Abstract
The kidney has a central role in the regulation of blood pressure, in large part through its role in the regulated reabsorption of filtered Na+. Epithelial Na+ channels (ENaCs) are expressed in the most distal segments of the nephron and are a target of volume regulatory hormones. A variety of factors regulate ENaC activity, including several aldosterone-induced proteins that are present within an ENaC regulatory complex. Proteases also regulate ENaC by cleaving the channel and releasing intrinsic inhibitory tracts. Polymorphisms or mutations within channel subunits or regulatory pathways that enhance channel activity may contribute to an increase in blood pressure in individuals with essential hypertension.
Keywords: Acid-sensing Ion Channels (ASIC), MAP Kinases (MAPKs), Protease, Serine/Threonine Protein Kinase, Sodium Channels, ENaC, Epithelial Sodium Channel, SGK1, Hypertension
Introduction
Role of Na+ in the Control of Blood Pressure
Hypertension, often labeled “the silent killer,” affects one in three adults in the United States and is a major independent risk factor for cardiovascular disease, stroke, and renal failure. Approximately 50% of hypertensive individuals are salt-sensitive (1), and dietary Na+ has a significant effect on blood pressure (2). These numbers are higher in African Americans and obese individuals.
The kidney has a primary role in the regulation of extracellular fluid volume and blood pressure by regulating the total body Na+ content. Given the facts that Na+ is largely excluded from cells due to the Na+,K+-ATPase and that the extracellular Na+ concentration is maintained at a constant level via water regulatory mechanisms, total body Na+ content is the main determinant of extracellular fluid volume. Changes in extracellular fluid volume will be reflected in changes in intravascular volume and blood pressure.
Mature human kidneys normally filter on the order of one pound of Na+ on a daily basis. The vast majority of filtered Na+ is reabsorbed in various nephron segments to achieve a rate of urinary Na+ excretion that matches rates of Na+ intake. It is in the distal nephron where fine-tuning of the absorption of filtered Na+ occurs. The epithelial Na+ channel (ENaC)2 is a key Na+ transporter in this segment of the nephron (3).
Aldosterone is one of the key hormones involved in the regulation of extracellular fluid volume and blood pressure, primarily through its effects on renal Na+ handling in the distal nephron (for review, see Ref. 4). Aldosterone is secreted from the adrenal cortex in response to a decrease in extracellular fluid volume or effective arteriolar volume, as well as in response to an increase in plasma [K+]. The distal nephron is an aldosterone target tissue, defined by the presence of mineralocorticoid receptors and β-hydroxysteroid dehydrogenase type II, an enzyme that degrades cortisol, a steroid hormone that binds to and activates mineralocorticoid receptors. The activation of mineralocorticoid receptors by aldosterone in the distal nephron eventually results in the activation of transport proteins that facilitate Na+ reabsorption (see below).
Increased Na+ reabsorption occurs in a number of disorders associated with hypertension. These include disorders associated with increased aldosterone secretion in the absence of the normal physiologic activators of secretion, disorders associated with inhibition or loss of function of β-hydroxysteroid dehydrogenase type II, and inherited disorders associated with activation of distal nephron Na+ transporters, such as Liddle syndrome and familial hyperkalemic hypertension (for reviews, see Refs. 5 and 6).
Liddle syndrome reflects gain-of-function mutations within ENaC that lead to hypertension and volume expansion (7–9). These mutations reside largely in the cytoplasmic C-terminal tails of the β- and γ-subunits of ENaC (7), resulting in loss of a key adaptor Pro-Tyr motif that interacts with Trp-Trp domains within Nedd4-2 (neural precursor cell-expressed, developmentally down-regulated 4-2), an E3 ubiquitin ligase. Nedd4-2-dependent ubiquitination of ENaC subunits targets channels for internalization from the plasma membrane (10). Once internalized, channels may be targeted for degradation (11–13). Alternatively, the release of ubiquitin by deubiquitinating enzymes allows channels to recycle to the plasma membrane (14). Loss of the Pro-Tyr motif leads to increased channel surface expression and channel processing by proteases that enhance its activity (see below).
Hypertension and associated sequelae are key clinical findings in individuals with Liddle syndrome and hyperaldosteronism. Although the primary etiology of hypertension in these disorders is well established, reflecting activation of renal Na+ reabsorption via ENaC and the Na+/Cl−-coupled cotransporter (NCC) with hyperaldosteronism, for the vast majority of individuals with hypertension, the etiology of hypertension is unknown. Both genetic and environmental factors such as Na+ intake contribute to an increase in blood pressure in this population with what is commonly referred to as essential hypertension.
Given the role of the kidney and, in particular, the role of ENaC in the regulation of extracellular fluid volume and blood pressure, it is quite likely that polymorphisms or mutations within the channel or within key cellular regulatory processes that lead to an increase in channel activity contribute to increased blood pressure in this population with essential hypertension. If so, this would suggest that therapy targeted at inhibiting ENaC or associated regulatory processes might be beneficial in the management of blood pressure in a subset of individuals with essential hypertension. Our minireview addresses some of the sites within the channel where polymorphisms or mutations would be predicted to increase channel activity, as well as several of the key regulatory mechanisms where polymorphisms or mutations within these pathways may result in increased channel activity.
ENaC Structure
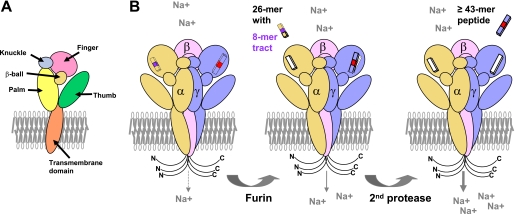
ENaC is an apically located Na+-selective ion channel expressed primarily in polarized epithelia of the distal nephron, lung, and distal colon. It is highly selective for Na+ and Li+ over K+ and is inhibited by K+-sparing diuretics such as amiloride (15, 16). In most tissues, ENaC is composed of three structurally related subunits, α, β, and γ. Each subunit has two membrane-spanning domains separated by a large extracellular region and short N- and C-terminal cytoplasmic tails (17, 18). The ENaC subunits are members of a family of ion channels that include the acid-sensing ion channels (ASICs). Channel subunits undergo post-translational modifications by terminal processing of N-glycans and by proteolytic cleavage at defined sites within their extracellular domains (see below) (19–23). The recently resolved crystal structure of ASIC1 has provided important insights into the structural organization of ENaCs (24). ASIC1 is a homotrimer, suggesting that ENaCs have an α1:β1:γ1 subunit stoichiometry. The extracellular region of ASIC1 is a highly ordered structure that resembles an outstretched hand containing a ball and has clearly defined domains termed wrist, finger, thumb, palm, knuckle, and β-ball. It is likely that the extracellular regions within ENaC subunits have a similar domain organization.
ENaC Gain-of-function Polymorphisms and Mutations
Polymorphisms or mutations that enhance ENaC activity have been described in cytoplasmic, membrane-spanning, and external sites within the channel. For example, two polymorphisms that have been described in the C-terminal tail of the α-subunit, C618F and A663T, are associated with enhanced ENaC activity, reflecting an increase in channel surface expression (25, 26). A T594M polymorphism in the C-terminal tail of the β-subunit was reported to be associated with enhanced cAMP activation and reduced protein kinase C-dependent inhibition of endogenous amiloride-sensitive Na+ currents in human lymphocytes (27, 28). These currents are presumably mediated by ENaC. However, similar levels of channel activity have been reported with the T594M polymorphism when expressed in heterologous systems (29).
The primary gate and cation selectivity filter of the channel are located within the membrane-spanning domains (15, 30–32). Selected mutations or modifications of introduced Cys residues at a key site within the gate, referred to as the degenerin site, activate ENaC (31–33). Mutations in the vicinity of this site also activate the channel (31). For example, a γ-subunit N530K mutation that activates ENaC has been reported in an individual with diabetic nephropathy (32).
The extracellular domains of ENaC subunits have an important role in the regulation of channel activity by external factors. For example, channels are inhibited by external Na+, a process referred to as Na+ self-inhibition (34). Na+ presumably binds to sites within the extracellular domains of the channel, leading to a reduction in channel open probability (35–38). Mutations have been described that are associated with loss of Na+ self-inhibition, resulting in an increase in channel activity, reflecting an increase in channel open probability. For example, mutations of a Met residue in the periphery of the thumb domain (Met438) (38) and mutation of a His residue in the finger domain (His239) (35) of the mouse γ-subunit led to loss of Na+ self-inhibition. A W493R mutant in the human α-subunit is also associated with loss of Na+ self-inhibition (39). External Cl− inhibits ENaC by enhancing Na+ self-inhibition. Mutations of residues within a putative Cl−-binding site in the thumb domain of the α-subunit (His418) and β-subunit (Arg388) of human ENaC activate the channel by reducing inhibition by external Cl− (40).
Are channel-activating polymorphisms associated with an increase in blood pressure in humans? This is a difficult question to address. The effects on blood pressure may be modest and may be dependent on other variables that influence blood pressure. The effects of these polymorphisms on blood pressure may be limited to specific populations, reflecting inherent genetic or environmental factors, including dietary Na+ consumption. Activation of ENaC may lead to compensatory changes in other renal Na+ transporters that may dampen any effects on blood pressure. Finally, a change in one allele may not be sufficient to exert an effect on blood pressure. Given these constraints, it has been challenging to demonstrate a link between ENaC polymorphisms and hypertension. For example, the βT594M polymorphism has been reported to be associated with increased blood pressure in an English population with African ancestry (41, 42), but an association was not observed in a South African population (43). It has been suggested that amiloride might help in controlling blood pressure in hypertensive individuals with the βT594M polymorphism (41, 44). Regarding the αA663T polymorphism, one study reported an association with blood pressure that was opposite of that predicted from functional studies (45, 46), whereas other studies found no correlation with blood pressure (47, 48).
In addition to affecting blood pressure, gain-of-function ENaC mutations may also affect airway surface liquid volume and mucociliary clearance. For example, individuals who are heterozygous for the ΔF508 mutation in CFTR (cystic fibrosis transmembrane conductance regulator) lack overt pulmonary symptoms. Mutations in ENaC have been reported in individuals with atypical cystic fibrosis who are heterozygous for the ΔF508 CFTR mutation (49, 50). For example, an activating ENaC mutation (αW493R) that has loss of Na+ self-inhibition has been described in this population (39, 49). At present, it is unclear whether individuals with atypical cystic fibrosis and activating ENaC mutations have elevated blood pressures.
ENaC Regulatory Pathways
ENaC is regulated by a variety of extrinsic and intrinsic factors through changes in plasma membrane abundance or inherent activity (for reviews, see Refs. 23, 51, and 52). Plasma membrane abundance is controlled primarily through changes in intracellular trafficking, and ENaC activity is controlled primarily through changes in open probability, which is profoundly influenced by proteolytic processing and by interactions with cytoplasmic domains with specific membrane acidic phospholipids. Regulatory effects on open probability and membrane abundance are not mutually exclusive and raise the interesting possibility that both could be operative and linked through trafficking-dependent events (53). Transcriptional regulation of ENaC subunit expression constitutes another important regulatory mechanism, particularly for consolidation of aldosterone-stimulated Na+ transport (reviewed in Ref. 54).
Renal Na+ reabsorption is highly controlled by the mineralocorticoid aldosterone, which acts primarily through the mineralocorticoid receptor to alter the transcription of a set of target genes (4). As with other steroid-regulated processes (55), two major classes of target genes have been identified: early and late. Early response genes appear to be required for initiation of the response, whereas late response genes participate in consolidation (56). The latter include components of the ion transport machinery itself, including ENaC and Na+,K+-ATPase subunits. The late response genes also encode regulatory proteins that likely act to limit the extent of the aldosterone response, such as activators of the MAPK cascade, including the epidermal growth factor receptor (57). Early response genes include primarily signaling molecules implicated in pathways that control ENaC activity and/or trafficking (4).
SGK1
The best characterized of these aldosterone-regulated genes is the serine/threonine kinase SGK1 (serum- and glucocorticoid-induced kinase-1) (4). To become competent to activate ENaC, SGK1 must itself undergo two activating phosphorylations: first by mTOR (mammalian target of rapamycin) (58) and second by PDK1 (PI3K-dependent kinase-1) (4). The molecular mechanisms of ENaC stimulation by activated SGK1 can be divided into three known categories: (i) post-translational effects on the E3 ubiquitin ligase Nedd4-2, (ii) post-translational Nedd4-2-independent effects, and (iii) transcription of gene products, i.e. αENaC. Nedd4-2 interacts with the C-terminal tails of ENaC subunits, decreases surface expression of the channel via channel ubiquitination, and hence inhibits Na+ currents (10). SGK1 physically interacts with Nedd4-2, phosphorylates and inhibits it (59, 60), and hence indirectly enhances cell-surface expression of ENaC (61, 62).
Nedd4-2-independent mechanisms of SGK1 stimulation of ENaC have also been proposed. SGK1 was shown to directly phosphorylate a serine residue in the intracellular C-terminal tail of αENaC, which directly activates channels at the cell surface (63). Recently, SGK1 has been implicated in the stimulation of ENaC via phosphorylation of WNK4 (with no lysine-4), a kinase mutated in familial hyperkalemic hypertension (64, 65). SGK1 may also indirectly enhance ENaC Po through effects on membrane-bound channel-activating serine proteases (53, 66). Furthermore, in addition to its effects on ENaC, SGK1 has been shown to stimulate the activity of the basolateral Na+,K+-ATPase, which separately increases ENaC-mediated Na+ transport (67, 68). The relative importance of these effects compared with the Nedd4-2-dependent inhibition has, however, not been determined (63).
A third mechanism of ENaC stimulation by SGK1 involves up-regulation of components of the Na+ transport machinery per se. SGK1 has been shown to regulate the expression of late aldosterone-responsive genes, primarily αENaC (69). Active SGK1 is an important mediator of aldosterone-sensitive αENaC transcription in vivo via inhibition of a transcriptional repression element, the Dot1a (disruptor of telomeric silencing alternative splice variant 1a)-Af9 (ALL1-fused gene from chromosome 9) complex (70). SGK1 phosphorylates Af9 and reduces interaction between Dot1a and Af9. This releases suppression of ENaC transcription by this complex. Thus, SGK1 not only acts on ENaC channels to rapidly enhance Na+ channel activity by an increase in active channels at the apical surface and an increase in Na+,K+-ATPase activity at the basolateral surface but also stimulates transcription of elements of the machinery for Na+ transport to promote a sustained response to aldosterone.
The ability of SGK1 to regulate in vivo renal Na+ reabsorption is well illustrated by the impaired Na+ retention of gene-targeted mice lacking functional SGK1 (71, 72). On a low Na+ diet, these mice have a significant decrease in blood pressure compared with their wild-type littermates (71). These sgk1−/− mice are also particularly resistant to the salt-sensitizing effect of hyperinsulinemia on blood pressure (73). SGK1 further mediates the stimulating effect of mineralocorticoids on salt appetite (74). Thus, SGK1 influences salt balance, and hence blood pressure, by affecting both Na+ intake and renal Na+ reabsorption. It is notable that two independent SGK1 knock-out models demonstrate the same key feature of failure to respond normally to NaCl restriction despite markedly increased aldosterone levels (71, 72), suggesting a mild form of the aldosterone resistance seen in the mineralocorticoid receptor knock-out mouse (75). Although some features of these two SGK1 knock-outs may diverge, evidence from both also supports the idea that both ENaC and NCC are regulated by SGK1 (72, 76). NCC is localized in kidney tubule segments upstream of ENaC (distal convoluted tubule) and mediates electroneutral uptake of NaCl. This transporter is regulated by aldosterone in a manner that appears to be controlled at least in part by WNK kinases (77). SGK1 phosphorylates WNK4 (64, 65), leading to the interesting speculation that SGK1 and WNK4 may act in concert to determine the degree to which Na+ is exchanged for K+ (which is favored when ENaC is the principal route for Na+ reabsorption) or alternatively is reabsorbed with Cl− without triggering K+ secretion (when NCC is used) (65, 78). It is notable in this context that SGK1 null mice on a high K+ diet are hyperkalemic despite markedly increased aldosterone (79). Finally, mouse knock-out models also have provided support for a role of SGK1 in regulating a variety of other processes, which could be implicated in the pathogenesis of metabolic syndrome and hypertension (80).
The connection between SGK1, salt balance, and blood pressure regulation in mice prompted the hypothesis that SGK1 might play a role in human blood pressure variation. Genetic studies in twins indeed revealed a specific SGK1 risk haplotype associated with moderately elevated blood pressure in individuals who simultaneously carry a homozygous genotype for a variant in intron 6 (I6CC) and a homozygous or heterozygous genotype for the C allele of a polymorphism in exon 8 (E8CC/CT) (81, 82). This SGK1 risk haplotype was later confirmed to be associated with hypertension in a large-scale population study of >4800 Swedish subjects (83).
GILZ1 and the ENaC Regulatory Complex
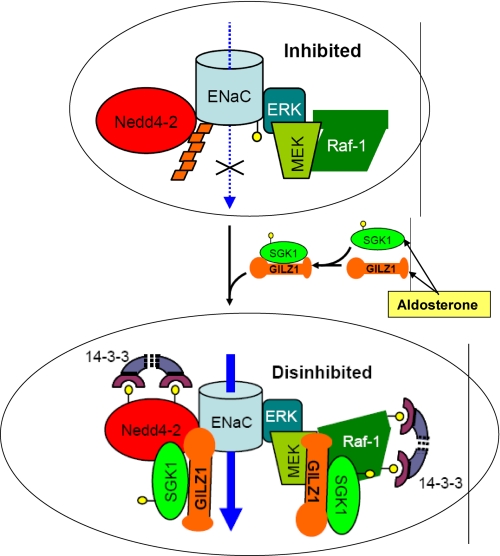
Renal ENaC function and renal mineralocorticoid action are only partially dependent on SGK1. Other aldosterone-regulated mediators provide stimulatory input into the system (84). Besides SGK1, aldosterone stimulates the expression of a number of other regulatory proteins, several of which impact on PI3K- or MAPK-dependent signaling (4). One of these is GILZ1 (glucocorticoid-induced leucine zipper protein 1), a small chaperone protein that has been found to stimulate ENaC surface expression by disrupting MAPK signaling (84, 85). ENaC is inhibited by the Raf-MEK-ERK1/2 MAPK pathway (57, 86, 87). GILZ1 physically interacts with and inhibits Raf-1 (88, 89), leading to ENaC activation. Remarkably, GILZ1 also physically interacts with Nedd4-2 and SGK1, and it synergizes with SGK1 in the inhibition of Raf-1 and Nedd4-2. All of these factors are associated with ENaC within an ENaC regulatory complex (ERC), the composition of which is controlled by GILZ1 (depicted schematically in Fig. 1) (90). In the absence of aldosterone with low levels of GILZ1 and SGK1, the complex is likely composed of Nedd4-2 and Raf-1, along with other components of the Raf-MEK-ERK signaling pathway that are known to be tightly associated with Raf-1. In the presence of aldosterone, GILZ1 recruits SGK1 into the complex. Together, they cooperatively inhibit Nedd4-2 and Raf-1 and synergistically stimulate ENaC surface expression and activity (Fig. 1) (90). It is interesting to note that GILZ1 has a strikingly different effect on FOXO3A, another important target of SGK1 that is implicated in triggering apoptosis. Like Nedd4-2 and Raf-1, FOXO3A is inhibited by SGK1. However, rather than cooperating with SGK1, GILZ1 blunts SGK1-mediated inhibition of FOXO3A (90). These divergent effects of GILZ1 are consistent with selective recruitment of SGK1 into the ERC and away from FOXO3A. Through this type of mechanism, pleiotropic signaling pathways (such as the PI3K and Raf-MEK-ERK1/2 cascades) are selectively “pressed into service” in the context of regulating a particular output (in this case, Na+ transport) without influencing others.
FIGURE 1.
Aldosterone regulation of ENaC activity: role of the ERC. In the “basal/no-hormone” state, ENaC is associated with and inhibited by an ERC, which contains Nedd4-2, as well as Raf-1, MEK, and ERK. ERK phosphorylates the channel (small yellow circles), which in turn stimulates recruitment of Nedd4-2 and hence channel ubiquitination (orange trapezoids). ENaC internalization, endocytic trafficking, and lysosome-mediated degradation are thus augmented. Aldosterone coordinately induces the expression of SGK1 and GILZ1. In turn, GILZ1 (a) recruits SGK1 to the ERC; (b) increases interaction of SGK1 with its substrates Nedd4-2 and Raf-1; (c) augments SGK1 inhibition of Nedd4-2 and Raf-1 within the ERC, which results in the recruitment of inhibitory 14-3-3 proteins; and (d) synergizes with SGK1 to selectively stimulate its ENaC-specific functions (90). The net effect is “dual disinhibition” of ENaC (by SGK1 and GILZ1), resulting in increased channel surface expression and activity. Blue arrows represent Na+ movement through the channel, whereas black arrows represent proteins recruited to the complex. The top ellipse shows ENaC in the “inhibited” state (endocytosis and degradation favored), whereas the bottom ellipse shows the activated or “disinhibited” state (favoring accumulation at the plasma membrane).
Proteases
Other factors have important roles in the activation of ENaC and have been highlighted in recent reviews (23, 51, 91, 92). For example, proteolytic cleavage has an important role in activating ENaC by cleaving at specific sites within the finger domains of the α- and γ-subunits and releasing intrinsic inhibitory tracts (Fig. 2) (23). Furin is a member of a family of proprotein convertases that reside primarily in the trans-Golgi network and process proteins transiting through the biosynthetic pathway, including ENaC. Furin cleaves the α-subunit twice, releasing a 26-residue inhibitory tract. In contrast, furin cleaves the γ-subunit at a single site. Subsequent cleavage of the γ-subunit at sites distal to the furin cleavage site also releases an inhibitory tract. The sequential release of inhibitory tracts from the α- and γ-subunits results in channels transitioning from a very low open probability to an increasingly high open probability.
FIGURE 2.
Activation of ENaC by proteases: the second-hit hypothesis. A, the five domains within the extracellular region of an ENaC subunit are illustrated. B, ENaC α-, β-, and γ-subunits are shown within a trimeric complex. Both the N and C termini are in the cytoplasm. Inhibitory tracts are present within the α- and γ-subunits and are released by proteases. The α-subunit is cleaved twice in its finger domain by the protease furin in the biosynthetic pathway, releasing a 26-residue fragment containing a key 8-mer inhibitory tract (shown in purple) (99, 100) and partially activating ENaC. Furin cleaves the γ-subunit at one site in the finger domain, leaving the inhibitory tract in place. A second cleavage of the γ-subunit in a post-biosynthetic compartment releases a 43-residue or larger peptide containing a minimal inhibitory tract (shown in red) and fully activates ENaC.
One of the proteases that cleaves the γ-subunit and activates ENaC is the serine protease plasmin (93, 94). This protease is not present in the nephron lumen under normal conditions. However, recent studies suggest that in the setting of glomerular diseases associated with proteinuria, plasminogen is filtered by the glomerulus and is converted to plasmin by urokinase that is present within the tubular lumen (93, 94). Activation of ENaC by plasmin in proteinuric states may contribute to the Na+ retention, extracellular volume expansion, and increased blood pressure that are observed in nephrotic syndrome. Activation of ENaC by proteases also likely contributes to the increase in channel activity in response to aldosterone, as well as in individuals with Liddle syndrome (22, 23, 53, 95).
Summary
It is important to understand how these multiple modes of regulation of expression, protein-protein interactions, and processing of ENaC are integrated to result in net membrane incorporation and activity of the channel. Essential to understanding the mechanistic basis of ENaC regulation by the ERC is knowledge of where the various interactions occur. Where do Raf-1 and Nedd4-2, on the one hand, and GILZ1 and SGK1, on the other, come into association with the channel? In which compartment(s) do they alter channel trafficking and potentially processing. Along these lines, it is of particular interest that although unprocessed α- and γ-subunits predominate in whole cell lysates, it is the processed forms that associate with GILZ1 (90). This is consistent with the idea that GILZ1 joins the ERC after ENaC has been processed within the biosynthetic pathway, e.g. in the trans-Golgi network (where furin-mediated processing occurs) (96), at the plasma membrane (where prostasin-dependent processing occurs) (16), or in early (97) or recycling (12) endosomes. It is also possible, but seems less likely, that GILZ1 has higher affinity for the processed forms. The molecular identities of players that facilitate this process and the specific subcellular compartments wherein these interactions occur are key areas for future research in ENaC regulation. It is interesting to speculate that other aldosterone-induced proteins that function as scaffolds, such as CNKSR3 (a member of the “connector enhancer of kinase suppressor of Ras” family of scaffolding proteins), might play a role in ERC formation.
Further studies are needed in the context of whole animals and eventually in humans to determine the relevance of polymorphisms and mutations within ENaC subunits and regulatory pathways in the population of individuals with essential hypertension. ENaC may have a central role in Na+ retention and volume expansion in individuals with nephrotic syndrome. It will be important to identify individuals at risk for developing ENaC-dependent hypertension, as well as individuals with ENaC-dependent hypertension who would likely benefit from therapy with K+-sparing diuretics.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK078679 (to R. S.), DK056695 and DK085101 (to D. P.), DK065161 (to T. R. K. and R. P. H.), and DK054354 (to T. R. K.). This is the third article in the “Biochemistry in Medicine: Hypertension Minireview Series.” This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- ENaC
- epithelial Na+ channel
- NCC
- Na+/Cl−-coupled cotransporter
- ASIC
- acid-sensing ion channel
- ERC
- ENaC regulatory complex.
REFERENCES
- 1.Weinberger M. H. (1996) Hypertension 27, 481–490 [DOI] [PubMed] [Google Scholar]
- 2.Meneton P., Jeunemaitre X., de Wardener H. E., MacGregor G. A. (2005) Physiol. Rev. 85, 679–715 [DOI] [PubMed] [Google Scholar]
- 3.Stokes J. B. (1999) Kidney Int. 56, 2318–2333 [DOI] [PubMed] [Google Scholar]
- 4.Bhalla V., Soundararajan R., Pao A. C., Li H., Pearce D. (2006) Am. J. Physiol. Renal Physiol. 291, F714–F721 [DOI] [PubMed] [Google Scholar]
- 5.Lifton R. P., Gharavi A. G., Geller D. S. (2001) Cell 104, 545–556 [DOI] [PubMed] [Google Scholar]
- 6.Lifton R. P., Wilson F. H., Choate K. A., Geller D. S. (2002) Cold Spring Harbor Symp. Quant. Biol. 67, 445–450 [DOI] [PubMed] [Google Scholar]
- 7.Hansson J. H., Nelson-Williams C., Suzuki H., Schild L., Shimkets R., Lu Y., Canessa C., Iwasaki T., Rossier B., Lifton R. P. (1995) Nat. Genet. 11, 76–82 [DOI] [PubMed] [Google Scholar]
- 8.Shimkets R. A., Warnock D. G., Bositis C. M., Nelson-Williams C., Hansson J. H., Schambelan M., Gill J. R., Jr., Ulick S., Milora R. V., Findling J. W., Canessa C. M., Rossier B. C., Lifton R. P. (1994) Cell 79, 407–414 [DOI] [PubMed] [Google Scholar]
- 9.Tamura H., Schild L., Enomoto N., Matsui N., Marumo F., Rossier B. C. (1996) J. Clin. Invest. 97, 1780–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. (1996) EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou R., Patel S. V., Snyder P. M. (2007) J. Biol. Chem. 282, 20207–20212 [DOI] [PubMed] [Google Scholar]
- 12.Lu C., Pribanic S., Debonneville A., Jiang C., Rotin D. (2007) Traffic 8, 1246–1264 [DOI] [PubMed] [Google Scholar]
- 13.Malik B., Yue Q., Yue G., Chen X. J., Price S. R., Mitch W. E., Eaton D. C. (2005) Am. J. Physiol. Renal Physiol. 289, F107–F116 [DOI] [PubMed] [Google Scholar]
- 14.Butterworth M. B., Edinger R. S., Ovaa H., Burg D., Johnson J. P., Frizzell R. A. (2007) J. Biol. Chem. 282, 37885–37893 [DOI] [PubMed] [Google Scholar]
- 15.Kellenberger S., Hoffmann-Pochon N., Gautschi I., Schneeberger E., Schild L. (1999) J. Gen Physiol. 114, 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossier B. C. (2004) Proc. Am. Thorac. Soc. 1, 4–9 [DOI] [PubMed] [Google Scholar]
- 17.Canessa C. M., Horisberger J. D., Schild L., Rossier B. C. (1995) Kidney Int. 48, 950–955 [DOI] [PubMed] [Google Scholar]
- 18.Canessa C. M., Merillat A. M., Rossier B. C. (1994) Am. J. Physiol. Cell Physiol. 267, C1682–C1690 [DOI] [PubMed] [Google Scholar]
- 19.Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., Kleyman T. R. (2003) J. Biol. Chem. 278, 37073–37082 [DOI] [PubMed] [Google Scholar]
- 20.Hughey R. P., Bruns J. B., Kinlough C. L., Kleyman T. R. (2004) J. Biol. Chem. 279, 48491–48494 [DOI] [PubMed] [Google Scholar]
- 21.Shi H., Asher C., Chigaev A., Yung Y., Reuveny E., Seger R., Garty H. (2002) J. Biol. Chem. 277, 13539–13547 [DOI] [PubMed] [Google Scholar]
- 22.Ergonul Z., Frindt G., Palmer L. G. (2006) Am. J. Physiol. Renal Physiol. 291, F683–F693 [DOI] [PubMed] [Google Scholar]
- 23.Kleyman T. R., Carattino M. D., Hughey R. P. (2009) J. Biol. Chem. 284, 20447–20451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 25.Samaha F. F., Rubenstein R. C., Yan W., Ramkumar M., Levy D. I., Ahn Y. J., Sheng S., Kleyman T. R. (2004) J. Biol. Chem. 279, 23900–23907 [DOI] [PubMed] [Google Scholar]
- 26.Tong Q., Menon A. G., Stockand J. D. (2006) Am. J. Physiol. Renal Physiol. 290, F821–F827 [DOI] [PubMed] [Google Scholar]
- 27.Su Y. R., Rutkowski M. P., Klanke C. A., Wu X., Cui Y., Pun R. Y., Carter V., Reif M., Menon A. G. (1996) J. Am. Soc. Nephrol. 7, 2543–2549 [DOI] [PubMed] [Google Scholar]
- 28.Cui Y., Su Y. R., Rutkowski M., Reif M., Menon A. G., Pun R. Y. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9962–9966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persu A., Barbry P., Bassilana F., Houot A. M., Mengual R., Lazdunski M., Corvol P., Jeunemaitre X. (1998) Hypertension 32, 129–137 [DOI] [PubMed] [Google Scholar]
- 30.Sheng S., Li J., McNulty K. A., Avery D., Kleyman T. R. (2000) J. Biol. Chem. 275, 8572–8581 [DOI] [PubMed] [Google Scholar]
- 31.Sheng S., Li J., McNulty K. A., Kieber-Emmons T., Kleyman T. R. (2001) J. Biol. Chem. 276, 1326–1334 [DOI] [PubMed] [Google Scholar]
- 32.Snyder P. M., Bucher D. B., Olson D. R. (2000) J. Gen. Physiol. 116, 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carattino M. D., Sheng S., Kleyman T. R. (2005) J. Biol. Chem. 280, 4393–4401 [DOI] [PubMed] [Google Scholar]
- 34.Fuchs W., Larsen E. H., Lindemann B. (1977) J. Physiol. 267, 137–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng S., Bruns J. B., Kleyman T. R. (2004) J. Biol. Chem. 279, 9743–9749 [DOI] [PubMed] [Google Scholar]
- 36.Bize V., Horisberger J. D. (2007) Am. J. Physiol. Renal Physiol. 293, F1137–F1146 [DOI] [PubMed] [Google Scholar]
- 37.Sheng S., Carattino M. D., Bruns J. B., Hughey R. P., Kleyman T. R. (2006) Am. J. Physiol. Renal Physiol. 290, F1488–F1496 [DOI] [PubMed] [Google Scholar]
- 38.Maarouf A. B., Sheng N., Chen J., Winarski K. L., Okumura S., Carattino M. D., Boyd C. R., Kleyman T. R., Sheng S. (2009) J. Biol. Chem. 284, 7756–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rauh R., Diakov A., Tzschoppe A., Korbmacher J., Azad A. K., Cuppens H., Cassiman J. J., Dötsch J., Sticht H., Korbmacher C. (2010) J. Physiol. 588, 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collier D. M., Snyder P. M. (2009) J. Biol. Chem. 284, 29320–29325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swift P. A., Macgregor G. A. (2004) Adv. Renal Replace. Ther. 11, 76–86 [DOI] [PubMed] [Google Scholar]
- 42.Baker E. H., Dong Y. B., Sagnella G. A., Rothwell M., Onipinla A. K., Markandu N. D., Cappuccio F. P., Cook D. G., Persu A., Corvol P., Jeunemaitre X., Carter N. D., MacGregor G. A. (1998) Lancet 351, 1388–1392 [DOI] [PubMed] [Google Scholar]
- 43.Nkeh B., Samani N. J., Badenhorst D., Libhaber E., Sareli P., Norton G. R., Woodiwiss A. J. (2003) Am. J. Hypertens. 16, 847–852 [DOI] [PubMed] [Google Scholar]
- 44.Baker E. H., Duggal A., Dong Y., Ireson N. J., Wood M., Markandu N. D., MacGregor G. A. (2002) Hypertension 40, 13–17 [DOI] [PubMed] [Google Scholar]
- 45.Ambrosius W. T., Bloem L. J., Zhou L., Rebhun J. F., Snyder P. M., Wagner M. A., Guo C., Pratt J. H. (1999) Hypertension 34, 631–637 [DOI] [PubMed] [Google Scholar]
- 46.Ambrosius W. T., Bloem L. J., Zhou L., Rebhun J. F., Snyder P. M., Wagner M. A., Guo C., Pratt J. H. (2003) Hypertension 41, 631–637 [DOI] [PubMed] [Google Scholar]
- 47.Wang X. F., Lu X. M., Lin R. Y., Wang S. Z., Zhang L. P., Qian J., Lu D. R., Wen H., Jin L. (2008) Kidney Blood Press. Res. 31, 268–273 [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama T., Kato N., Ishinaga Y., Yamori Y., Yazaki Y. (2001) Hypertens. Res. 24, 515–521 [DOI] [PubMed] [Google Scholar]
- 49.Azad A. K., Rauh R., Vermeulen F., Jaspers M., Korbmacher J., Boissier B., Bassinet L., Fichou Y., des Georges M., Stanke F., De Boeck K., Dupont L., Balascáková M., Hjelte L., Lebecque P., Radojkovic D., Castellani C., Schwartz M., Stuhrmann M., Schwarz M., Skalicka V., de Monestrol I., Girodon E., Férec C., Claustres M., Tümmler B., Cassiman J. J., Korbmacher C., Cuppens H. (2009) Hum. Mutat. 30, 1093–1103 [DOI] [PubMed] [Google Scholar]
- 50.Mutesa L., Azad A. K., Verhaeghe C., Segers K., Vanbellinghen J. F., Ngendahayo L., Rusingiza E. K., Mutwa P. R., Rulisa S., Koulischer L., Cassiman J. J., Cuppens H., Bours V. (2009) Chest 135, 1233–1242 [DOI] [PubMed] [Google Scholar]
- 51.Bhalla V., Hallows K. R. (2008) J. Am. Soc. Nephrol. 19, 1845–1854 [DOI] [PubMed] [Google Scholar]
- 52.Sheng S., Johnson J. P., Kleyman T. R. (2008) in The Kidney, Physiology and Pathophysiology (Alpern R. J., Hebert S. C. eds) 4th Ed., pp. 743–768, Elsevier Inc., Philadelphia [Google Scholar]
- 53.Knight K. K., Olson D. R., Zhou R., Snyder P. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2805–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearce D., Kleyman T. R. (2007) J. Clin. Invest. 117, 592–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King-Jones K., Thummel C. S. (2005) Nat. Rev. Genet. 6, 311–323 [DOI] [PubMed] [Google Scholar]
- 56.Verrey F. (1999) Am. J. Physiol. Renal Physiol. 277, F319–F327 [DOI] [PubMed] [Google Scholar]
- 57.Grossmann C., Freudinger R., Mildenberger S., Krug A. W., Gekle M. (2004) Am. J. Physiol. Renal Physiol. 286, F1226–F1231 [DOI] [PubMed] [Google Scholar]
- 58.Lu M., Wang J., Jones K. T., Ives H. E., Feldman M. E., Yao L. J., Shokat K. M., Ashrafi K., Pearce D. (2010) J. Am. Soc. Nephrol. 21, 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Münster C., Chraïbi A., Pratt J. H., Horisberger J. D., Pearce D., Loffing J., Staub O. (2001) EMBO J. 20, 7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snyder P. M., Olson D. R., Thomas B. C. (2002) J. Biol. Chem. 277, 5–8 [DOI] [PubMed] [Google Scholar]
- 61.Alvarez de la Rosa D., Canessa C. M. (2003) Am. J. Physiol. Cell Physiol. 284, C404–C414 [DOI] [PubMed] [Google Scholar]
- 62.Alvarez de la Rosa D., Zhang P., Náray-Fejes-Tóth A., Fejes-Tóth G., Canessa C. M. (1999) J. Biol. Chem. 274, 37834–37839 [DOI] [PubMed] [Google Scholar]
- 63.Diakov A., Korbmacher C. (2004) J. Biol. Chem. 279, 38134–38142 [DOI] [PubMed] [Google Scholar]
- 64.Ring A. M., Leng Q., Rinehart J., Wilson F. H., Kahle K. T., Hebert S. C., Lifton R. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4025–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rozansky D. J., Cornwall T., Subramanya A. R., Rogers S., Yang Y. F., David L. L., Zhu X., Yang C. L., Ellison D. H. (2009) J. Clin. Invest. 119, 2601–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vuagniaux G., Vallet V., Jaeger N. F., Hummler E., Rossier B. C. (2002) J. Gen Physiol. 120, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alvarez de la Rosa D., Gimenez I., Forbush B., Canessa C. M. (2006) Am. J. Physiol. Cell Physiol. 290, C492–C498 [DOI] [PubMed] [Google Scholar]
- 68.Zecevic M., Heitzmann D., Camargo S. M., Verrey F. (2004) Pflugers Arch. 448, 29–35 [DOI] [PubMed] [Google Scholar]
- 69.Boyd C., Náray-Fejes-Tóth A. (2005) Am. J. Physiol. Renal Physiol. 288, F505–F512 [DOI] [PubMed] [Google Scholar]
- 70.Zhang W., Xia X., Reisenauer M. R., Rieg T., Lang F., Kuhl D., Vallon V., Kone B. C. (2007) J. Clin. Invest. 117, 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wulff P., Vallon V., Huang D. Y., Völkl H., Yu F., Richter K., Jansen M., Schlünz M., Klingel K., Loffing J., Kauselmann G., Bösl M. R., Lang F., Kuhl D. (2002) J. Clin. Invest. 110, 1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fejes-Tóth G., Frindt G., Náray-Fejes-Tóth A., Palmer L. G. (2008) Am. J. Physiol. Renal Physiol. 294, F1298–F1305 [DOI] [PubMed] [Google Scholar]
- 73.Huang D. Y., Boini K. M., Friedrich B., Metzger M., Just L., Osswald H., Wulff P., Kuhl D., Vallon V., Lang F. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R935–R944 [DOI] [PubMed] [Google Scholar]
- 74.Vallon V., Huang D. Y., Grahammer F., Wyatt A. W., Osswald H., Wulff P., Kuhl D., Lang F. (2005) Am. J. Physiol. Regul. Integr. Comp. Physiol. 289, R395–R401 [DOI] [PubMed] [Google Scholar]
- 75.Berger S., Bleich M., Schmid W., Cole T. J., Peters J., Watanabe H., Kriz W., Warth R., Greger R., Schütz G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9424–9429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vallon V., Schroth J., Lang F., Kuhl D., Uchida S. (2009) Am. J. Physiol. Renal Physiol. 297, F704–F712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang C. L., Angell J., Mitchell R., Ellison D. H. (2003) J. Clin. Invest. 111, 1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kahle K. T., Ring A. M., Lifton R. P. (2008) Annu. Rev. Physiol. 70, 329–355 [DOI] [PubMed] [Google Scholar]
- 79.Huang D. Y., Wulff P., Völkl H., Loffing J., Richter K., Kuhl D., Lang F., Vallon V. (2004) J. Am. Soc. Nephrol. 15, 885–891 [DOI] [PubMed] [Google Scholar]
- 80.Lang F., Artunc F., Vallon V. (2009) Curr. Opin Nephrol. Hypertens. 18, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Busjahn A., Aydin A., Uhlmann R., Krasko C., Bähring S., Szelestei T., Feng Y., Dahm S., Sharma A. M., Luft F. C., Lang F. (2002) Hypertension 40, 256–260 [DOI] [PubMed] [Google Scholar]
- 82.Busjahn A., Luft F. C. (2003) Cell Physiol. Biochem. 13, 51–58 [DOI] [PubMed] [Google Scholar]
- 83.von Wowern F., Berglund G., Carlson J., Månsson H., Hedblad B., Melander O. (2005) Kidney Int. 68, 2164–2172 [DOI] [PubMed] [Google Scholar]
- 84.Soundararajan R., Zhang T. T., Wang J., Vandewalle A., Pearce D. (2005) J. Biol. Chem. 280, 39970–39981 [DOI] [PubMed] [Google Scholar]
- 85.Muller O. G., Parnova R. G., Centeno G., Rossier B. C., Firsov D., Horisberger J. D. (2003) J. Am. Soc. Nephrol. 14, 1107–1115 [DOI] [PubMed] [Google Scholar]
- 86.Falin R., Veizis I. E., Cotton C. U. (2005) Am. J. Physiol. Cell Physiol. 288, C1003–C1011 [DOI] [PubMed] [Google Scholar]
- 87.Nicod M., Michlig S., Flahaut M., Salinas M., Fowler Jaeger N., Horisberger J. D., Rossier B. C., Firsov D. (2002) EMBO J. 21, 5109–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ayroldi E., Zollo O., Macchiarulo A., Di Marco B., Marchetti C., Riccardi C. (2002) Mol. Cell. Biol. 22, 7929–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ayroldi E., Zollo O., Bastianelli A., Marchetti C., Agostini M., Di Virgilio R., Riccardi C. (2007) J. Clin. Invest. 117, 1605–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soundararajan R., Melters D., Shih I. C., Wang J., Pearce D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7804–7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Butterworth M. B. (2010) Biochim. Biophys. Acta, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rossier B. C., Stutts M. J. (2009) Annu. Rev. Physiol. 71, 361–379 [DOI] [PubMed] [Google Scholar]
- 93.Passero C. J., Mueller G. M., Rondon-Berrios H., Tofovic S. P., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 36586–36591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Svenningsen P., Bistrup C., Friis U. G., Bertog M., Haerteis S., Krueger B., Stubbe J., Jensen O. N., Thiesson H. C., Uhrenholt T. R., Jespersen B., Jensen B. L., Korbmacher C., Skøtt O. (2009) J. Am. Soc. Nephrol. 20, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Masilamani S., Kim G. H., Mitchell C., Wade J. B., Knepper M. A. (1999) J. Clin. Invest. 104, R19–R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 97.Wang H., Traub L. M., Weixel K. M., Hawryluk M. J., Shah N., Edinger R. S., Perry C. J., Kester L., Butterworth M. B., Peters K. W., Kleyman T. R., Frizzell R. A., Johnson J. P. (2006) J. Biol. Chem. 281, 14129–14135 [DOI] [PubMed] [Google Scholar]
- 98.Ziera T., Irlbacher H., Fromm A., Latouche C., Krug S. M., Fromm M., Jaisser F., Borden S. A. (2009) FASEB J. 23, 3936–3946 [DOI] [PubMed] [Google Scholar]
- 99.Carattino M. D., Sheng S., Bruns J. B., Pilewski J. M., Hughey R. P., Kleyman T. R. (2006) J. Biol. Chem. 281, 18901–18907 [DOI] [PubMed] [Google Scholar]
- 100.Carattino M. D., Passero C. J., Steren C. A., Maarouf A. B., Pilewski J. M., Myerburg M. M., Hughey R. P., Kleyman T. R. (2008) Am. J. Physiol. Renal Physiol. 294, F47–F52 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.