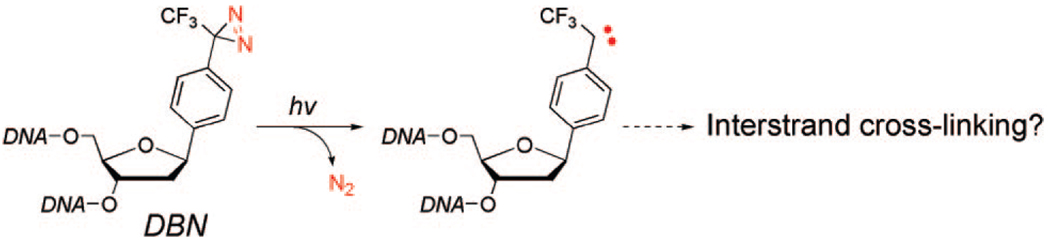
A common strategy for cancer therapy is to induce DNA damage subsequent apoptosis of cancer cells. Some of the most effective anticancer agents in clinical use, such as cyclophosphamide, mitomycin C, and cisplatin, exert their effects by creating DNA interstrand cross-links.1–4 We report here the design and synthesis of a diazirine-based nucleoside analogue (DBN, Figure 1) that can be incorporated into DNA by solid phase oligonucleotide synthesis. The DBN-containing dsDNA efficiently forms an interstranded cross-link upon photoirradiation.
Figure 1.
A diazirine-based nucleoside analogue (DBN) that would form a DNA interstrand cross-link under photoirradiation.
Diazirine derivatives have been widely used to investigate the structure and function of nucleic acids, proteins, and various macromolecular complexes.5–18 These compounds possess several noticeable advantages over other photoaffinity groups: first, they can be rapidly photolyzed at wavelengths (350–360 nm) beyond the absorbance region of most biomacromolecules; second, they form highly reactive carbenes upon photolysis, which readily cross-link to various functional groups including the inert aliphatic C–H bonds; third, the cross-linking reaction with carbenes produces carbon-based bonds that are stable under typical experimental conditions; finally, the diazirine derivatives are sterically less hindered than other photoactive groups and possess excellent chemical stability to be handled under moderate laboratory illumination.
Various approaches have been developed to tether the aryl(trifluoromethyl) diazirine group to the base (pyrimidine7–12 or purine13,14) or the ribose part 15,16 of nucleoside analogues. All these approaches, however, lead to extrahelical attachment of the diazirine group to the DNA double helices. We envisioned that direct installation of an aryl(trifluoromethyl)diazirine group to the ribose may provide an intrahelical attachment of this group to the DNA duplex (Figure 1). Upon UV irradiation the diazirine group forms a carbene intermediate that could cross-link to the opposite strand. In addition, the lack of steric hindrance of DBN may allow it to be recognized by DNA modification or repair proteins to facilitate protein-nucleic acid photocross-linking.
We developed a synthetic scheme that allows the production of the corresponding phosphoramidite at a reasonable efficiency (Scheme 1). The starting materials furanoid glycal 119,20 and 3-(4-iodophenyl)-3-(trifluoromethyl)-3H-diazirine 210,21,22 were prepared from commercially available thymidine and 1,4-diiodobenzene, respectively. The critical C-glycoside bond formation was achieved by β-anomeric selectivity using Heck chemistry23,24 under Jeffery’s conditions.25 The free nucleoside analogue 5 was then obtained from 3 by deprotection of the tert-butyldimethylsilyl group and reduction with sodium triacetoxyborohydride.24 Two subsequent steps following standard procedures were carried out to prepare the 5′-O-dimethoxytrityl, 3′-O-phosphoramidite derivative 7 that can be used for the solid phase oligonucleotide synthesis.
Scheme 1.
Synthesis of Diazirine-Based Nucleoside Analoguea
a Reagents and conditions: (a) Pd(OAc)2, nBu4N+Cl−, Cy2NCH3, DMF, 60 °C (46%); (b) nBu4N+F−, THF/HOAc, rt (92%); (c) Na(OAc)3BH, CH3CN/HOAc, −10 °C (56%); (d) DMTrCl, DMAP, pyridine, 0 °C (71%); (e) NC(CH2)2OP(Cl)N(iPr)2, DIPEA, CH2Cl2, 0 °C (84%).
A trimer T-B(DBN)-T was synthesized and deprotected with concentrated ammonia at room temperature overnight. The MALDI-TOF mass spectrum of the deprotected product gave three peaks (Figure S1), which correspond to the mass of the trimer, the mass of the trimer with the loss of N2, and the mass of the trimer reacted with the matrix, respectively. When the deprotected product was irradiated (near-UV, hv > 300 nm) in water for 10 min, its MALDI-TOF mass spectrum gave a new peak corresponding to the product of the carbene reacted with water, together with the disappearance of the previous three peaks (Figure S2). This observation clearly indicated the presence of the diazirine group in the trimer.
A 15-mer oligonucleotide sequence B1 (5′-ATG AAC CBG GAA AAC-3′) was synthesized and purified following the same protocol. It was annealed with complementary strands with A, T, G, or C opposite B, respectively. The melting temperatures (Tm) of the dsDNAs (in 10 mM Tris-HCl, pH 7.4, 100 mM NaCl) were measured by differential scanning calorimetry (Table 1). With the exception of the B:A pair (entry 1, Table 1), in which case the melting behavior was not detected, the other three pairs (entries 2, 3, and 4, Table 1) lowered the Tm values by 18–21% (10–12 °C) compared to the normal base pairs (entries 5 and 6, Table 1). This relatively mild reduction of Tm indicated that the dsDNAs with DBN basically keep the confirmation as the normal dsDNA.27,28 The diazirine group may participate in base pairing, which needs to be investigated structurally in the future.
Table 1.
Thermodynamic Properties of dsDNAs
| entry | dsDNA | sequences | ΔH (kcal · mol−1) |
ΔS (cal · K−1 · mol−1) |
Tm (°C) |
|---|---|---|---|---|---|
| 1 | (B:A) | 5′-ATG AAC CBG GAA AAC-3′ 3′-TAC TTG GAC CTT TTG-5′ |
n.a. | n.a. | n.a. |
| 2 | (B:T) | 5′-ATG AAC CBG GAA AAC-3′ 3′-TAC TTG GTC CTT TTG-5′ |
63.0 | 200 | 45 |
| 3 | (B:G) | 5′-ATG AAC CBG GAA AAC-3′ 3′-TAC TTG GGC CTT TTG-5′ |
47.5 | 147 | 48 |
| 4 | (B:C) | 5′-ATG AAC CBG GAA AAC-3′ 3′-TAC TTG GCC CTT TTG-5′ |
66.4 | 198 | 46 |
| 5 | (T:A) | 5′-ATG AAC CTG GAA AAC-3′ 3′-TAC TTG GAC CTT TTG-5′ |
- | - | 55 |
| 6 | (C:G) | 5′-ATG AAC CCG GAA AAC-3′ 3′-TAC TTG GGC CTT TTG-5′ |
96.8 | 296 | 58 |
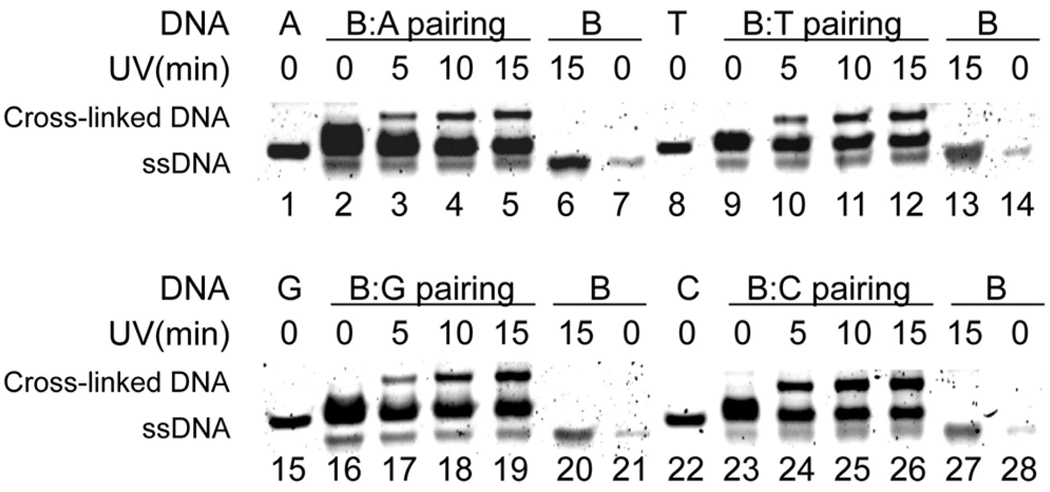
The four dsDNAs 1–4 (Table 1) were treated with near-UV irradiation at various time intervals at 0 °C (30 µM dsDNA in 10 mM Tris-HCl, pH 7.4, and 100 mM NaCl), then denatured, and analyzed by PAGE. The appearance of low mobility bands showed the formation of the interstranded cross-linked products (lanes 3–5, 10–12, 17–19, and 24–26, Figure 2), while no trace amount of such products were detected without UV irradiation (lanes 2, 9, 16, and 23, Figure 2). The cross-linked upper bands were purified by PAGE and confirmed by MALDI-TOF MS (Figures S4–S8). The DNA interstrand cross-linking efficiency varies with different bases opposite B. While the B:A pair gave the lowest yield, the yield of the B:C pair is quite high. Two 42-mer dsDNAs with B opposite T were subjected to photocross-linking. The bands were excised from the gel, 32P-labeled, and subjected to hydroxyl radical footprinting, which indicated that DBN can cross-link to multiple nearby bases on the opposite strand but mostly to the 5′ and 3′ adjacent bases (of T) on the complementary strand (Figure S9).28
Figure 2.
Interstrand photocross-link for the DBN paired with A, T, G, and C, respectively (30 µM). B represents the 15-mer ssDNA containing DBN; A, T, G, and C represent the complementary ssDNA strands shown in Table 1.
The experiments also indicate that the cross-linking reaction by diazirine is complete within ~10 min. A prolonged irradiation time did not seem to give significant improvements on the cross-linking efficiency (Figure 2). This is expected as the diazirine is typically photolyzed rapidly and the lifetime of its photolysis product carbine is on a nanosecond time scale.
In summary, we have developed a nucleoside analogue that efficiently forms interstrand cross-linking in dsDNA upon near-UV irradiation. This nucleoside analogue may find broad applications in biotechnology such as probing nucleic acid–nucleic acid and protein–nucleic acid interactions and in phototherapy. Further studies of the details of the photocross-linking of this and analogous nucleosides and their applications are currently ongoing in our laboratory.
Supplementary Material
Acknowledgment
We are grateful to Dr. P. R. Chen, Mr. U. K. Shigdel, and Dr. J. Zhang for assistance and discussions. This research is supported by the W. M. Keck Foundation, the Arnold and Mabel Beckman Foundation, the Research Corporation, and a Camille Dreyfus Teacher Scholar Award.
Footnotes
Supporting Information Available: A detailed experimental section, Figures S1–S9. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hurley LH. Nat. ReV. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Lippard SJ. Nat. ReV. Drug Discovery. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 3.Wang P, Liu RP, Wu XJ, Ma HJ, Cao XP, Zhou P, Zhang JY, Weng XC, Zhang XL, Qi J, Zhou X, Weng LH. J. Am. Chem. Soc. 2003;125:1116–1117. doi: 10.1021/ja029040o. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimura Y, Ito Y, Fujimoto K. Nucleic Acids Symp. Ser. 2004:81–82. doi: 10.1093/nass/48.1.81. [DOI] [PubMed] [Google Scholar]
- 5.Tomohiro T, Hashimoto M, Hatanaka Y. Chem. Rec. 2005;5:385–395. doi: 10.1002/tcr.20058. [DOI] [PubMed] [Google Scholar]
- 6.Hatanaka Y, Sadakane Y. Curr. Top. Med. Chem. 2002;2:271–288. doi: 10.2174/1568026023394182. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi T, Saneyoshi M. Nucleic Acids Res. 1996;24:3364–3369. doi: 10.1093/nar/24.17.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi T, Suyama K, Narita K, Kohgo S, Tomikawa A, Saneyoshi M. Nucleic Acids Res. 1997;25:2352–2358. doi: 10.1093/nar/25.12.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sergiev PV, Lavrik IN, Wlasoff VA, Dokudovskaya SS, Dontsova OA, Bogdanov AA, Brimacombe R. RNA. 1997;3:464–475. [PMC free article] [PubMed] [Google Scholar]
- 10.Topin AN, Gritsenko OM, Brevnov MG, Gromova ES, Korshunova GA. Nucleosides Nucleotides. 1998;17:1163–1175. [Google Scholar]
- 11.Tate JJ, Persinger J, Bartholomew B. Nucleic Acids Res. 1998;26:1421–1426. doi: 10.1093/nar/26.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persinger J, Sengupta SM, Bartholomew B. Mol. Cell. Biol. 1999;19:5218–5234. doi: 10.1128/mcb.19.7.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zofall M, Bartholomew B. Nucleic Acids Res. 2000;28:4382–4390. doi: 10.1093/nar/28.21.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbfinger E, Gorochesky K, Lévesque SA, Beaudoin AR, Sheihet L, Margel S, Fischer B. Org. Biomol. Chem. 2003;1:2821–2832. doi: 10.1039/b303425a. [DOI] [PubMed] [Google Scholar]
- 15.Sergiev P, Dokudovskaya S, Romanova E, Topin A, Bogdanov A, Brimacombe R, Dontsova O. Nucleic Acids Res. 1998;26:2519–2525. doi: 10.1093/nar/26.11.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liebmann M, Di Pasquale F, Marx A. ChemBioChem. 2006;7:1965–1969. doi: 10.1002/cbic.200600333. [DOI] [PubMed] [Google Scholar]
- 17.Suchanek M, Radzikowska A, Thiele C. Nat. Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 18.Shigdel UK, Zhang J, He C. Angew. Chem. Int. Ed. 2008;47:90–93. doi: 10.1002/anie.200703625. [DOI] [PubMed] [Google Scholar]
- 19.Larsen E, Jørgensen PT, Sofan MA, Pedersen EB. Synthesis-Stuttgart. 1994:1037–1038. n/a. [Google Scholar]
- 20.Cameron MA, Cush SB, Hammer RP. J. Org. Chem. 1997;62:9065–9069. [Google Scholar]
- 21.Brunner J, Senn H, Richards FM. J. Biol. Chem. 1980;255:3313–3318. [PubMed] [Google Scholar]
- 22.Hatanaka Y, Hashimoto M, Kurihara H, Nakayama H, Kanaoka Y. J. Org. Chem. 1994;59:383–387. [Google Scholar]
- 23.Daves GD., Jr Acc. Chem. Res. 1990;23:201–206. [Google Scholar]
- 24.Lee AHF, Kool ET. J. Am. Chem. Soc. 2006;128:9219–9230. doi: 10.1021/ja0619004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffery T. Tetrahedron. 1996;52:10113–10130. [Google Scholar]
- 26.Kool ET. Acc. Chem. Res. 2002;35:936–943. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 27.Millican TA, Mock GA, Chauncey MA, Patel TP, Eaton MAW, Gunning J, Cutbush SD, Neidle S, Mann J. Nucleic Acids Res. 1984;12:7435–7453. doi: 10.1093/nar/12.19.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wombacher R, Jäschke A. J. Am. Chem. Soc. 2008;130:8594–8595. doi: 10.1021/ja802931q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.