagmaister et al., 2006). Transgenic animals containing a construct with the entire 2 kb hlh-8 intron 1 expressed gfp in the hmc, the vulval muscles (vms), and the enteric muscles (Fig. 2, 3A). This construct did not express in the M lineage or in the coelomocytes. A series of increasingly smaller, overlapping constructs was used to isolate a minimal enhancer region sufficient to drive expression in mesodermal tissue (Fig. 3B). A 503 nucleotide fragment that expressed gfp in all analyzed tissues was identified (pSM7(E1E2); Fig. 3B). pSM7(E1E2) contained two E-boxes referred to here as E1 and E2 (Fig. 3A). pSM7(E1E2) was divided into two constructs, pSM9(E1a) and pSM10(E2a), containing either E1 or E2 and each had expression in a subset of the tissues (Fig. 3B). Constructs made from smaller portions of pSM7(E1E2) revealed two 163 nucleotide regions, with either E1 or E2, which were sufficient to drive gfp in some of the tissues where hlh-8 is expressed (pSM14(E1b) and pSM15(E2b); Fig. 3C). Altogether, E1 or E2 containing constructs were able to express gfp and those with E2 alone expressed gfp in more tissues in a higher frequency of animals than those with E1 alone (Fig. 3B,C).
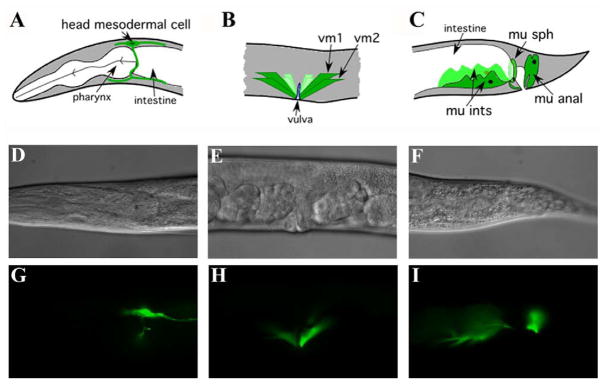
Figure 2. Intron 1 sequences control expression of hlh-8 in differentiated mesodermal tissues.
(A–C) Schematic representation of the tissues where hlh-8 is expressed from intron elements. (D–F) Nomarski and (G–I) GFP images of tissues. gfp expression is in (A, D, G) the head mesodermal cell, (B, E, H) the vulval muscles, (C, F, I) and the four enteric muscles.
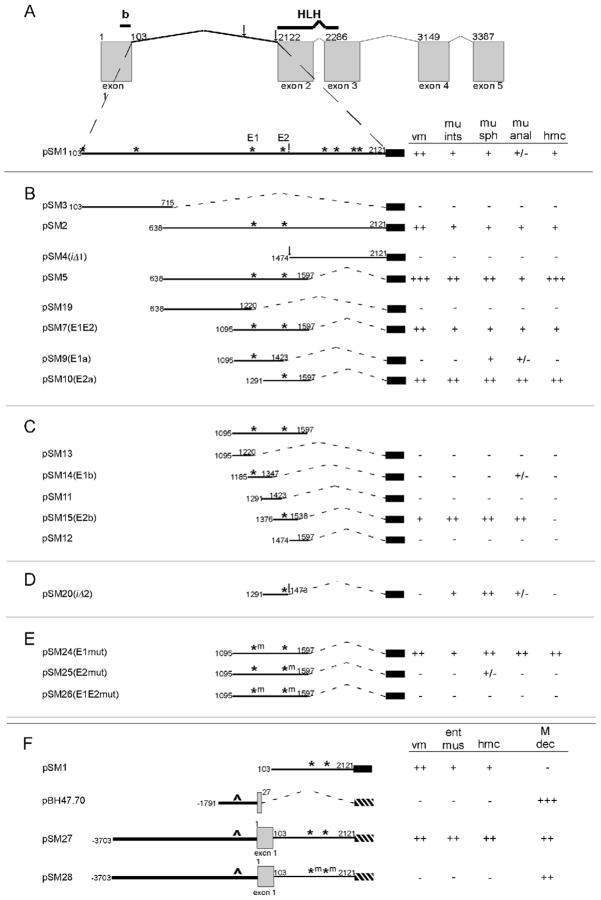
Figure 3. Two E boxes in intron 1 are necessary for expression of hlh-8::gfp in differentiated tissues.
Different regions of intron 1 were inserted into the egl-18::gfp minimal promoter construct (black rectangle) or reporters were made using gfp cDNA (hashed rectangle). GFP activity was scored in the vulval muscles (vm), enteric muscles (mu ints, mu sph, mu anal or ent mus), and head mesodermal cell (hmc). The gfp expression is reported based on the percentage of animals expressing the gfp: +++ (90–100%); ++ (60–89%); + (20–59%), +/− (7–19%), − (0–6%). (A) Line drawing of the hlh-8 locus. The first nucleotide of all exons and intron 1 are numbered above. The hlh-8(iΔ) 5′ deletion break points are indicated by vertical arrows (see Fig. 7B for sequence at 3′ end). pSM1 includes the entirety of intron 1 and all E boxes are indicated (asterisks). (B) Four sets of constructs were made to isolate a smaller region of DNA that retained expression. E1 and E2 are indicated with asterisks. (C) pSM7(E1E2) was divided into 5 smaller fragments. Expression was weakly retained in those fragments that contain the E1 E box, pSM14(E1b), and strongly retained in those with the E2 E box, pSM15(E2b). (D) pSM20(iΔ2) is a 3′ deletion of pSM10 where the hlh-8(iΔ) deletion starts. (E) Site-Directed Mutagenesis (m) of E1 and E2. (F) Expression of gfp was additionally scored in the undifferentiated cells of the M lineage (M dec). Constructs that contained only hlh-8 intron 1 DNA, did not express in the M lineage, pSM1. The pBH47.70 reporter contains the promoter and the first nine amino acids of hlh-8 (Harfe et al., 1998b). Elements in the promoter of hlh-8 (depicted with ^) control expression in the M lineage, pBH47.70. pSM27 contains the promoter, exon 1, and the complete DNA of intron 1 and expressed gfp in both differentiated and undifferentiated cells. E1 and E2 are mutated in pSM28 and expression was lost in the differentiated cells, but remained in the undifferentiated cells.
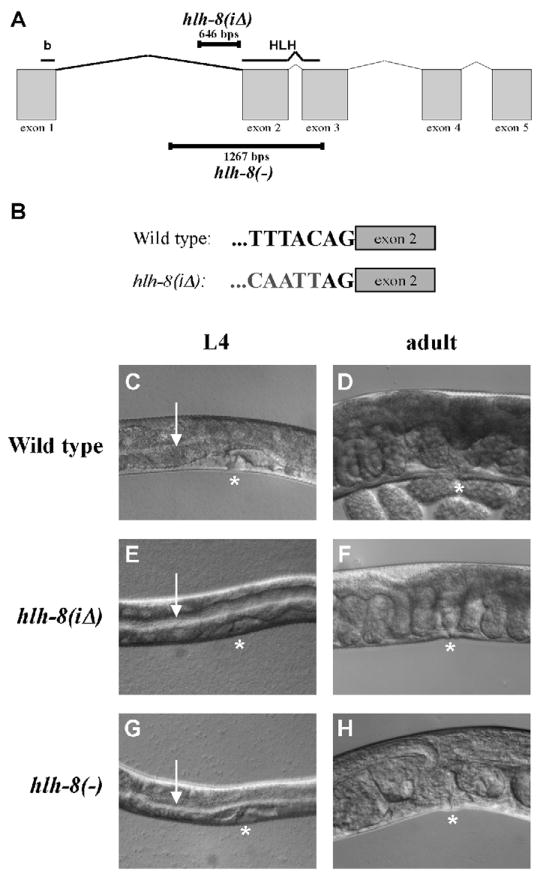
An interesting allele of hlh-8, whose phenotype is described below, contains a 646-nucleotide deletion of the 3′ region of intron 1 (Fig. 3A). To predict the expression pattern for the hlh-8(tm726) locus, referred to as hlh-8(iΔ), additional gfp reporters were examined. The DNA deleted in hlh-8(iΔ) animals was not sufficient to drive gfp expression (pSM4(iΔ1); Fig. 3B). A modified construct of pSM10(E2a) was made that removed the deleted nucleotides in the hlh-8(iΔ) locus (pSM20(iΔ2); Fig. 3D). Interestingly, the DNA that is adjacent to E2 but is absent in hlh-8(iΔ) animals was necessary for strong expression in nearly all tissues (compare pSM10(E2a) to pSM20(iΔ2); Fig. 3B,D). Therefore, it is unlikely that the DNA deleted in the hlh-8(iΔ) locus contained any elements that were sufficient for expression, but appears important for conferring strong expression from E2-containing constructs.
E1 and E2 E boxes regulate hlh-8 expression
To examine the contributions of E1 and E2 to gfp expression, Site-Directed Mutagenesis (SDM) was performed to mutate E1 and E2 in pSM7(E1E2). The E boxes were changed from CATCTG to AATCAG, which is expected to eliminate E box function (Karp and Greenwald, 2003). Mutating E1 did not affect the reporter from being activated in all scored tissues although the expression levels increased in the enteric muscles and hmc: from 46% of animals to 55% for mu int, 35% to 74% for mu sph, 22% to 66% for mu anal and 56% to 82% for hmc (compare pSM7(E1E2) to pSM24(E1mut); Fig. 3B,E). This expression pattern is similar to pSM10(E2a) that contains E2 and lacks E1 where 71% of animals expressed in mu int, 69% in mu sph, 84% in mu anal and 68% in hmc (compare pSM10(E2a) to pSM24(E1mut); Fig. 3B,E). Mutation of E2 resulted in gfp expression only in the anal sphincter (pSM25(E2mut); Fig. 3E). Furthermore, when both E1 and E2 are mutated, gfp expression was absent (pSM26(E1E2mut); Fig. 3E). Therefore, SDM confirmed that E1 and E2 are necessary for full expression of gfp in the hmc, vms and enteric muscles and E2 is more important than E1 for this function.
E1 and E2 elements are necessary for expression in differentiated tissues
Our reporter construct data taken together with previously reported data (Harfe et al., 1998b) suggested that the intron elements controlled expression in differentiated tissues, whereas promoter elements controlled expression in the undifferentiated M lineage (pSM1 and pBH47.70; Fig. 3F). To confirm this hypothesis, we engineered gfp lines that contained both the native hlh-8 promoter and the intron elements. A gfp construct, pSM27, that included 3703 bp upstream of the hlh-8 ATG, exon 1, and intron 1, had expression in the M lineage, enteric muscles, vms, and hmc. However, when the E1 and E2 sites were abolished by changing the E box nucleotides to AATCAG, only expression in the M lineage persisted (pSM27 and pSM28; Fig 3F). These results confirm that E1 and E2 are necessary for expression of hlh-8::gfp in differentiated tissues.
Extensive homology of a portion of intron 1 exists between distantly-related nematodes
A nucleotide comparison analysis was performed to determine the degree of conservation between the first intron of hlh-8 in C. elegans with the first intron of hlh-8 homologs in four distantly-related nematode species (Fig. 4). Interestingly, there was a long 470-nucleotide (1128–1597) region of C. elegans intron 1 homology across the varying species. When this C. elegans sequence was compared with the other species, C. elegans had 74% identity with C. brenneri, 72% identity with C. briggsae, 57% identity with C. japonica and 74% identity with C. remanei. However, when all five sequences were aligned there was 32% nucleotide identity. In support of the importance of the two intron E boxes, E1 and E2 were perfectly conserved in all five distantly-related species (Fig. 4). Furthermore, the last 124 nucleotides of the homologous region are deleted in the hlh-8(iΔ) mutant discussed below. However, this region is not shown in the alignment since there is no more than 3 bp in a row that are conserved among all five species.
Figure 4. Conservation of intron 1 between distantly related nematodes.
Alignment of a portion of hlh-8 intron 1 in C. elegans and homologs found in C. brenneri, C. japonica, C. briggsae and C. remanei. Shown is the DNA of intron 1 that had overlapping conservation between the five species. Black shading indicates all five nucleotides from each species are identical. Red boxes mark the location of the E1 and E2 E boxes. The purple line indicates the nucleotides deleted in the hlh-8(iΔ) mutation. Further downstream, there was no extensive homology among the five species so that alignment is not shown.
CeTwist and CeE/DA proteins bind to E1 and E2 E boxes in vitro
An in vitro Electrophoretic Mobility Shift Assay (EMSA) was used to ask if CeTwist and/or its known binding partner, CeE/DA, were able to bind to E1 and E2. Radiolabeled 20mers containing single E boxes were incubated with purified, recombinant CeTwist and/or CeE/DA for the EMSA. In addition to E1 and E2, two additional E boxes were tested in this assay (Fig. 5A,B). The control (Tw) E box was used as a negative control because it does not confer gfp expression (pSM3; Fig. 3B). The E2 (arg-1) E box is found in the well-characterized promoter region of the CeTwist target gene, arg-1, and was used as a positive control in this experiment (Zhao et al., 2007). Furthermore, E1 (Tw), E2 (Tw) and E2 (arg-1) all have the same E box sequence, CATCTG (Fig. 5B). Interestingly, CeTwist homodimers and CeTwist/CeE/DA heterodimers bound with greater affinity to E2 (Tw) than E1 (Tw) (Fig. 5C). Quantification demonstrated that CeTwist homodimers bound 4.5 fold more to E2 (Tw) than E1 (Tw) and CeTwist/CeE/DA heterodimers bound 3.6 fold more E2 (Tw) than E1 (Tw). More CeTwist/CeE/DA heterodimers preferentially bound to the probes than CeTwist homodimers when increasing amounts of CeE/DA protein were added. Importantly, the proteins were not able to bind to the Control (Tw) E box 20mer, nor to mutant E box probes (Fig. 5C). The EMSA data corresponded with the gfp expression data since E2 had greater affinity for CeTwist and CeE/DA than E1, and E2 constructs led to broader tissue expression than E1 constructs (Fig. 3B,C,E, 5C).
hlh-8 undergoes autoregulation through E2
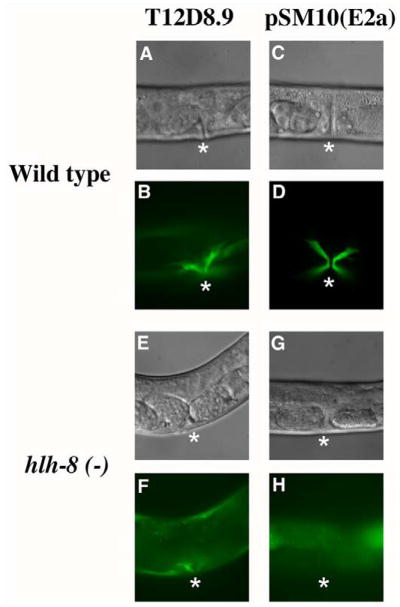
Since both CeTwist homodimers and CeTwist/CeE/DA heterodimers bound E1 and E2 in vitro, it was important to address whether these proteins were required for hlh-8 expression in vivo. The presumptive null mutant, hlh-8(nr2061), referred to here as hlh-8(−), was used to address whether hlh-8 can control its expression through the intron. The hlh-8(−) animals contain a large 1267 nucleotide deletion that removes 95% of the DNA responsible for coding the helix-loop-helix domain and are egg-laying deficient (Egl) and constipated (Con) due to the improper development of the vms and enteric muscles, respectively (Corsi et al., 2000). However, when reporter constructs of non-CeTwist target genes in the vms are introduced into the hlh-8(−) background, vulval muscle-like cells are observed. The vulval muscle-like cells are able to make connections to the body wall as seen with myo-3::gfp, but are not formed properly and the mutant animals do not lay any eggs (Corsi et al., 2000). The gene, T12D8.9 is a non-target gene that is expressed in vulval muscle-like cells of hlh-8(−) animals (Fox et al., 2007; Wang et al., 2006). A gfp reporter construct from T12D8.9 or pSM10(E2a) (Fig. 3B) was crossed into hlh-8(−) animals. The extrachromosomal T12D8.9::gfp reporter was expressed in the vms of 97% of wild-type young adults (n=30) and in vulval muscle-like cells of 100% of hlh-8(−) young adults (n=24). In a wild-type background, pSM10(E2a) was expressed in the vm cells of 92% of young adults (n=66). However, there was no expression of pSM10(E2a) in the vulval muscle-like cells of hlh-8(−) young adults (n=59) (Fig. 6). Therefore, the expression of pSM10(E2a) in the vms depends upon the presence of wild-type CeTwist molecules.
Figure 6. hlh-8 is autoregulated through E2.
Nomarski and GFP images of vulval region of wild-type (A–D) and hlh-8(−) animals (E–H) with reporter constructs of a non-target gene of CeTwist, T12D8.9 (A, B, E, F) and pSM10(E2a) (C, D, G, H). GFP expression was lost in vulval muscle-like cells in hlh-8(−) animals with pSM10(E2a) reporter in (H), but not with the T12D8.9 reporter in (F). Asterisks mark the vulval opening where vms or vm-like cells are located.
Regulation of hlh-8 by CeE/DA
Since a null mutation of the gene that encodes for CeE/DA, hlh-2, has not been isolated, hlh-2 RNAi was performed to investigate whether CeE/DA is also responsible for hlh-8 expression. CeE/DA is required early in embryogenesis and to circumvent this requirement, synchronized L1 animals carrying the pSM10(E2a) transgene were fed bacteria expressing either hlh-2 dsRNA or an empty control vector (Kamath et al., 2000). Previously, it was shown that hlh-2 RNAi treated animals are sterile with a protruding vulva (Pvl) (Kamath et al., 2000; Karp and Greenwald, 2004). Hence, the gfp pattern of RNAi-treated animals was scored in conjunction with those phenotypes to ensure the animals had a sufficient decrease in CeE/DA. In 58% of the hlh-2 RNAi-treated animals (n=86), the gfp was expressed in the vms, compared to 92% of the control animals (n=92). The hmc and enteric muscles are already born and expressing the gfp at the time of treatment so this experiment would only detect whether CeE/DA was required for maintenance rather than initial expression in these cells. The hmc expression decreased from 91% in control animals to 84% in treated animals and expression in the enteric muscles decreased from 100% in control to 93% in treated animals. The decrease of gfp expression in hlh-2 RNAi-treated animals was not as dramatic as seen in hlh-8(−) animals and may reflect the difference in protein decrease with the CeE/DA knock-down technique versus the CeTwist knock-out approach. Alternatively, there may be a partial requirement of CeE/DA in hlh-8 transcriptional regulation. Nonetheless, the RNAi experiments revealed an important role for CeE/DA in hlh-8 regulation in the vms.
An hlh-8 intron mutant has a subset of hlh-8(−) defects
To further investigate the control of hlh-8 expression, hlh-8(iΔ) animals containing a 646-nucleotide intron 1 deletion mutation were characterized. The 3′ break point of the deletion preserves the AG of the splicing acceptor site adjacent to exon 2, known to be required for splicing in a variety of genes (Aroian et al., 1993; Fig. 7A,B). The hlh-8(iΔ) animals were Con and able to lay embryos, but not at a wild-type rate, thus leading to embryos becoming stacked within the uterus (Semi-Egl) and an overall lower brood size (Fig. 7E,F). hlh-8(iΔ) animals laid an average of 26, 50, and 12 embryos on day 1, 2, and 3 of adulthood, respectively, and had an average brood size of 90 progeny (n=20). In comparison, wild-type animals laid 77, 177, and 24 embryos on day 1, 2, and 3 of adulthood, respectively, and had an average brood size of 278 progeny (n=10). In addition, 72% of hlh-8(iΔ) animals developed either a Pvl or an everted vulva (Evl) phenotype within 5 days of adulthood (n=46). Wild-type and hlh-8(−) animals were not Pvl or Evl within the first 5 days of adulthood (n>100). Altogether, hlh-8(iΔ) animals were Con, Semi-Egl, and Pvl/Evl, in contrast to hlh-8(−) animals that were Con and Egl (Fig. 7C–H).
Figure 7. hlh-8(iΔ) animals are constipated and partially egg-laying defective.
(A) Line drawing of the genomic region of hlh-8. The regions that encode the basic and helix-loop-helix domains are indicated above. The nucleotides deleted in hlh-8(iΔ) and the null allele hlh-8(−) are indicated with black bars. (B) 3′ sequence of intron 1 adjacent to exon 2 in wild-type and hlh-8(iΔ) animals. Grey nucleotides represent DNA 5′ of the deletion break point. (C–H) Nomarski images with asterisks marking the vulva location. (C, E, G) L4 larvae. Arrow indicates the lumen of the intestine. Note the expanded lumen in E and G. (D, F, H) Adults. (F) In hlh-8(iΔ) animals, embryos are overlapping each other as they are backing up in the uterus. (H) In hlh-8(−) animals developing late-stage embryos can be seen within the hermaphrodite.
To test the expression of CeTwist target genes, arg-1, ceh-24, and egl-15, in hlh-8(iΔ) animals, gfp reporter constructs were employed. The promoter regions and expression patterns of these three genes have been well characterized (Harfe et al., 1998b; Corsi et al., 2000; Zhao et al., 2007). In the hlh-8(−) animals, no gfp was expressed in any of the three gfp reporters (Corsi et al., 2000). Similarly, in hlh-8(iΔ) animals, arg-1::gfp and NdEbox::gfp (ceh-24) were not expressed in the animals (n>100). Conversely, egl-15::gfp was expressed in the vms in 15% of the animals (n=131). In wild-type animals, egl-15::gfp continued to express for at least 2 days of adulthood (n=32). However, of the hlh-8(iΔ) animals that did express the construct, the gfp prematurely turned off in 74% of those animals (n=19) (Table 1). As with the other characterized phenotypes, hlh-8(iΔ) animals were not as severe as hlh-8(−) animals, in that they were able to partially activate one of the CeTwist downstream targets.
Table 1.
GFP expression pattern of CeTwist downstream gene reporters and coelomocyte reporter in wild-type and hlh-8 mutant animals.
| Reportera | Genotype | GFP pattern |
|||
|---|---|---|---|---|---|
| vms | Ent Mus | hmc | cc | ||
| arg-1::gfp | |||||
| Wild type | +c | + | + | − | |
| hlh-8 (−) | − | − | − | ||
| hlh-8 (iΔ) | − | − | − | ||
| egl-15::gfp | |||||
| Wild type | + | − | − | − | |
| hlh-8 (−) | − | ||||
| hlh-8 (iΔ) | +/−d | ||||
| Ndebox::gfp | |||||
| Wild type | + | − | − | − | |
| hlh-8 (−) | − | ||||
| hlh-8 (iΔ) | − | ||||
| Intrinsic cc::gfp | |||||
| Wild type b | − | − | − | + | |
| hlh-8 (−) | +/−e | ||||
| hlh-8 (iΔ) | + | ||||
Integrated reporters: arg-1::gfp and egl-15::gfp are downstream targets of hlh-8. Ndebox::gfp is a transcriptional reporter of ceh-24, also a downstream target of hlh-8. Intrinsic cc::gfp is expressed in all six coelomocytes (cc) including the two that arise from the M lineage.
n value was greater than 100 for each category with the exception of Wild type (n=33) and hlh-8 (iΔ) (n=46) animals being scored with the intrinsic cc::gfp reporter.
Symbols used: +, reporter expression present in animals; −, no gfp expression; +/−, non-wild-type expression
15% of animals expressed GFP; 74% of those turned off prematurely as compared to wild-type animals which had persistent gfp expression past day 2 of adulthood.
23% of animals did not have 6 coelomocytes as found in wild-type.
We were also interested in characterizing the pattern of the M lineage in these animals. To accomplish this, a reporter that is expressed in the M lineage and a reporter that marks the coelomocytes as an output of proper M lineage division and differentiation were employed. M lineage patterning and differentiation is tightly controlled in C. elegans to become body wall muscles, coelomocytes and sex muscles, including vms, which are derived from sex myoblasts (SMs) (Fig. 1). An hlh-8 promoter gfp reporter is expressed in the M cell and descendants prior to cell differentiation (pBH47.70, Fig. 3F). This non-rescuing construct was used to view M patterning and division in different genetic backgrounds: wild-type, hlh-8(−), and hlh-8(iΔ) (Table 2). Animals were scored for the dorsal/ventral division of the first M cell division and the number and division of SMs. hlh-8(iΔ) animals displayed M patterning and differentiation defects, but not as frequently as the defects as in hlh-8(−) animals (Table 2). To examine differentiation from the M lineage, we counted the number of coelomocytes in the mutant animals. The posterior 2 of the 6 C. elegans coelomoctyes are derived from the M lineage. An intrinsic coelomocyte marker was used to express gfp in all 6 coelomocytes (Yanowitz et al., 2004). This construct revealed no significant difference in the number of coelomocytes in hlh-8(iΔ) animals(n=46) compared to wild type (n=33). However in hlh-8(−) animals only 77% of the time there were the correct number of 6 coelomocytes (n=130) (Table 2). Therefore, it is unlikely the intron 1 deletion in hlh-8(iΔ) animals is affecting the differentiation of these cells.
Table 2.
M lineage descendants in wild-type and hlh-8 mutant animals.
| Genotype | M lineage pattern in animalsa | ||
|---|---|---|---|
| D/V division of M cell | 2 SM-like cells | Division of SM-like cells | |
| Wild type | 100% (72)b | 100% (22) | 100% (26) |
| hlh-8 (−) | 30% (98) | 32% (38) | 100% (27) |
| hlh-8 (iΔ) | 79% (97) | 48% (44) | 92% (53) |
Animals expressed an integrated hlh-8::gfp that contained a promoter and no coding sequences.
n values of animals scored are in parentheses.
Splicing defects in hlh-8(iΔ) animals
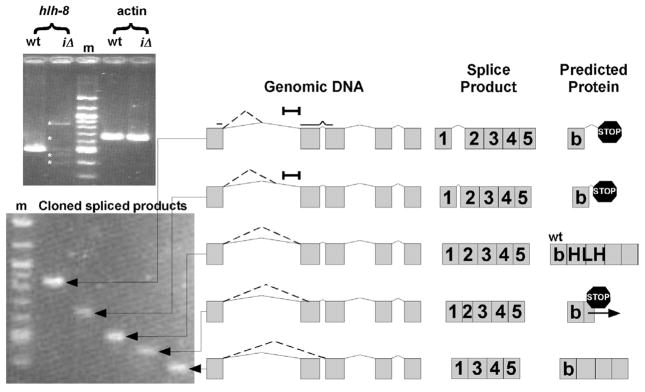
Due to the incomplete penetrance of the hlh-8(iΔ) phenotype and the position of the intron deletion, we investigated whether there could be splicing defects at the hlh-8 locus in the mutant animals. RT-PCR revealed five spliced products from the hlh-8(iΔ) locus that were sequenced to determine the genomic location of splicing (Fig. 8). The two larger molecular weight products were caused by splicing into intron 1. Protein formation is not predicted to occur from these mRNAs due to stop codons in all three reading frames of the intron. The third spliced product was generated through wild-type splicing. The hlh-8(iΔ) animals had a decreased level of the wild-type product when normalized against actin transcript levels. The remaining two splice products corresponded to splicing occurring into exon 2 or directly to the wild-type exon 3 acceptor site. Splicing into exon 2 led to a frameshift followed by a stop codon and thus, is not predicted to form a functional protein. However, the smallest molecular weight transcript does not cause a frameshift and potentially could result in a protein product that contained the intact basic domain, but not the majority of the helix-loop-helix domain. Altogether, the hlh-8(iΔ) animals had splicing defects that caused an overall decrease in hlh-8 mRNA and four alternative splice products (Fig. 8). The decrease in CeTwist is likely to contribute to the phenotype of the hlh-8(iΔ) animals.
Figure 8. Splicing defects in hlh-8(iΔ) animals.
RT-PCR revealed 4 alternate spliced products (asterisks) of hlh-8 in hlh-8(iΔ) animals (Top Gel). Individual clones of spliced fragments (Bottom Gel). The schematics on the right are drawn to scale. ‘Genomic DNA’ indicates where splicing occurs in each fragment (dotted line). Exons are represented by gray boxes and introns by solid lines. A black bar indicates the region of DNA removed in hlh-8(iΔ) animals. ‘Spliced Product’ designates the various splice products determined from sequencing data. ‘Predicted Protein’ depicts the polypeptide that results from each of the spliced products. The third splice product is the wild-type product, identified by ‘wt’. Premature stop codons (stop signs) and the frameshift (horizontal arrow) are indicated.
DISCUSSION
In this study, it was shown that two conserved E boxes in the first intron of hlh-8, E1 and E2, were necessary to drive gfp expression in the enteric muscles, hmc, and vms (Fig. 3). Furthermore, in vitro and in vivo results showed that E2 was more critical for the expression of gfp in these tissues (Fig. 3, 5) and that hlh-8 undergoes autoregulation through E2 (Fig. 6). Additionally, hlh-2 RNAi revealed an important role for CeE/DA in the expression of hlh-8 through E2. Furthermore, characterization of the hlh-8(iΔ) allele revealed attenuated phenotypes when compared to hlh-8(−) animals (Fig. 7, Table 1, 2).
DNA adjacent to E2 is critical for expression in the vms and hmc
This study revealed that DNA in addition to E1 and E2 is important to enhance expression of gfp in specific tissues. Intriguingly, the 3′ DNA adjacent to E2 was important for expression in the vms and hmc. Specifically, a construct that contained E2 and 133 nucleotides of the adjacent 3′ DNA had high expression in all tissues (pSM10(E2a); Fig. 3B). However, removing 60 bps from the 3′ region resulted in no hmc expression and decreased vm expression from 87% to 22% of the animals examined (pSM15(E2b); Fig. 3C). In contrast, expression in the enteric muscles was unchanged in these animals compared to those that harbored the longer E2 3′ DNA (pSM10(E2a) vs. pSM15(E2b); Fig. 3B,C). A construct that removed an additional 65 nucleotides of the 3′ DNA that corresponds to the remainder of the deleted nucleotides of the hlh-8(iΔ) allele, was not expressed in the vms nor the hmc but retained enteric muscle expression (pSM20(iΔ2); Fig. 3D). These constructs clearly emphasize the importance of the E2 3′ flanking DNA that is removed in hlh-8(iΔ) animals.
There are two possibilities to explain the importance of the E2 3′ flanking DNA in expression in the vms and hmc. First, the sequence may be critical for CeTwist dimers to properly bind. Changing the three nucleotides immediately flanking the E boxes in the promoter of a CeTwist target, ceh-24, disrupts the activity of the E boxes (Harfe et al., 1998b). Furthermore, the in vitrogel shift assay results demonstrated that the seven flanking nucleotides are important for CeTwist-containing dimers to bind, since both E1 and E2 had the same E box sequence, CATCAG, yet the dimers bound with differing affinities (Fig. 5). However, the additional DNA of the constructs used in this study that affected the activity of E2 was at least nine nucleotides away from E2 (pSM20(iΔ2); Fig. 3D). CeTwist dimer selection may explain the tissue-specific expression dependence on the additional 3′ DNA. Perhaps, CeTwist homodimers control expression in the enteric muscles and binding of this dimer to the E boxes is not sensitive to the 3′ DNA sequence. On the other hand, CeTwist/CeE/DA heterodimers may control hmc and vms expression and this dimer binding is dependent on the 3′ sequence. Second, the E2 3′ flanking DNA may contain elements required for a tissue-specific co-factor to bind, allowing CeTwist dimers to bind or to function properly. This type of tissue-specific regulation has previously been proposed for arg-1, adownstream target of CeTwist that is expressed in the hmc, vms, and enteric muscles (Zhao et al., 2007). Three E boxes and another element, called a GT box, located in the upstream promoter region of arg-1 are responsible for distinct aspects of tissue-specific expression. Specifically, the GT element is important for expression in the hmc and vms, but does not influence the expression of arg-1 in the enteric muscles (Zhao et al., 2007). Although there were no GT or other known elements in the hlh-8 first intron, there were sequences 3′ of E2 that were completely conserved among nematode species and could represent binding sites for other transcription factors (Fig. 4). Multiple transcription factors are expressed in the M lineage and the differentiated cells where CeTwist functions (Reece-Hoyes et al., 2007) and could be contributing to hlh-8 regulation as well. It will be important to explore this region of DNA as either a binding element for a CeTwist co-factor or as an important sequence for proper CeTwist dimer binding. The deletion of these potential elements could also be contributing to the hypomorphic phenotypes of the hlh-8(iΔ) animals (see model below).
E1 may have repressor and enhancer activity
We also uncovered a potential repressor role for E1. The repressor activity was observed when comparing the construct that contains both E1 and E2, to the SDM construct with the mutated E1 or to the construct that contains E2 and lacks E1 (pSM7(E1E2); pSM24(E1mut); pSM10(E2a); Fig. 3B,E). When E1 is disrupted or not present, the expression level in the enteric muscles and hmc increased. Interestingly, the DNA directly upstream of E1 is highly conserved between all 5 Caenorhabditis species, and may contain a binding site for an additional factor (Fig. 4). To explore the possibility of a co-repressor element in the conserved DNA, the TESS program was used to search multiple databases for factors that bind to consensus sequences (http://www.cbil.upenn.edu/tess). However, we did not find any transcription factor binding sites. Therefore, this conserved portion of DNA could correspond to a binding site for a new factor or represent a non-consensus site.
Human Twist gene regulation
The concentration of Twist molecules is critical to control because inappropriate up-regulation of human Twist is implicated as a key factor for the metastasis of tumors and coding region mutations cause Saethre-Chotzen syndrome, an autosomal dominant disorder (Wilkie, 1997; Yang et al., 2004). In the human Twist gene there are three E boxes in the 2 kb upstream region, three in intron 1, one flanking intron 1 and exon 2, one in exon 2, and four in the 2 kb downstream region. The E box flanking intron 1 and exon 2 has the specific E1/E2 sequence in the reverse orientation, CAGATG. A recent comprehensive study of bHLH factors in C. elegans has shown that of the 16 possible E boxes examined, CeTwist and CeE/DA preferentially bind to three E boxes, including the hlh-8 E1 and E2 sequence CATCTG (Grove et al., 2009). It is plausible that the transcriptional regulation is conserved and human Twist undergoes autoregulation through the CAGATG or a different E box. In fact, only a fourth of the patients in a recent study diagnosed with Saethre-Chotzen syndrome had a mutation in the coding region of the Twist gene (Stenirri et al., 2007). Perhaps, disruption of an element outside of the Twist coding region could explain the phenotype of these Saethre-Chotzen syndrome patients. Furthermore, mutations in Twist regulatory elements could also provide a molecular basis for other craniosynostotic disorders.
The hlh-8(iΔ) phenotype may be due to a decreased level of wild-type CeTwist and also disruption of E2 3′ DNA
hlh-8(iΔ) animals had a less severe phenotype than the presumptive hlh-8 null mutants. Two important findings may explain the hlh-8(iΔ) phenotype. First, RT-PCR unveiled splicing defects in hlh-8(iΔ) animals due to disruption of the intron 1 splicing acceptor site leading to four aberrant splice products and a decrease of wild-type CeTwist mRNA (Fig. 8). Second, the proximity of the 5′ deletion breakpoint of the hlh-8(iΔ) mutation to E2 may interfere with the autoregulation of hlh-8 leading to a decreased amount of CeTwist in differentiated tissues (Fig. 3D). In both scenarios, the lower level of CeTwist would be predicted to cause less severe phenotypes including partial egg-laying defects, attenuated M lineage patterning defects, and partial gene target activation in comparison to hlh-8(−) animals that are missing functional CeTwist protein. Additionally, the partial vm target gene activity in hlh-8(iΔ) may be due to the unique regulation of arg-1 and ceh-24 versus egl-15. Indeed, previous studies have shown animals that are heterozygous for a semi-dominant E29K mutation in the basic domain of CeTwist do not express egl-15 but do express arg-1 (Corsi et al, 2002) further emphasizing the unique response of the genes to the level of wild-type CeTwist.
Model for regulating hlh-8 expression
The elements necessary for hlh-8 expression in the undifferentiated M lineage cells and coelomocytes have been identified previously in the promoter region (Harfe et al., 1998b). Furthermore, it has previously been reported that Hox factors are responsible for activating the hlh-8 promoter (Liu and Fire, 2000). This prior information about hlh-8 fits well with the new gene regulation discovery from this study.
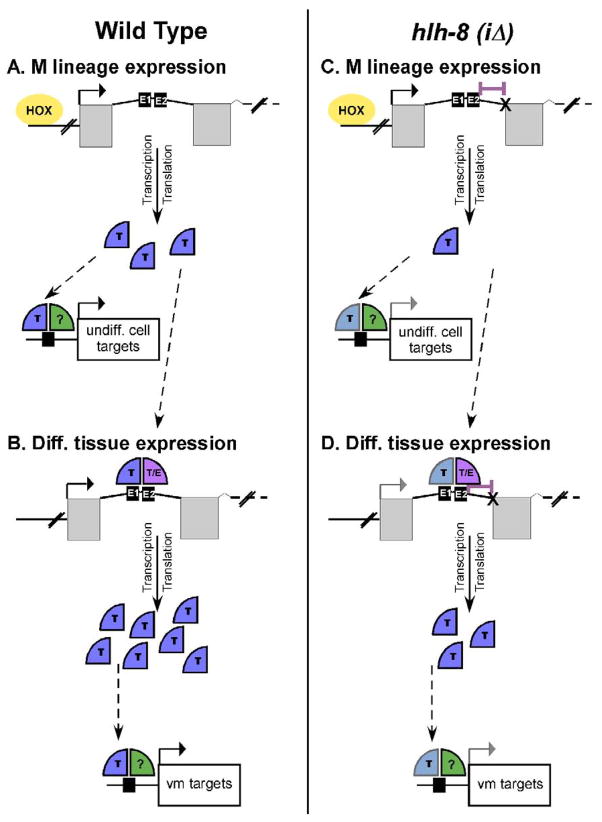
We propose a model in which the Hox factors bind to the hlh-8 promoter and are responsible for the expression of a moderate level of CeTwist in the animal. This moderate level of CeTwist molecules is sufficient for early M lineage development (Fig. 9A). Once a threshold of CeTwist molecules accumulates, autoregulation of CeTwist occurs through the E boxes in the first intron, which increases the concentration of CeTwist molecules in the tissues. This higher level of CeTwist is required for proper development of the vms (Fig. 9B). This model fits well with the fact that the vms are derived from M lineage cells (Fig. 1). However, if E1 and E2 are strictly autoregulatory elements, then additional unidentified elements must initially be responsible for expression of CeTwist in the enteric muscles and the hmc. Furthermore, it is possible that certain target genes may require a higher level of CeTwist for either activation or full expression. Evidence for this possibility comes from hlh-8(iΔ) animals (Fig 9C,D). These nematodes had a lower level of CeTwist and were able to partially activate downstream targets of CeTwist. Additionally, even though vms were made in hlh-8(iΔ) animals, they did not function properly, which caused a semi-Egl phenotype. Thus, a threshold of CeTwist was required for development of vms and certain target gene activation, but a higher level was needed for proper vm function and the expression of other CeTwist target genes (Fig. 9C,D). A better understanding of exactly how individual target genes are regulated may help in understanding the importance of CeTwist concentration for specific function and tissue development.
Figure 9. Model for the transcriptional regulation of CeTwist.
(Left Panel) Wild-type animals. (Right Panel) hlh-8(iΔ) animals. (A) Hox Factors (HOX) cause expression of CeTwist (T) earlier in development. CeTwist and an unknown dimer partner (?) activate target genes (black arrows). (B) CeTwist homodimers or CeTwist/CeE/DA heterodimers autoregulate hlh-8 through intron 1 (T, T/E), leading to an increase of CeTwist molecules which activate vm target genes. (C) Splicing defects (X) in hlh-8(iΔ) animals (deletion indicated with purple bar) cause an initial decrease in CeTwist molecules resulting in a decrease in target gene activation in undifferentiated cells (grey arrow). (D) The lower starting CeTwist concentration coupled with the deletion of the 3′ nucleotides adjacent to E2 causes inefficient autoregulation in the M lineage leading to a decrease in vm target gene activation in hlh-8(iΔ) animals (grey arrow).
To clarify the CeTwist transcription regulation model, it will be important to find additional elements in the intron that are responsible for contributing to the spatial expression of hlh-8 controlled by E1 and E2. Mutational and RNAi analysis of the factors that bind to these elements will not only elucidate the transcriptional regulation of CeTwist, but also unlock CeTwist’s relationship with other transcription factors. Furthermore, an understanding of CeTwist homodimer and CeTwist/CeE/DA heterodimer individual activities will provide added support for the model.
The CeTwist regulation model presented here is strengthened by the fact autoregulation has been reported to affect the temporospatial expression of other bHLH factors such as PTF1a, a non-Twist family factor. Interestingly, PTF1a regulates itself in through E boxes found in the promoter region of its own gene. The autoregulatory element is shown to have a maintenance role in PTF1a expression in the early epithelium precursors and also later in development to maintain a superinduction of PTF1a for the differentiation program of the polarized acinar cells of the pancreas (Masui et al., 2008). However, the CeTwist autoregulation is unique because it does not share the maintenance role that PTF1 autoregulation controls. E1 and E2 elements seem to be important for controlling expression of CeTwist in distinct cells at specific developmental time points. Specifically, elements in the promoter region control expression of CeTwist in sex myoblast descendants, which are precursors to the sex muscles, up to the point of differentiation. In contrast, E1 and E2 of intron 1 are responsible for CeTwist expression in differentiated vms. This control mechanism could represent a way to alter the levels of CeTwist and thereby switch which target genes are regulated in undifferentiated versus differentiated cells by CeTwist. Furthermore, this regulation of CeTwist may portray an important universal control mechanism for transcription factors that play more than one role in the same cell at different points in development.
Acknowledgments
We thank David Eisenmann, Javier Wagmaister, Michael Krause, Shohei Mitani and the Japanese National BioResource Project for strains and reagents. We also thank Michael Krause, Thomas Brodigan, Tetsunari Fukushige, Elizabeth McGinn, and Peng Wang for technical support. We are grateful to Michael Krause, Andy Golden, Michael Mullins and John Golin for insightful comments on the manuscript. This project was supported by Grants K22DE14541 and R15DE018519 from the National Institute of Dental and Craniofacial Research at the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aroian RV, Levy AD, Koga M, Ohshima Y, Kramer JM, Sternberg PW. Splicing in Caenorhabditis elegans does not require an AG at the 3′ splice acceptor site. Mol Cell Biol. 1993;13:626–637. doi: 10.1128/mcb.13.1.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. Twist1 dimer selection regulated cranial suture patterning and fusion. Dev Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- Corsi AK, Kostas SA, Fire A, Krause M. Caenorhabditis elegans Twist plays an essential role in non-striated muscle development. Development. 2000;127:2041–2051. doi: 10.1242/dev.127.10.2041. [DOI] [PubMed] [Google Scholar]
- Corsi AK, Brodigan TM, Jorgensen EM, Krause M. Characterization of a dominant negative C. elegans Twist mutant protein with implications for human Saethre-Chotzen syndrome. Development. 2002;129:2761–2772. doi: 10.1242/dev.129.11.2761. [DOI] [PubMed] [Google Scholar]
- Fox RM, Watson JD, Von Stetina SE, McDermott J, Brodigan TM, Fukushige T, Krause M, Miller DM., III The embryonic muscle transcriptome of Caenorhabditis elegans. Genome Biology. 2007;8:188. doi: 10.1186/gb-2007-8-9-r188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gort EH, Van Haaften G, Groot AJ, Plasterk RH, Shvarts A, Suijkerbuijk KP, Van Laar T, Van der Wall E, Raman V, Van Diest PJ, Tijsterman M, Vooijs M. The TWIST1 oncogene is a direct target of hypoxia-inducible factor-2α. Oncogene. 2007:1–10. doi: 10.1038/sj.onc.1210795. [DOI] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJM. A Multiparameter Network Reveals Extensive Divergence between C. elegans bHLH Transcription Factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Branda CS, Krause M, Stern MJ, Fire A. MyoD and the specification of the muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development. 1998a;125:2479–2488. doi: 10.1242/dev.125.13.2479. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Fire A. Muscle and nerve-specific regulation of a novel NK-2 class homeodomain factor in Caenorhabditis elegans. Development. 1998;125:421–429. doi: 10.1242/dev.125.3.421. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Vas Gomes A, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998b;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR Fusion-Based Approach to Create Reporter Gene Constructs for Expression Analysis in Transgenic C. elegans. BioTechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008;30:965–979. doi: 10.1002/bies.20823. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biology. 2000;2:1–10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X, Greenwald I. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 2003;17:3100–3111. doi: 10.1101/gad.1160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X, Greenwald I. Multiple roles for the E/Daughterless ortholog HLH-2 during C. elegans gonadogenesis. Dev Biol. 2004;272:460–469. doi: 10.1016/j.ydbio.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Krause M, Park M, Zhang JM, Yuan J, Harfe B, Xu SQ, Greenwald I, Cole M, Paterson B, Fire A. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. 1997;124:2179–2189. doi: 10.1242/dev.124.11.2179. [DOI] [PubMed] [Google Scholar]
- Liu J, Fire A. Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development. 2000;127:5179–5190. doi: 10.1242/dev.127.23.5179. [DOI] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky LS, Matsudarira P, Baltimore D, Darnell J. Molecular Cell Biology. 4. New York: W. H. Freeman; 2000. [Google Scholar]
- Masui T, Swift GH, Hale MA, Meredith DM, Johnson JE, MacDonald RJ. Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol Cell Biol. 2008;28:5458–5468. doi: 10.1128/MCB.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJ, Vidal M, Hope IA. Insight into transcription factor gene duplication from Caenorhabditis elegans promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Hamada S, Kimura K, Kanno A, Hirota M, Umino J, Fujibuchi W, Masamune A, Tanaka N, Miura K, Egawa S, Motoi F, Unno M, Vonderhaar BK, Shimosegawa T. Up-regulation of MSX2 enhances the malignant phenotype and is associated with Twist 1 expression in human pancreatic cancer cells. Am J Pathol. 2008;172:926–939. doi: 10.2353/ajpath.2008.070346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenirri S, Restagno G, Ferrero GB, Alaimo G, Sbaiz L, Mari C, Genitori L, Maurizio F, Cremonesi L. Integrated strategy for fast and automated molecular characterization of genes involved in craniosynostosis. Clinical Chemistry. 2007;53(10):1767–1774. doi: 10.1373/clinchem.2007.089292. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic lineages of the nematode Caenorhabditis elegans. Nucl Acids Res. 1977;18:2033–2036. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Wagmaister JA, Miley GR, Morris CA, Gleason JE, Miller LM, Kornfeld K, Eisenmann DM. Identification of cis-regulatory elements from the C. elegans Hox gene lin-39 required for embryonic expression and for regulation by the transcription factors LIN-1, LIN-31 and LIN-39. Dev Biol. 2006;297:550–565. doi: 10.1016/j.ydbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhao J, Corsi AK. Identification of novel target genes of CeTwist and CeE/DA. Dev Biol. 2006;293:486–498. doi: 10.1016/j.ydbio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Wilkie AO. Craniosynostosis: genes and mechanisms. Hum Mol Genet. 1997;6:1647–1656. doi: 10.1093/hmg/6.10.1647. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yanowitz JL, Shakir MA, Hedgecock E, Hutter H, Fire A, Lundquist EA. UNC-39, the C. elegans homolog of the human myotonic dystrophy-associated homeodomain protein Six5, regulates cell motility and differentiation. Dev Biol. 2004;272:389–402. doi: 10.1016/j.ydbio.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Zhao J, Wang P, Corsi AK. The C. elegans Twist target gene, arg-1, is regulated by distinct E box promoter elements. Mech Dev. 2007;124:377–389. doi: 10.1016/j.mod.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]