Abstract
Although it has been established that Atg6/Beclin 1, the phosphatidylinositol 3-kinase (PI3K) Vps34, and associated proteins have direct or indirect roles in autophagic pathways in both mammals and yeasts, the elucidation of these roles and the proteins required for them is ongoing. The involvement of the Beclin 1-binding protein, UVRAG, has been a particular source of disagreement. We found that PpAtg6 is required for all autophagic pathways that have been identified in the yeast Pichia pastoris, as well as for the carboxypeptidase Y (PpCPY) vacuolar protein sorting pathway. We localized PpAtg6 to the phagophore assembly site (PAS) and observed its continued presence at that site as the isolation membrane grew from it and matured into a pexophagosome. PpUvrag, however, was required for proper PpCPY sorting, but not for any autophagic pathway. Rather, the defects in all autophagic pathways observed when PpUvrag was overexpressed support its presence in a complex that competes with the PI3K complex required for autophagy.
Keywords: phagophore, PI3K, Atg6, Vps34, Uvrag, isolation membrane, autophagy, pexophagy, VPS pathway
Introduction
The term “autophagy” refers to a number of intracellular trafficking and degradative processes, most of which result in the delivery of either nonselective or specific cytosolic cargoes (proteins or organelles) to the vacuole (in yeasts) or lysosomes (in higher eukaryotes). In higher eukaryotes, autophagy proteins and pathways are implicated in such varied processes as cell differentiation, innate immunity, organismal development and tumor suppression (reviewed in refs. 1–3).
Many of the advances in the understanding of autophagy proteins and pathways in higher eukaryotes stem from studies in yeasts. The autophagic processes that have been most extensively studied in yeasts are: macroautophagy, cytoplasm-tovacuole targeting (Cvt) and pexophagy (reviewed in refs. 4–6). Macroautophagy is induced above a basal level when cells are starved for nitrogen or amino acids. Macroautophagy (hereafter referred to as “autophagy”) proceeds through the expansion of a membrane sac termed the phagophore or isolation membrane, which leads to the formation of double-membraned vesicles called autophagosomes that engulf cytoplasm nonspecifically and then fuse with the vacuole to deliver their contents (autophagic bodies) for degradation. The proposed site for autophagosome formation is the phagophore assembly site (PAS). The PAS is still poorly understood, but it can be defined as the site of convergence of the isolation membrane (or phagophore) and the core machinery proteins. Following the expansion of the phagophore, most core machinery proteins, except for Atg8, are excluded from the completed autophagosome.7, 8 The Cvt pathway is morphologically similar to autophagy, but is a constitutive, biosynthetic pathway. Cvt vesicles specifically envelop aminopeptidase I (Ape1) and α-mannosidase for delivery to the vacuole, where those enzymes perform their function. Pexophagy is induced under conditions that do not require a high level of peroxisomal enzyme activity for growth and involves the specific delivery of peroxisomes to the vacuole for degradation. In P. pastoris, two pexophagy pathways have been observed: macropexophagy and micropexophagy. Macropexophagy is induced when cells are shifted from methanol to ethanol as a carbon source and proceeds by a mechanism (involving pexophagosomes) morphologically similar to the macroautophagy and Cvt pathways. In contrast, micropexophagy, which is induced when cells are shifted from methanol to glucose, involves the direct engulfment of peroxisomes by extended vacuolar sequestering membranes (VSMs) of an invaginating vacuole, which are proposed to fuse with a micropexophagy apparatus (MIPA) that forms a lid on the cup-shaped VSMs.
Among the core machinery proteins required for many autophagic pathways in yeast is Vps34,9 a class III phosphatidylinositol 3-kinase (PI3K), producing phosphatidylinositol 3-phosphate (PtdIns3P) from phosphatidylinositol (PtdIns).10 Also required for autophagic pathways in S. cerevisiae are three other proteins that associate with Vps34 to form PI3K complex I: Vps15, Atg14 and Vps30/Atg6.9
A second complex in S. cerevisiae (PI3K complex II) contains the same components as complex I, except that Vps38 replaces Atg14. PI3K complex II produces endosomal PtdIns3P, which is required for proper localization of the retromer complex. The retromer complex includes two components, Vps5 and Vps17, with PX domains that bind PtdIns3P and mediates proper trafficking of CPY via the CPY receptor, Vps10. This trafficking is referred to as the Vacuolar Protein Sorting (VPS) pathway.11
One of the components shared by PI3K complexes I and II is Atg6/Vps30, a highly conserved protein from yeast to mammals. Recently, the human ortholog of Atg6, Beclin 1, was shown to bind UVRAG (protein encoded by an ultraviolet radiation resistance-associated gene). The consensus that UVRAG binds Beclin 1 has not extended to UVRAG's function. Liang and colleagues found that UVRAG interacts with the class C Vps complex, and thus promotes not only autophagosome formation via its interaction with Beclin 1, but also autophagosome maturation and endocytic trafficking by recruiting membrane fusion machinery.12 Takahashi and colleagues proposed that UVRAG promotes autophagy by binding to Beclin 1 and bringing along a second interaction partner, Bif-1, which has a BAR domain to promote the membrane curvature required for autophagosome formation.13 Other studies, however, suggest that UVRAG is the mammalian ortholog of Vps38, competing with the mammalian ortholog of Atg14 to bind Beclin 1 and making no contribution to autophagic flux.14 Most recently, UVRAG was found in a separate complex (or complexes) from Atg14L, sometimes in association with Rubicon, a negative regulator of autophagy.15
Because most of the PI3K complex proteins (Atg6, Vps15 and Vps34) and UVRAG have orthologs in P. pastoris, we investigated the involvement and function of those orthologs in the autophagy, Cvt, and pexophagy pathways in P. pastoris. While we found that PpAtg6, PpVps15 and PpVps34 are required for all of the autophagic pathways we tested, as well as for PpCPY sorting, we found scant evidence that PpUvrag is required for any autophagy-related pathway. Instead, PpUvrag is a component of another PI3K complex (probably PI3K complex II) that competes with PI3K complex I for shared components. We also could localize PpAtg6 (and, by extension, PI3K complex I) at the PAS during the elongation and maturation of the phagophore and were able to document the steps in the biogenesis of the isolation membrane from the PAS. Surprisingly, PpVps17, a component of the retromer complex, involved in PpCPY sorting and not in autophagy, was localized at the PAS when autophagy was induced.
Results
Identification of the components of PI3K complexes
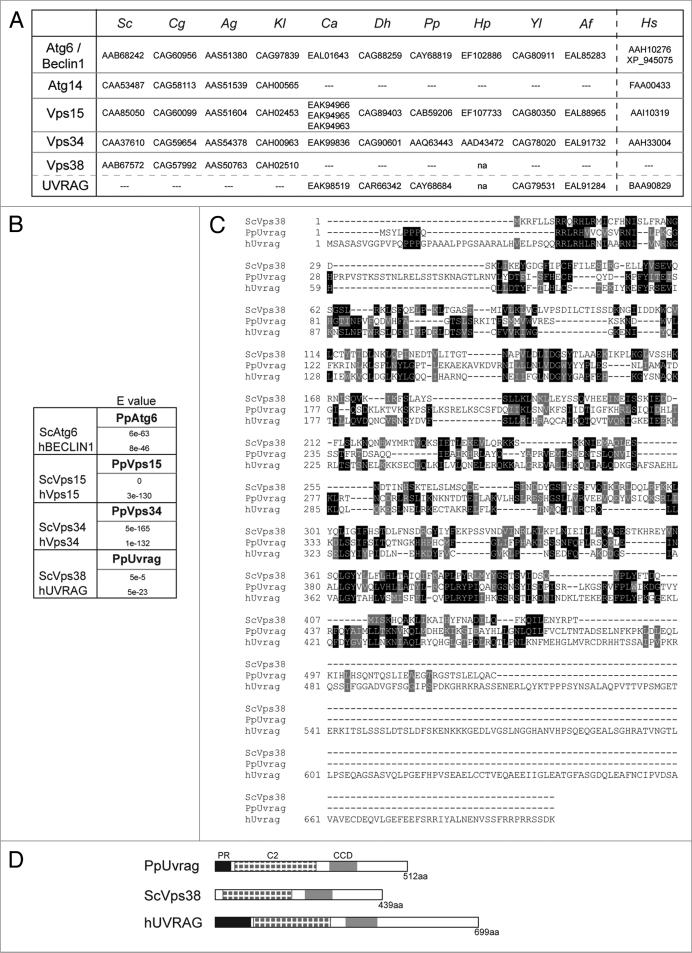
In P. pastoris, Vps15 is the only component of the PI3K complexes described to date,16 and only its requirement for pexophagy was studied. We identified homologs of Atg6 and Vps34 (Fig. 1A and B) in P. pastoris by querying the ERGO database (Integrated Genomics, Chicago, IL) and/or GenBank.17 PpVps34, PpVps15 and PpAtg6 (the three core proteins present in both PI3K complexes I and II) are all similar in domain structure to their human and S. cerevisiae orthologs (Fig. S1), which reflects how highly conserved across species are the three proteins.
Figure 1.
Identification of P. pastoris PI3K complex components. (A) Adapted table containing GenBank accession numbers of orthologs of PI3K complex subunits and UVRAG-like proteins in different species.19 Sc: S. cerevisiae, Cg: Candida glabrata, Ag: Ashbya gossypii, Kl: Kluyveromyces lactis, Ca: Candida albicans, Dh: Debaryomyces hansenii, Pp: P. pastoris, Hp: H. polymorpha, Yl: Yarrowia lipolytica, Af: Aspergillus fumigatus, Hs: H. sapiens, na, not analyzed. (B) e values (indicating statistical significance) of homology between P. pastoris and other yeast and human proteins obtained using bl2seq from NcBI. (c) Amino acid alignment of S. cerevisiae Vps38, P. pastoris Uvrag and H. sapiens UVRAG prepared using clustal-W. Identical residues are indicated with black boxes, and similar residues with grey boxes. (D) Domain comparison of UVRAG and Vps38 proteins.
Our BLAST searches did not, however, yield an obvious ortholog in P. pastoris to the fourth member of either PI3K complex I (Atg14) or complex II (Vps38). According to our search (RPSBLASTP),18 the P. pastoris protein most similar to Atg14 did contain a putative structural-maintenance-of-chromosomes domain (SMC prok B-like domain), similar to that found in mammalian Atg14. It also contained both N-terminal and C-terminal regions with predicted coiled-coil domain (CCDs). Only N-terminal CCDs are found in Atg14. In analyzing whether this putative Atg14 had homologs in other yeast, we found a strong similarity with several putative Atg25 proteins,19 including HpAtg25.20 We noticed, as described earlier,19 that when Atg25 was present in a yeast, Atg14 was absent and vice versa. Unfortunately, we found no evidence that this PpAtg25-like protein was part of the PI3K complex. It shared none of the expected functions of Atg14 and exhibited some functions of HpAtg25 (Fig. S2). Our analysis of the P. pastoris genome thus revealed no ortholog of Atg14.
Although we did not find an obvious ortholog of ScVps38, we did find an ortholog of the mammalian UVRAG, recently postulated, by some, as the mammalian Vps38 (Fig. 1A, B and D). PpUvrag exhibits modest sequence and structural homology to ScVps38 (Fig. 1B-D). At 512 amino acids, PpUvrag is shorter than hUVRAG, but longer than ScVps38. All three proteins have a C2 calcium-dependent membrane targeting domain (C2) and a central CCD (Fig. 1D). Additionally, the PpUvrag and hUVRAG share a proline-rich N-terminal (PR) domain. Some yeasts contain Vps38-like proteins, while others have UVRAG-like proteins, but these two proteins are never present in the same organism (Fig. 1A). Sequence homology and domain conservation indicates that PpUvrag is more closely related to hUVRAG than to ScVps38.
Role of PI3K complex proteins and PpUvrag in autophagy, autophagy-related and the VPS pathways
One advantage of P. pastoris as an organism in which to study autophagy is the presence, not only of autophagy, but also of readily assayed autophagy-related pathways that appear to require the same core proteins.6,21,22 Thus, to elucidate the function of PpUvrag and other proteins of interest, we assayed the autophagy, Cvt and pexophagy pathways in mutant strains. Further, based on our preliminary experiments and suggestions that UVRAG might be an ortholog of Vps38, we also assayed the PpCPY pathway. Control strains included the autophagy mutants Ppatg8Δ and/or Ppatg9Δ, the pexophagy mutant Ppatg30Δ, and the retromer and CPY sorting mutant Ppvps17Δ.23–25 Vps17 has not been studied in P. pastoris (GenBank accession #CAY69447) but is 30% identical to ScVps17 and, like ScVps17, contains a PX domain followed by two CCDs.
Autophagy
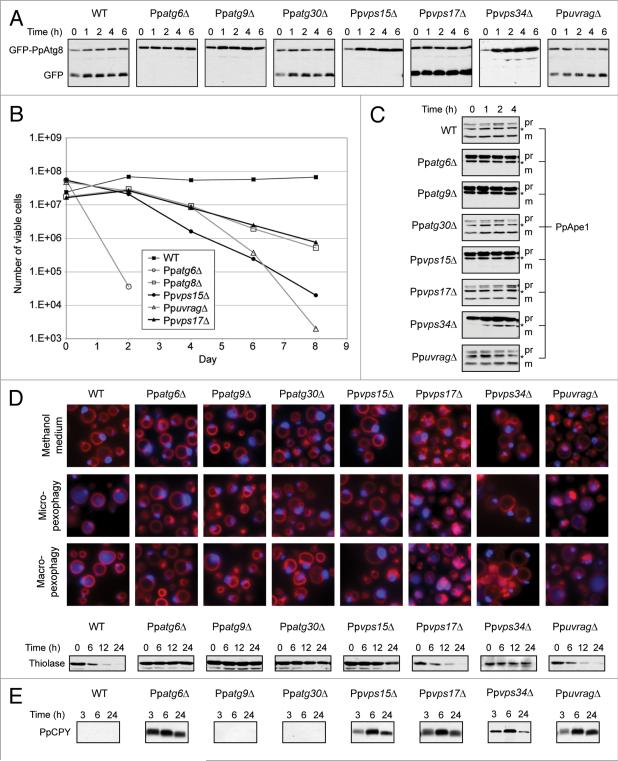
We used two assays to determine whether the autophagy pathway was functioning normally in strains: a GFP-PpAtg8 cleavage assay and a nitrogen starvation cell viability assay. These assays both pointed to a defect in autophagy for Ppatg6Δ, Ppvps15Δ and Ppvps34Δ (Fig. 2A and B), but produced seemingly contradictory results for PpuvragΔ and Ppvps17Δ.
Figure 2.
PpuvragΔ has a PpCPY sorting defect but is proficient for autophagy pathways, and PpAtg6, PpVps15 and PpVps34 are required for both PpCPY sorting and all types of autophagy. (A) Detection of GFP-PpAtg8 cleavage as an indicator of autophagy. WT and mutant cells were transformed to express GFP-PpAtg8 from the PpATG8 promoter. Experiment was performed as described in Materials and Methods. (B) Cellviability following nitrogen starvation. Cells were shifted from YPD to sD-N medium on day 0 and periodically assayed for their ability to form colonies on YPD plates. (c) Detection of PpApe1 processing. The same protein samples used in the GFP-PpAtg8 cleavage assay were analyzed for PpApe1 maturation. Pr: unprocessed PpApe1 form, m: processed PpApe1 form and *: cross-reacting band. (D) Pexophagy assays. Fluorescence microscopy: cells were grown in methanol medium for 15 h and shifted to glucose medium for 3 h to induce micropexophagy or ethanol medium for 6 h to induce macropexophagy. Peroxisomes were labeled with BFP-sKL and vacuolar membranes with FM 4-64. Biochemical experiment: cells were grown in oleate for 15 h and shifted to nitrogen starvation conditions. At the indicated time points, protein samples were analyzed as described in Materials and Methods. (e) PpCPY sorting assay. Extracellular PpCPY-hA of cells growing in YPD, analyzed as described in Materials and Methods.
Autophagy results in the trafficking of GFP-Atg8 to the yeast vacuole lumen.26 In the lumen, the Atg8 moiety is quickly cleaved and degraded, while the GFP moiety is more stable. Thus, the appearance of free GFP is a measure of autophagy. In WT, Ppatg30Δ, Ppvps17Δ and PpuvragΔ cells, autophagy induced by nitrogen starvation led to a small induction of GFP-PpAtg8 (compare 0 h and 1 h), enhanced proteolysis of GFP-PpAtg8 and the accumulation of free GFP (Fig. 2A). No free GFP was observed in the autophagy mutant Ppatg9Δ. Ppatg6Δ, Ppvps15Δ and Ppvps34Δ cells were similar to the autophagy mutant, showing no free GFP during the 6 h of nitrogen starvation. These results indicate that while PpAtg6, PpVps15 and PpVps34 are necessary for autophagy, PpUvrag is not. Interestingly, in the retromer mutant, Ppvps17Δ, GFP-PpAtg8 processing was comparable to WT and PpuvragΔ cells, but more of the GFP moiety accumulated than expected.
The premise of the cell viability assay is that protein recycling by autophagy is required for nitrogen-starved cells to remain viable. While the number of viable WT cells remained relatively stable throughout the eight-day duration of the assay, less than 0.1% of the Ppatg6Δ cells retained viability after just two days of nitrogen starvation (Fig. 2B). This assay produced evidence of direct or indirect PpUvrag involvement in autophagy, in that approximately 0.01% of PpuvragΔ cells remained viable following eight days of nitrogen starvation, comparable to Ppvps15Δ. Standing alone, this result seems inconsistent with the result of the GFP-Atg8 cleavage assay. Interestingly, however, Ppvps17Δ cells also exhibited a defect in cell viability, slightly less severe than PpuvragΔ cells, but comparable to Ppatg8Δ. Since Vps17 is involved in protein sorting to the vacuole, it should not be surprising that it indirectly affects nutrient recycling from the vacuole, and that its absence may therefore compromise a cell's viability after starvation. This suggested the possibility that PpUvrag may also be involved in vacuolar protein sorting, rather than directly in autophagy.
Cvt pathway
To study the Cvt pathway, we assayed the maturation of its main cargo, PpApe1 (Fig. 2C). During growth conditions (0 h), PpApe1 was processed normally in WT, Ppatg30Δ, Ppvps17Δ and PpuvragΔ cells, whereas Ppatg6Δ, Ppvps15Δ and Ppvps34Δ as well as the control, Ppatg9Δ, showed defects in processing PpApe1 to its mature form. Therefore, PpUvrag is not necessary for the Cvt pathway, whereas PpAtg6, PpVps15 and PpVps34 are essential.
Pexophagy
Pexophagy was assayed by fluorescence microscopy for peroxisomes induced by methanol and by western blotting of a peroxisomal protein (thiolase) for peroxisomes induced by oleate (Fig. 2D). The fluorescence microscopy assay involved tracking whether peroxisome-targeted BFP (BFP-SKL) moved to the vacuole when cells were shifted from methanol medium (which induces peroxisome biogenesis) to a carbon source that induced either micropexophagy (glucose) or macropexophagy (ethanol). Induction of peroxisomes (labeled in blue) was normal in all strains tested (Fig. 2D). The Ppvps17Δ and PpuvragΔ cells exhibited fragmented vacuolar structures prior to pexophagy induction. Following induction of either micropexophagy or macropexophagy, WT, Ppvps17Δ and PpuvragΔ cells displayed blue fluorescence in the vacuole lumen, indicative of normal pexophagy. In contrast, Ppatg6Δ, Ppatg9Δ, Ppatg30Δ, Ppvps15Δ and Ppvps34Δ cells displayed blue fluorescence only in peroxisomes, reflecting defects in both micropexophagy and macropexophagy.
As seen in S. cerevisiae and in P. pastoris,27 shifting oleate-grown cells to nitrogen starvation induced pexophagy. Thiolase was degraded normally in WT, Ppvps17Δ and PpuvragΔ cells, but Ppatg6Δ, Ppatg9Δ, Ppatg30Δ, Ppvps15Δ and Ppvps34Δ cells sustained thiolase levels at least up to 24 h. These results show that, while PpAtg6, PpVps15 and PpVps34 are required for pexophagy (as also observed in S. cerevisiae), PpVps17 and PpUvrag are not.
VPS pathway
Since the behavior of Ppvps17Δ and PpuvragΔ cells was similar in all the assays described above, we also examined the possibility that, like ScVps38 and ScVps17, PpUvrag is required for proper sorting of CPY. In the absence of any component of the CPY sorting pathway, CPY is secreted to the extracellular medium instead of normal vacuolar targeting. We tagged PpCPY with an HA epitope at its C-terminus and we assayed the presence of PpCPY in the extracellular medium by western blot (Fig. 2E). As expected, WT, Ppatg9Δ and Ppatg30Δ did not secrete PpCPY, whereas the mutants affecting subunits of PI3K complex II (Ppatg6Δ, Ppvps15Δ and Ppvps34Δ) and the retromer (Ppvps17Δ) showed significant amounts of secreted PpCPY after 3 h. PpuvragΔ cells also mislocalized and secreted PpCPY, as would be expected for a vps38 mutant. These data prove that PpUvrag is required for the vacuolar sorting of PpCPY, and in its absence PpCPY is mis-sorted.
Thus, in our functional assays of autophagic and VPS pathways, Ppatg6Δ, Ppvps15Δ and Ppvps34Δ cells exhibited defects in all pathways tested, but PpuvragΔ cells behaved like the Ppvps17Δ cells, affected only in PpCPY sorting and in cell viability upon nitrogen starvation.
PpUvrag and PpAtg6 localization, and role in localization of a PtdIns3P-binding protein, PpAtg24
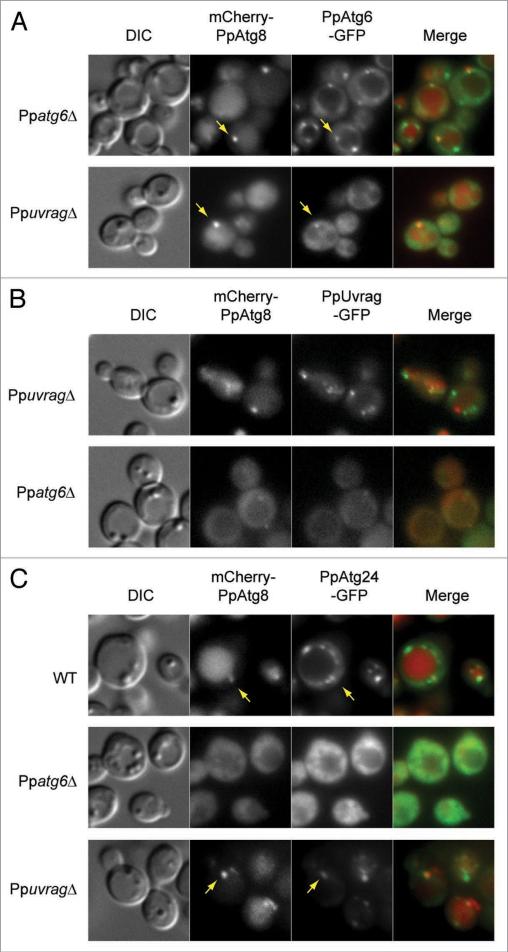
In S. cerevisiae cells, Atg6, like several other autophagy proteins, partially localizes to a perivacuolar structure called the PAS. This PAS localization, along with that of the other components of PI3K complex I, requires Atg14.28 Conversely, the endosomal localization of Atg6 and the other components of PI3K complex II requires Vps38.28 We examined the localization of PpAtg6-GFP, expressed under its own promoter (the fusion protein complemented the survival defect of Ppatg6Δ [data not shown]) in Ppatg6Δ and PpuvragΔ cells after 90 min of nitrogen starvation (Fig. 3A). PpAtg6-GFP was distributed on the vacuole membrane and as a single or a few bright dots around the vacuole when expressed in Ppatg6Δ (Fig. 3A, upper) or in WT cells (data not shown). The PpAtg6 dot colocalized with the PAS marker, mCherry-PpAtg8. In the absence of PpUvrag, most of the PpAtg6 mislocalized to the cytoplasm, but a weak PpAtg6 localization was detected in a dot-like structure, which colocalized with PpAtg8 at the PAS (Fig. 3A, lower). This localization resembles that of ScAtg6 in Scvps38Δ cells.28
Figure 3.
PpUvrag and PpAtg6 depend on each other for normal localization and both are required for normal localization of endosomal PpAtg24 protein. Cells were grown in sD medium for 15 h, shifted to sD-N for 90 min and analyzed using fluorescence microscopy. Arrow indicates colocalization with PAS marker (mcherry-PpAtg8). (A) Mislocalization of PpAtg6-GFP in PpuvragΔ cells. PpAtg6-GFP and mcherry-PpAtg8 were localized in Ppatg6Δ and PpuvragΔ cells. (B) Mislocalization of PpUvrag-GFP in Ppatg6Δ cells. PpUvrag-GFP under control of its endogenous promoter and mcherry-PpAtg8 were localized in PpuvragΔ and Ppatg6Δ cells. (c) Mislocalization of PpAtg24-GFP in Ppatg6Δ and PpuvragΔ cells. PpAtg24-GFP and mcherry-PpAtg8 were localized in WT, Ppatg6Δ and PpuvragΔ cells.
The mammaliam UVRAG and S. cerevisiae Vps38 proteins have been localized to endosomes.14,15,29 In addition, hUVRAG has been localized to autophagosomes.30 To study PpUvrag localization, we expressed PpUvrag-GFP under its own promoter in PpuvragΔ cells (the fusion protein complemented the survival defect of PpuvragΔ [data not shown]). The distribution of PpUvrag-GFP resembled that of PpAtg6-GFP, with several punctate structures around the vacuole, but almost none of these punctate structures colocalized with the PAS (Fig. 3B, upper). In the absence of PpAtg6, PpUvrag-GFP as well as PpAtg8 fully mislocalized to the cytosol (Fig. 3B, lower). In contrast, in S. cerevisiae, the localization of PI3K complex proteins, including Vps38, does not depend on Atg6.28
Although there is disagreement about the function of UVRAG in autophagy,12,14,30,31 there is no doubt that UVRAG, along with Beclin 1/Atg6, is part of one or more PI3K complexes. Such complexes produce PtdIns3P, which recruits proteins involved in different functions. We localized a PtdIns3P-binding protein, PpAtg24 (Fig. 3C), whose S. cerevisiae ortholog is on endosomal membranes and at the PAS.32,33 In P. pastoris, Atg24 was C-terminally tagged with GFP, expressed under its own promoter (the fusion protein complemented the pexophagy defect of Ppatg24Δ [data not shown]) and localized in WT, Ppatg6Δ and PpuvragΔ cells (Fig. 3C). PpAtg24 was present in several puncta around the vacuole and at the vacuole membrane in WT cells. Colocalization with the PpAtg8 in WT cells confirmed that one of the PpAtg24 puncta was the PAS. In the Ppatg6Δ mutant, PpAtg24 was fully mislocalized to the cytosol, together with PpAtg8. In the absence of PpUvrag, PpAtg24 localized in fewer perivacuolar dots than in WT and the vacuolar membrane localization was lost, but its PAS localization was preserved. These data show that the localization of the PtdIns3P-binding protein, PpAtg24, to punctate non-PAS and PAS structures, requires the presence of PpAtg6. In contrast, in the absence of PpUvrag, while most of the PpAtg24 is mislocalized, it still localizes to the PAS structures, indicating that in the absence of PpUvrag, some PtdIns3P is still generated at the PAS.
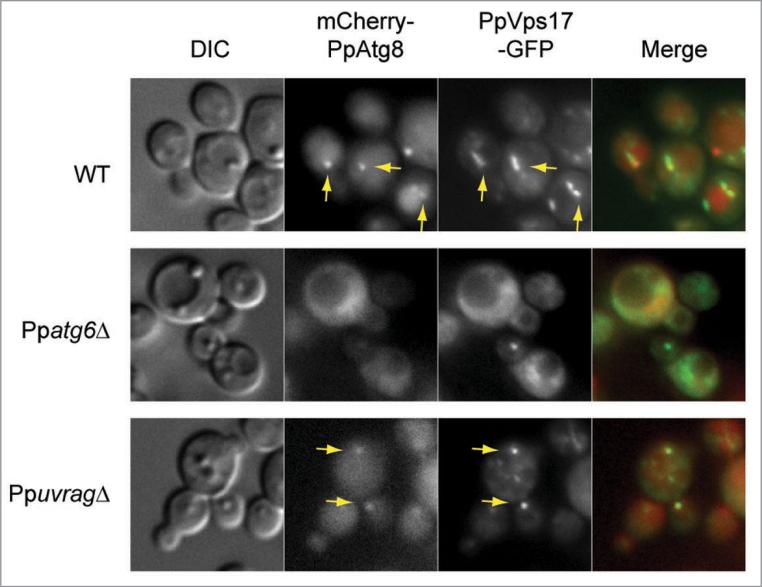
A portion of the retromer protein, PpVps17, is localized at the PAS, independent of PpUvrag
We localized a second PtdIns3P-binding protein, PpVps17, after 90 min of nitrogen starvation, in WT, Ppatg6Δ and PpuvragΔ cells (Fig. 4). The S. cerevisiae ortholog is on endosomal membranes.11 PpVps17 was C-terminally tagged with GFP and expressed under its own promoter (the fusion protein complemented the survival defect of Ppvps17Δ [data not shown]). The fusion protein was present in several puncta around the vacuole in WT cells and colocalization with the PpAtg8 in WT cells indicated that one punctum was the PAS. In the Ppatg6Δ mutant, PpVps17 was fully mislocalized to the cytosol, together with PpAtg8. In the absence of PpUvrag, most of PpVps17 mis-localized to the cytosol, but similar to PpAtg24, some PpVps17 was still at the PAS. Since the retromer subunits, Vps17 and Vps5, of S. cerevisiae interact with Atg27, a membrane protein whose absence causes a partial defect in all autophagy pathways and affects the cycling of Atg9, as well as its localization to the PAS and peripheral compartments,34 we wonder whether the retromer plays some role in recycling Atg9 and/or Atg8 from the pexophagosome. Further work is needed to test this hypothesis.
Figure 4.
The retromer protein, PpVps17, localizes partially at the PAS and this is independent of PpUvrag. Cells were grown in sD medium for 15 h, shifted to sD-N for 90 min and analyzed using fluorescence microscopy. PpVps17-GFP and mcherry-PpAtg8 were localized in WT, Ppatg6Δ and PpuvragΔ cells. Arrow indicates colocalization with PAS marker (mcherry-PpAtg8).
Overexpressing PpUvrag produces defects in autophagic pathways
Because our results seemed consistent with PpUvrag being a component, not of PI3K complex I, but of some other complex containing PpAtg6, we hypothesized that overexpressing PpUvrag might interfere with the formation of PI3K complex I by competing for the cell's limited supply of PpAtg6 or some other component of PI3K complex I. We expected that overexpressing PpUvrag might produce defects in autophagic pathways.
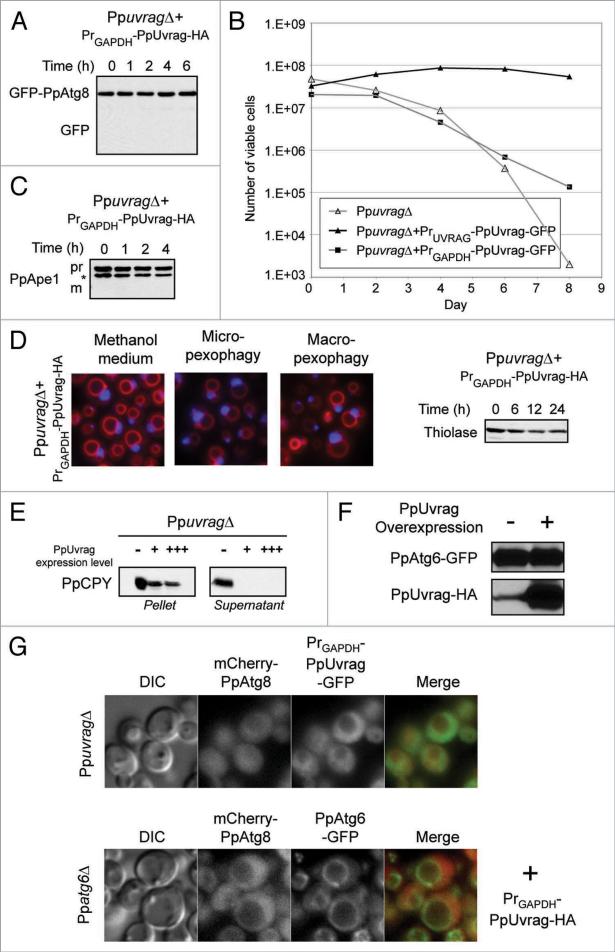
We constructed strains in which tagged versions of PpUvrag were overexpressed from the GAPDH promoter in PpuvragΔ cells (Fig. 5) and subjected them to the same battery of autophagic pathway assays as our other strains (Fig. 2). The results indicate that overexpression of PpUvrag produces defects in all of the autophagic pathways assayed. In the PpAtg8 trafficking assay for autophagy, GFP-PpAtg8 was not cleaved under starvation conditions (Fig. 5A); in the cell viability assay, less than 1% of cells remained viable after eight days of nitrogen starvation (Fig. 5B); in the Cvt assay, Ape1 was not processed (Fig. 5C); and in the pexophagy assays; peroxisomes induced by oleate or methanol medium were not degraded in pexophagy-inducing conditions (Fig. 5D). Consistent with a competition hypothesis, we found that overexpressed PpUvrag was sufficiently functional to eliminate PpCPY secretion in PpuvragΔ cells (Fig. 5E).
Figure 5.
Overexpression of PpUvrag impairs all autophagy pathways, but not PpCPY sorting. Cells expressing PpUvrag under the control of either the endogenous (PrUVRAG) or the GAPDH promoters (PrGAPDH) were tested for: (A) autophagy pathway; (B) cell viability upon nitrogen starvation; (C) Cvt pathway function [*: cross-reacting band], (D) pexophagy pathway; (E) VPS pathway, the cells were grown in YPD for 6 h at 30°c and analyzed by detection of PpCPY-HA in the cells (Pellet) and in the extracellular medium (supernatant) by western blotting of PpuvragΔ cells expressing different levels of PpUvrag (−: no PpUvrag, +: PpUvrag expressed from its own promoter, +++: PpUvrag express from the GAPDH promoter); (F) PpAtg6 stability, immuno-detection of PpAtg6-GFP in cells expressing PpUvrag-HA from its own promoter [−] and the GAPDH promoter [+]; (G) PpAtg6 and PpUvrag localization in cells expressing high levels of PpUvrag, PpAtg6, PpAtg8 and PpUvrag were localized in cells overexpressing PpUvrag after 90 min of nitrogen starvation. [+] indicates overexpression of PpUvrag-HA fusion protein.
Because cells overexpressing PpUvrag functionally resemble Ppatg6 mutant cells, we thought the overexpression might be affecting the stability or localization of PpAtg6. Expression of a PpAtg6-GFP fusion was assayed by using anti-GFP antibodies in immunoblots and by fluorescence microscopy. Normal levels were found in both cases (Fig. 5F and G). Overexpression of PpUvrag did have an impact on its own localization, producing a largely cytosolic localization, similar to the effect this had on PpAtg8 localization (Fig. 5G). PpUvrag overexpression seemed not to affect PpAtg6 localization, but PpAtg6 localization at the PAS could not be confirmed because of the mislocalization of the PAS marker, PpAtg8 (Fig. 5G).
Phagophore formation
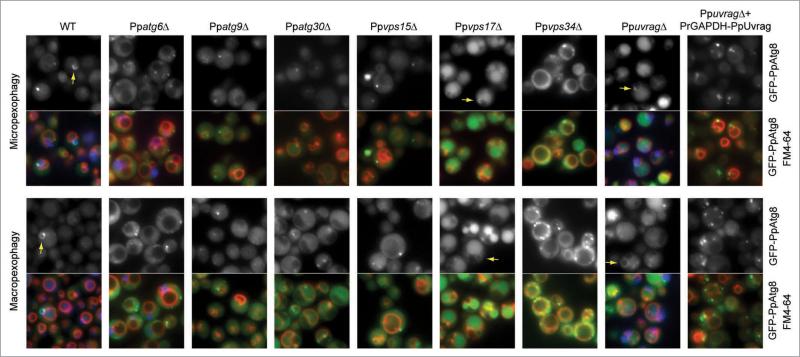
In yeast, when autophagy is induced (by starvation or rapamycin treatment), PpAtg8 redistributes from a diffuse pattern in the cytoplasm to the PAS. Although this structure is poorly defined, it is considered to be a potential organizing center for formation of the sequestering vesicles and probably where the phagophore is formed. In S. cerevisiae, a distinction between the PAS, phagophore and autophagosomes has not been possible by fluorescence microscopy. However, in P. pastoris, during pexophagy, the isolation membrane is much bigger and it can easily be differentiated from the PAS. This has led to the proposal that during pexophagy in P. pastoris, the phagophore originates from the PAS to form the MIPA and the pexophagosome,22 but this hypothesis remained untested.
We analyzed GFP-PpAtg8-labeled structures formed during pexophagy (Fig. 6). During micropexophagy and macropexophagy, the MIPA and pexophagosomes, respectively, were detected in WT, Ppvps17Δ, and PpuvragΔ cells, consistent with their proficiency in pexophagy. In addition, Ppvps17Δ, and to a lesser extent PpuvragΔ, cells, showed a strong accumulation of GFP fluorescence in the lumens of their fragmented vacuoles in every condition tested (SM, SM to SD/SE/SD-N, SD, SD to SD-N). In contrast, in the pexophagy-deficient Ppatg9Δ and Ppatg30Δ cells, most of the GFP-PpAtg8 was cytosolic, and the MIPA and pexophagosomes were absent, although occasional rare GFP-PpAtg8 dots were found. Ppatg6Δ, Ppvps15Δ, Ppvps34Δ cells, and cells overexpressing PpUvrag, did not form any MIPA or pexophagosomes, suggesting that the PI3K complex I is necessary for the generation of the phagophore. Although, GFP-PpAtg8 localized to some dot-like structures, these structures could not transition into the phagophore without the subunits of PI3K complex I.
Figure 6.
Failure to form isolation membranes in Ppatg6Δ, Ppvps15Δ, Ppvps34Δ cells and in cells overexpressing PpUvrag. WT and mutant cells expressing GFP-PpAtg8 were grown in methanol medium for 15 h and shifted to glucose medium for 30 min to induce micropexophagy and ethanol medium for 1 h to induce macropexophagy. The GFP-PpAtg8 localization was observed by fluorescence microscopy. The vacuole was stained with FM4-64. Peroxisomes are labeled with BFP-sKL in WT, Ppatg6Δ and PpuvragΔ. Arrow indicates isolation membranes labeled with GFP-PpAtg8.
PI3K complex localization during the elongation of the phagophore
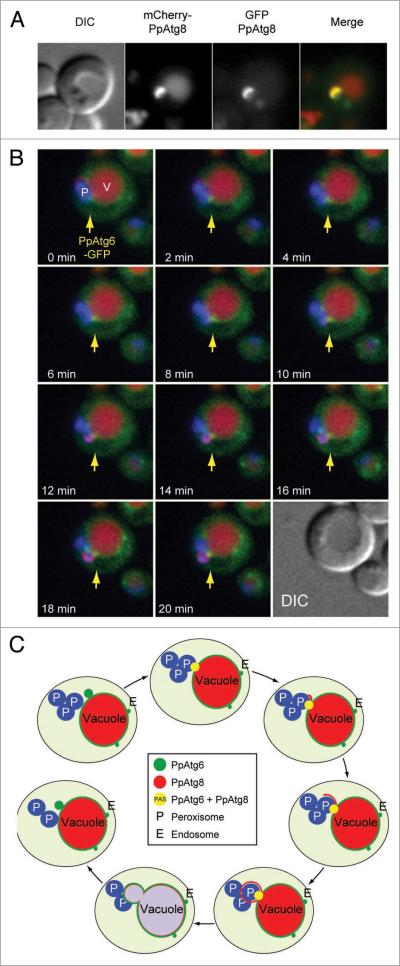
We followed the localization of PpAtg6 (and by extension the PI3K complex) during pexophagy of methanol-induced peroxisomes, taking advantage of the large size of the phagophore to distinguish PAS localization from the subsequent biogenesis of an isolation membrane (pexophagosome or MIPA). Phagophores could be visualized by using mCherry-PpAtg8 or GFP-PpAtg8. One difference between the two fusion proteins is that mCherry-PpAtg8 was more abundant inside the vacuole, probably because GFP loses its fluorescence in the acidic environment inside the vacuole, whereas mCherry does not.35 In the control experiment coexpressed mCherry-PpAtg8 and GFP-PpAtg8 labeled the same phagophores (Fig. 7A).
Figure 7.
Visualization of steps in phagophore biogenesis from the PAS. (A) colocalization of mcherry-PpAtg8 and GFP-PpAtg8 in WT cells during pexophagy induced by shifting from methanol medium to nitrogen starvation medium. (B) Time lapse microscopy of cells expressing PpAtg6-GFP, mcherry-PpAtg8 and BFP-sKL. Cells were grown in methanol medium for 15 h and pexophagy was induced by shift to nitrogen starvation medium for 1 h prior to the start of time-lapse microscopy. Also see videos 2 and 3. Arrow indicates PpAtg6 localization at the PAS. (c) schematic view of isolation membrane formation and localization of PpAtg6 and PpAtg8 during this process.
To track the localization of PpAtg6 relative to the forming isolation membrane, Ppatg6Δ cells were transformed with mCherry-PpAtg8 and PpAtg6-GFP (both expressed from their endogenous promoters), and macropexophagy was induced by subjecting methanol-grown cells to nitrogen starvation. We induced macropexophagy because pexophagosome formation mimics a general autophagy mechanism better than MIPA formation. Furthermore, the assay was carried out in nitrogen starvation conditions because more isolation membranes were detected than in ethanol-induced macropexophagy [Video 1]. After one hour of nitrogen starvation, cells were subjected to in vivo time-lapse microscopy with a temperature-controlled chamber and objective. Peroxisomes were surrounded by the phagophore one-by-one and a complete pexophagosome was formed in around 20 min (Fig. 7B, video 2 and 3), consistent with macropexophagy.
From this real-time microscopy, we could distinguish several steps in pexophagosome formation (Fig. 7C): (1) PpAtg6 localizes to several perivacuolar dots; (2) PpAtg8 starts to localize in one of these structures, defining this as the PAS; (3) the isolation membrane begins to elongate from the PAS (where PpAtg8 and PpAtg6 colocalize); (4) the isolation membrane engulfs one peroxisome, creating a pexophagosome; and (5) PpAtg6 stays at the PAS during the entire process.
PI3K complex interactions
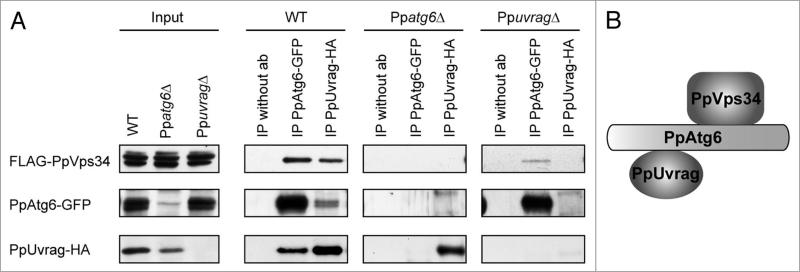
Studies of the protein-protein interactions in PI3K complexes have indicated differences in structure between the S. cerevisiae and mammalian complexes. In yeast, Vps38 or Atg14 provides the link between Atg6 and Vps34. In mammals, Beclin1/Atg6 is actually the link between UVRAG/Vps38 or Barkor/Atg14 and Vps34.30,36 To elucidate how these proteins interact in P. pastoris (Fig. 8A), we constructed tagged fusion proteins of PpVps34, PpAtg6 and PpUvrag, each expressed from its endogenous promoter. PpVps34 was tagged with two FLAG sequences at its N-terminus, PpAtg6 was tagged with GFP at its C-terminus, and PpUvrag was tagged with HA at its C-terminus. In WT cells, immunoprecipitation using an antibody against GFP pulled down abundant levels of PpAtg6-GFP and together with it, FLAG-PpVps34 and PpUvrag-HA. Similarly, immunoprecipitation of PpUvrag-HA brought down FLAG-PpVps34 and PpAtg6-GFP. In the absence of antibody, no nonspecific proteins were detected. These co-immunoprecipitations demonstrate that these proteins are present in the same complex.
Figure 8.
PpUvrag interacts with PpVps34 through PpAtg6. (A) Co-immunoprecipitation study was performed with: WT cells expressing FLAG-PpVps34, PpAtg6-GFP and PpUvrag-HA; Ppatg6Δ cells expressing FLAG-PpVps34 and PpUvrag-HA; and PpuvragΔ cells expressing FLAG-PpVps34 and PpAtg6-GFP. Cells were grown in sD for 15 h and shifted to SD-N for 1 h. Immunoprecipitation was performed as described in Materials and Methods. (B) schematic view of PpAtg6, PpVps34 and PpUvrag interactions.
To separate indirect from direct interactions, we performed the same co-immunoprecipitation in the absence of PpAtg6 or PpUvrag. In the absence of PpUvrag, the interaction between PpAtg6-GFP and FLAG-PpVps34 was still detectable, suggesting that PpAtg6 interacts with PpVps34 either directly or through other proteins not tested (e.g., an Atg14 ortholog). In Ppatg6Δ cells, although PpUvrag-HA and FLAG-PpVps34 were both detected at the same levels as in the WT, the interaction between them was not, suggesting that the link between PpUvrag and PpVps34 is indirect, probably through PpAtg6 (Fig. 8B). Nevertheless, because the stability of PpUvrag was somewhat lower in the absence of PpAtg6 relative to the WT strain, we cannot rigorously exclude the possibility that a direct interaction between PpUvrag and PpVps34 was missed because the complex, or a particular component, is unstable.
Discussion
Role of PI3K and Uvrag in autophagy-related and VPS pathways
Our finding that PpAtg6, PpVps15 and PpVps34 are required for the autophagy, Cvt, micropexophagy and macropexophagy pathways adds P. pastoris to the list of species in which these proteins are required for general autophagy. Further, the requirement of PpAtg6, PpVps15 and PpVps34 for all autophagy-related pathways (in addition to the VPS pathway) in P. pastoris reinforces the notion that these proteins lie at the core of the autophagic machinery.
The precise functional role of UVRAG, a binding partner for Beclin 1, remains controversial in mammals. One area of disagreement concerns whether UVRAG expression enhances autophagic flux.12,30,31,37 Our assays clearly indicate that PpUvrag is not required for any of the autophagic pathways described in P. pastoris.
Both PpuvragΔ and Ppvps17Δ cells share a defect in PpCPY sorting to the vacuole, but have fully functional autophagic pathways. It seems likely that the inviability upon nitrogen starvation of cells lacking PpUvrag or PpVps17 might be the result of the inability of these cells to deliver proteins to the vacuole, causing secondary defects in the vacuolar degradation or nutrient recycling pathways.
In many respects, our findings are consistent with the proposition that Uvrag is, at least in P. pastoris, an ortholog of Vps38. Like Vps38, it localizes to numerous, presumably endosomal, puncta, but not the PAS and is required for proper sorting of CPY, but not for any autophagic pathway. It binds PpVps34, albeit indirectly through PpAtg6 (rather than directly as in S. cerevisiae). Consistent with this interaction of PpUvrag with PpAtg6, PpUvrag is mislocalized in the absence of PpAtg6, and is required for the localization of PpAtg6 at most cytosolic puncta, but not at the PAS. PpAtg24, similar to PpAtg6, is not mislocalized from the PAS in the absence of PpUvrag.
Further support for the notion that PpUvrag binds PpAtg6 in a complex distinct from the PI3K complex required for autophagy comes from our finding that overexpression of PpUvrag produces cells that are defective in all autophagic pathways. This is the phenotype that one might expect from overexpressing a protein that competes with autophagy-related proteins for Atg6-binding sites. The suggestion that PpUvrag is found in a nonautophagic PI3K complex is related to recent findings in mammalian cells that the mammalian orthologs of Atg6, Vps34 and Vps15 are found in at least two distinct complexes, of which one (with UVRAG) promotes endocytosis and autophagosome maturation by fusion with endosomes, and the other (with Atg14/Barkor/Atg14L) promotes autophagosome formation.15
A putative role for Vps17 at the PAS
Serendipitously, we found that the PpVps17 localized to the PAS in WT and PpuvragΔ cells during autophagy. This was interesting as we, and others, have shown that autophagy is unaffected in the absence of Vps17. Despite this fact, the rate of GFP-PpAtg8 processing during autophagy is faster than in WT (or PpuvragΔ) cells. GFP appeared prematurely in the vacuole even during vegetative growth. The PAS localization of PpVps17 and the premature GFP-PpAtg8 accumulation in the vacuole suggest that this retromer subunit could have some retrograde function in autophagy.
PI3K complex 1 involvement in stages of the biogenesis of the phagophore
Most studies of the role of the PI3K complex I during autophagy have revealed its role in the initiation of autophagosome formation (nucleation). In the absence of Atg14, PAS localization of several Atg proteins is disturbed.38 As a consequence of the essential role of PI3K in the early stage of autophagosome formation, the role of PI3K complex I during later stages, such as elongation of the isolation membrane, has not been directly demonstrated.
Indications of the possible role of PI3K complex I in elongation arise from the observation that, in yeast, PtdIns3P is localized not only at the PAS (as expected from the localization of Atg14), but also on the phagophore.39 Furthermore, Atg5 is dependent on Atg14 for its correct localization, and in mammals, Atg5 is localized to the outer face of expanding isolation membrane, suggesting a role for complex I in elongation.38,40
Using time-lapse microscopy, we showed that during the nucleation stage, PpAtg6 accumulates in one perivacuolar dot, where, after a couple of minutes, PpAtg8 also appears. This structure where both proteins colocalize is the PAS. Next, the phagophore starts to elongate from the PAS, until it forms a complete pexophagosome which then fuses with the vacuole. PpAtg6 (and therefore the PI3K complex I) remains at the PAS during elongation until the commencement of fusion of the phagophore. This suggests that the PI3K complex I plays a role not only during nucleation, but also in the elongation and maybe in the fusion of the tips of phagophore or the fusion of pexophagosome with the vacuole membrane. These studies define more precisely, in yeast, the role of PI3K complex I in the biogenesis of the isolation membrane and indicate that the PtdIns3P present on the phagophore flows from the PAS.
Materials and Methods
Strains and plasmids
Strains and plasmids used in this study are listed in Tables 1 and 2, respectively. All strains were constructed by electroporation of linear DNA into competent cells of the parent strain.41
Table 1.
strains
| Description | Strain | Genotype | Reference |
|---|---|---|---|
| GS115 | his4 | 42 | |
| GS200 | his4, arg4 | 43 | |
| WT | PPY12 | his4, arg4 | 44 |
| Ppatg6Δ | Srdm006 | PPY12 atg6Δ∷KAN | This study |
| Ppatg8Δ | Sjcf257 | PPY12 atg8Δ∷ZEO | 25 |
| Ppatg9Δ | R19 | GS115 atg9Δ∷ZEO | 24 |
| Ppatg30Δ | Sjcf44 | PPY12 atg30Δ∷ZEO | 25 |
| Ppvps15Δ | OP5 | GS200 vps15Δ∷ARG4 | 16 |
| Ppvps17Δ | Srdm122 | PPY12 vps17Δ∷KAN | This study |
| Ppvps34Δ | Sjcf56 | PPY12 vps34Δ∷ZEO | This study |
| PpuvragΔ | Srdm083 | PPY12 uvragΔ∷ZEO | This study |
| Ppatg25-likeΔ | Sjcf1231 | PPY12 atg25-likeΔ∷KAN | This study |
Table 2.
Plasmids
| Plasmid | Promoter | Fusion protein | Integration locus | Selectable marker |
|---|---|---|---|---|
| pJCF208 | ATG8 | GFP-PpAtg8 | HIS4 | HIS4 |
| pJCF477 | ATG8 | mCherry-PpAtg8 | ARG4 | ARG4 |
| pJCF143 | AOX1 | BFP-SKL | AOX1 | Blasticidin |
| pRDM054 | AOX1 | BFP-SKL | AOX1 | Hygromycin |
| pJCF94 | CPY | PpCPY-HA | CPY | Zeocin |
| pJCF427 | CPY | PpCPY-HA | CPY | Kanamycin |
| pRDM037 | ATG6 | PpAtg6-GFP | HIS4 | HIS4 |
| pRDM058 | UVRAG | PpUvrag-GFP | HIS4 | HIS4 |
| pRDM066 | GAPDH | PpUvrag-GFP | HIS4 | HIS4 |
| pJCF426 | UVRAG | PpUvrag-HA | HIS4 | HIS4 or Zeocin |
| pJCF464 | GAPDH | PpUvrag-HA | HIS4 | HIS4 or Hygromycin |
| pJCF210 | ATG24 | PpAtg24-GFP | HIS4 | HIS4 |
| pRDM069 | VPS17 | PpVps17-GFP | HIS4 | HIS4 |
| pRDM034 | VPS34 | FLAG-PpVps34 | ARG4 | ARG4 |
| pJCF474 | ATG25-like | PpAtg25-like-MYC | HIS4 | HIS4 or Hygromycin |
| pJCF472 | ATG25-like | PpAtg25-like-GFP | HIS4 | HIS4 |
Media and culture growth conditions
Media used to grow strains include: YPD (2% glucose, 2% bactopeptone and 1% yeast extract); YNB (0.17% yeast nitrogen base without amino acids and ammonium sulfate; 0.5% ammonium sulfate); glucose medium or SD (YNB; 0.079% complete supplement mixture [CSM] {4500-0 × 2, BIO 101}; 0.05% yeast extract; 2% glucose); nitrogen starvation medium or SD-N (0.17% yeast nitrogen base without amino acids and ammonium sulfate; 2% glucose); ethanol medium or SE (YNB; 0.079% CSM; 0.05% yeast extract; 0.5% ethanol); oleate medium (YNB; 0.079% CSM; 0.05% yeast extract; 0.0125% Tween-80; 0.5% oleate) and methanol medium or SM (YNB; 0.079% CSM; 0.05% yeast extract; 0.5% methanol). All cultures were grown at 30°C.
Nitrogen starvation cell viability assay
Cells were grown in YPD medium to an optical density at 600 nm (OD600) near 1, washed twice in sterile, double-distilled H2O, resuspended in SD-N medium at an OD600 near 0.5, and incubated on a shaker at 30°C. At the indicated time points, 100 μl aliquots were taken from each culture and used to prepare serial 10-fold dilutions in YNB. Appropriate dilutions were plated in triplicate (200 μl per plate) on YPD agar plates. Colonies were counted 2 to 3 days after plating. The dilution yielding between 30 and 400 colonies per plate was selected for counting of colonies. The survival data were plotted as the average number of viable cells (based on average colony counts) remaining per OD600 equivalent of the original culture (one OD600 equivalent is the number of cells required for one ml of culture with an OD600 = 1).
GFP-PpAtg8 cleavage assay
Cells were grown in SD medium to an OD600 near 1, washed twice in sterile, double-distilled H2O, and resuspended in SD-N medium at an OD600 near 2. At each time point, a 1 ml sample was removed. Cells were harvested by centrifugation (with medium removed by aspiration) and resuspended in 300 μl of a phosphate buffer (pH 7.5). 100 μl of 50% TCA was added. Samples were then mixed and stored at −80°C for more than an hour. Protein samples were then washed twice in 80% acetone and dissolved in 100 μl of 1% SDS/0.1 N NaOH buffer, and 20 μl of 6× SDS sample buffer was added. The samples were resolved by 12% SDS-PAGE gels and analyzed by western blotting using a monoclonal anti-GFP antibody (1181446001, Roche Applied Science).
PpApe1 processing assay
Cells were grown in SD medium to an OD600 near 1, washed twice in sterile, double-distilled H2O, and resuspended in SD-N medium at an OD600 near 2. At each time point, a 1 ml sample was removed and processed as described above with respect to the GFP-PpAtg8 cleavage assay, except that the polyclonal anti-PpApe1 antibody was used instead of the anti-GFP antibody.21
Biochemical studies of pexophagy
Peroxisomes were induced by incubation of cells in oleate medium (starting OD600 of 0.5) for 15 to 16 h, washed twice in sterile, double-distilled H2O and transferred to SD-N medium at an OD600 of 2, to induce pexophagy. Cells from 1 ml of culture samples were collected by centrifugation after 0, 6, 12 and 24 h. Crude extracts were prepared by TCA precipitation as described above (GFP-PpAtg8 cleavage assay). The samples were resolved by 10% SDS-PAGE gels and analyzed by western blotting using antibody against the peroxisomal protein, thiolase.
Biochemical studies of VPS pathway
Cells were grown in YPD medium to an OD600 of 1, then shifted to fresh YPD medium and incubated at 30°C. At each time point, a 500 μl sample was removed and cells were harvested by centrifugation. The pellets were resuspended in 300 μl of a phosphate buffer (pH 7.5) and proteins were precipitated with 100 μl of 50% TCA as described before. The proteins of the supernatant (extracellular medium) were precipitated with 1.2 ml of 100% cold acetone and stored at −80°C for more than an hour. Proteins were centrifuged for 15 min at 21,000 ×g for 15 min, dissolved in 80 μl of 1% SDS/0.1 N NaOH buffer, and 16 μl of 6× SDS sample buffer was added. The samples were resolved by 12% SDS-PAGE gels and analyzed by western blotting using a monoclonal anti-HA antibody (Roche Applied Science, 11583816001).
Fluorescence microscopy
Cells were grown initially in YPD medium to an OD600 near 1, then shifted to methanol, oleate or glucose medium depending the experiment and incubated overnight (15 to 16 h) at 30°C. When FM4-64 (Invitrogen, T-3166) was used, 5 μl/ml of a 1 mg/ml stock solution in dimethyl sulfoxide was added, together with 3.7 μl/ml of a 1 M HEPES buffer (pH 8.0), after resuspension of cells in SM medium. A motorized fluorescence microscope (Axioscope 2 MOT; Carl Zeiss MicroImaging, Jena, Germany) coupled with a monochrome digital camera (Axiocam MRm; Carl Zeiss MicroImaging, Jena, Germany) was used to capture images, which were processed using AxioVision software (Carl Zeiss MicroImaging, Jena, Germany).
Time-lapse microscopy
Cells were grown initially in YPD medium to an OD600 near 1, then shifted to methanol (starting OD600 of 0.5) and incubated overnight (15 to 16 h) at 30°C. Cells were harvested, washed, and resuspended into SD-N medium at an OD600 of 2, and incubated 1 h at 30°C to induce pexophagy. Small aliquots of cells were applied to microscopy slides coated with poly-L-Lysine solution (P8920, Sigma-Aldrich). The slide coated with cells was placed into a temperature-controlled chamber (FCS3 Closed Chamber System for Upright Microscopes, Bioptechs, Butler, USA), which, together with a temperature controlled 100× objective (Bioptechs Objective Heater, Bioptechs, Butler, USA), was maintained at 30°C. Images were acquired in a fully automated fashion at 2 min intervals using AxioVision software, Axiocam MRm camera and Axioscope 2 MOT microscope (Carl Zeiss MicroImaging, Jena, Germany).
Co-immunoprecipitation studies
Cells were grown overnight in glucose medium and transferred for 30 min on SD-N medium before extraction. Fifty OD600 equivalents of yeast cells were washed in phosphate-buffered saline (pH 7.4) and lysed with glass beads (vortexed 5 times for 2 min at 4°C) in 1 ml IP lysis buffer (50 mM HEPES-NaOH [pH 8], 0.15 M NaCl, 1% Triton, 10% glycerol, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Sigma-Aldrich, P8215]) and centrifuged (500 ×g, 3 min). The membrane protein solubilization was performed together with a pre-clearing step. For this purpose 100 μl of GammaBind G Sepharose (GE Healthcare, 17-0885-01) was added to the supernatant and incubated at 4°C for 30 min with rotation. Next the sample was centrifuged (21,000 ×g, 15 min) and the supernatant was incubated overnight with the selected primary antibody, rotating at 4°C. After the overnight incubation, 100 μl of GammaBind G Sepharose was added to the supernatant with primary antibody and incubated at 4°C for 1 h with rotation. Beads were washed three times (5 ml) with the IP lysis buffer for 5 min. Bound protein was eluted with 100 μl of sample buffer, resolved by SDS-PAGE, and visualized by immunoblotting. Loading was as follows: input, 0.2 OD600 equivalent and immunopurified proteins (IP), 7.5 OD600 equivalent.
Supplementary Material
Acknowledgements
This work was supported by an NIH grant GM69373 to S.S. We thank R. Tsien (University of California, San Diego) for the kind gift of mCherry.
Abbreviations
- PAS
phagophore assembly site
- MIPA
micropexophagy apparatus
- VPS
vacuolar protein sorting
- CPY
carboxypeptidase Y
- Ape1
aminopeptidase I
- Cvt
cytoplasm-to-vacuole targeting
- VSM
vacuolar sequestering membrane
- PI3K
class III phosphatidylinositol 3-kinase
- PtdIns
phosphatidylinositol
- PtdIns3P
phosphatidylinositol 3-phosphate
- Atg
autophagy-related
Footnotes
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–8. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- 6.Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 8.Xie Z, Nair U, Klionsky DJ. Dissecting autophagosome formation: the missing pieces. Autophagy. 2008;4:920–2. doi: 10.4161/auto.6692. [DOI] [PubMed] [Google Scholar]
- 9.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 11.Burda P, Padilla SM, Sarkar S, Emr SD. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci. 2002;115:3889–900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- 12.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–87. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumori-genesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 16.Stasyk OV, van der Klei IJ, Bellu AR, Shen S, Kiel JA, Cregg JM, Veenhuis M. A Pichia pastoris VPS15 homologue is required in selective peroxisome autophagy. Curr Genet. 1999;36:262–9. doi: 10.1007/s002940050499. [DOI] [PubMed] [Google Scholar]
- 17.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:205–10. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–16. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 20.Monastyrska I, Kiel JA, Krikken AM, Komduur JA, Veenhuis M, van der Klei IJ. The Hansenula polymorpha ATG25 gene encodes a novel coiled-coil protein that is required for macropexophagy. Autophagy. 2005;1:92–100. doi: 10.4161/auto.1.2.1832. [DOI] [PubMed] [Google Scholar]
- 21.Farre JC, Vidal J, Subramani S. A cytoplasm to vacuole targeting pathway in P. pastoris. Autophagy. 2007;3:230–4. doi: 10.4161/auto.3905. [DOI] [PubMed] [Google Scholar]
- 22.Farre JC, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21:522–30. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukaiyama H, Oku M, Baba M, Samizo T, Hammond AT, Glick BS, et al. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 2002;7:75–90. doi: 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- 24.Chang T, Schroder LA, Thomson JM, Klocman AS, Tomasini AJ, Stromhaug PE, Dunn WA., Jr PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol Biol Cell. 2005;16:4941–53. doi: 10.1091/mbc.E05-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–76. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- 27.Nazarko TY, Polupanov AS, Manjithaya RR, Subramani S, Sibirny AA. The requirement of sterol glucoside for pexophagy in yeast is dependent on the species and nature of peroxisome inducers. Mol Biol Cell. 2007;18:106–18. doi: 10.1091/mbc.E06-06-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–39. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 30.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 31.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–6. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hettema EH, Lewis MJ, Black MW, Pelham HR. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–57. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yen WL, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007;18:581–93. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 36.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 37.Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534–6. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 39.Obara K, Noda T, Niimi K, Ohsumi Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008;13:537–47. doi: 10.1111/j.1365-2443.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- 40.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cregg JM, Russell KA. Transformation. Methods Mol Biol. 1998;103:27–39. doi: 10.1385/0-89603-421-6:27. [DOI] [PubMed] [Google Scholar]
- 42.Cregg JM, Barringer KJ, Hessler AY, Madden KR. Pichia pastoris as a host system for transformations. Mol Cell Biol. 1985;5:3376–385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterham HR, de Vries Y, Russel KA, Xie W, Veenhuis M, Cregg JM. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol Cell Biol. 1996;16:2527–36. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gould SJ, McCollum D, Spong AP, Heyman JA, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–28. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.