Abstract
Mycobacterium tuberculosis utilizes unique strategies to survive amid the hostile environment of infected host cells. Infection-specific expression of a unique mycobacterial cell surface antigen that could modulate key signaling cascades can act as a key survival strategy in curtailing host effector responses like oxidative stress. We demonstrate here that hypothetical PE_PGRS11 ORF encodes a functional phosphoglycerate mutase. The transcriptional analysis revealed that PE_PGRS11 is a hypoxia-responsive gene, and enforced expression of PE_PGRS11 by recombinant adenovirus or Mycobacterium smegmatis imparted resistance to alveolar epithelial cells against oxidative stress. PE_PGRS11-induced resistance to oxidative stress necessitated the modulation of genetic signatures like induced expression of Bcl2 or COX-2. This modulation of specific antiapoptotic molecular signatures involved recognition of PE_PGRS11 by TLR2 and subsequent activation of the PI3K-ERK1/2-NF-κB signaling axis. Furthermore, PE_PGRS11 markedly diminished H2O2-induced p38 MAPK activation. Interestingly, PE_PGRS11 protein was exposed at the mycobacterial cell surface and was involved in survival of mycobacteria under oxidative stress. Furthermore, PE_PGRS11 displayed differential B cell responses during tuberculosis infection. Taken together, our investigation identified PE_PGRS11 as an in vivo expressed immunodominant antigen that plays a crucial role in modulating cellular life span restrictions imposed during oxidative stress by triggering TLR2-dependent expression of COX-2 and Bcl2. These observations clearly provide a mechanistic basis for the rescue of pathogenic Mycobacterium-infected lung epithelial cells from oxidative stress.
Keywords: Cyclooxygenase (COX) Pathway, Multifunctional Protein, Oxidative Stress, p38 MAPK, PI 3-Kinase, Toll-like Receptors (TLR)
Introduction
Mycobacterium tuberculosis exhibits diverse clever strategies to survive inside the hostile environment of host cells (1). The variable efficacy of Mycobacterium bovis bacillus Calmette-Guérin vaccine, emergence of multidrug-resistant and extensively drug-resistant strains, and coinfection of HIV and mycobacteria in patients have culminated in the immediate need to identify unique targets as well as develop new therapeutic intervention strategies for tuberculosis disease (2, 3). In this perspective, functional characterization of enzymes or antigenic proteins that possess enzymatic domains catalyzing important metabolic functions assumes critical importance. In this regard, current study attempts to understand molecular details on how cell wall-associated proline-glutamic acid (PE)3 family members of M. tuberculosis could modulate host cellular pathways and their functions, which could impart survival amid hostile host effector functions such as oxidative stress. PE antigens along with proline-proline-glutamic acid (PPE) represent 10% of the coding capacity of the M. tuberculosis genome and are characterized by a conserved PE or PPE domain near the N terminus with substantial variation in the C terminus of the antigens (4, 5). Besides being uniquely restricted to mycobacteria, PE family proteins are suggested to have critical roles in the pathogenesis of tuberculosis and in modulation of host innate and adaptive immunity (6–9). Expression profiling studies have demonstrated infection-specific expression of several PE genes in host cells, and polymorphism in the C-terminal PGRS region has been implicated in antigenic variation with subsequent roles in evasion from recognition by host immunity (10, 11). A significant number of PE_PGRS antigens associate with the cell wall and are exposed on the surface of the bacterium; thus, they are effectively trafficked out from the phagolysosomal platform into intracellular compartments as well as to the extracellular milieu (10, 12, 13). Nevertheless, precise pathophysiological attributes of PE_PGRS antigens remain the focus of extensive research. In the current study, we demonstrate that PE_PGRS11 (Rv0754), a prototype hypothetical PE_PGRS antigen, is a hypoxia-responsive gene and encodes a functional phosphoglycerate mutase. Enforced expression of PE_PGRS11 by a replication-deficient recombinant adenovirus or recombinant Mycobacterium smegmatis imparted resistance to alveolar epithelial cells against H2O2-induced oxidative stress. PE_PGRS11-induced resistance to oxidative stress involved extensive participation and signaling cross-talk among members of the phosphoinositide 3-kinase (PI3K)-ERK1/2-NF-κB signaling axis. The PE_PGRS11-induced signaling required Toll-like receptor 2 (TLR2), which culminates in the expression of cyclooxygenase-2 (COX-2) and Bcl2. Importantly, in addition to its in vivo expression during infection, mycobacterial cell surface association of PE_PGRS11 played a novel role in survival of mycobacteria under oxidative stress. These results implicate PE_PGRS11 as an immunodominant antigen that plays a crucial role in modulating alveolar epithelial cell fate decisions under oxidative stress.
EXPERIMENTAL PROCEDURES
Cell Line and Bacterial Culture
The human type II alveolar epithelial cell line A549 (obtained from the National Centre for Cell Sciences, Pune, India) was cultivated in DMEM supplemented with 10% heat-inactivated FBS (Sigma-Aldrich). M. bovis BCG 1173P2 was grown to mid-log phase in Middlebrook 7H9 plus albumin-, dextrose-, and catalase-supplemented medium. Batch cultures were aliquoted and stored at −70 °C. Representative vials were thawed and enumerated for viable colony-forming units (cfu) on Middlebrook 7H10 agar (Difco) supplemented with oleic acid, albumin, dextrose, and catalase. Single cell suspensions of mycobacteria were obtained by short pulses of sonication (Sonics). M. smegmatis mc2155 was cultured in Middlebrook 7H10 agar (Difco) supplemented with oleic acid, albumin, dextrose, and catalase.
Reagents and Antibodies
General laboratory chemicals were purchased from Sigma-Aldrich or Merck. The anti-Ser65 phospho-4EBP1, anti-4EBP1, anti-Thr180/Tyr182 phospho-p38 MAPK, anti-p38 MAPK, anti-Thr202/Tyr204 phospho-ERK1/2, anti-ERK1/2, anti-phospho-Tyr458/Tyr199 p85, anti-p85, and anti-NF-κB p65 were purchased from Cell Signaling Technology. Anti-Bcl2 was obtained from Santa Cruz Biotechnology. Anti-β-actin antibody (AC-15) and anti-PGE2 were procured from Sigma-Aldrich. Anti-COX-2 and anti-proliferating cell nuclear antigen were obtained from Calbiochem.
Cloning, Mutagenesis, Expression, and Purification of PE_PGRS11
The PE_PGRS11 gene was PCR-amplified from M. tuberculosis H37Rv genomic DNA using the following gene-specific primers: 5′-CGGGATCCATGTCATTTGTGATCGTGGCG-3′ (forward) and 5′-CCCAAGCTTTCATGGGATCAGGCTGGGCAG-3′ (reverse). The amplified PCR product was ligated into the pGEMT-Easy vector (Promega), and the recombinant clones carrying the PE_PGRS11 gene insert were confirmed by DNA sequencing. The PE_PGRS11 gene insert from pGEMT-Easy vector was subcloned into pRSETA vector (Invitrogen) to generate pRSETA-PE_PGRS11 for protein expression and purification. The phosphoglycerate mutase domain of PE_PGRS11 was PCR-amplified from pRSETA-PE_PGRS11 plasmid using domain-specific primers, 5′-CGCGGATCCATGCAGATCGTCATC-3′ (forward) and 5′-CCCAAGCTTTCACGGATGGGGGTCGAC-3′ (reverse), followed by ligation into pGEMT-Easy vector and further subcloning into pRSETA vector. Putative active site mutants of PE_PGRS11 protein were generated by site-directed mutagenesis using the megaprimer inverse PCR method. The forward primer comprised the desired mutation, and respective reverse primers were used to generate the megaprimer. The megaprimer was used in turn to amplify the entire plasmid. The sequences of the primers (forward and reverse) used to incorporate mutations spanning the residues R289A, H290A, G291A, and S348A in the putative ligand binding motif of PE_PGRS11 are illustrated in supplemental Fig. S1. All the mutations were confirmed by DNA sequence analysis. Furthermore, Escherichia coli BL21 cells carrying the appropriate recombinant plasmid were induced with isopropyl β-d-thiogalactopyranoside, and His-tagged recombinant proteins were purified with a nickel-nitrilotriacetic acid column (Qiagen).
Generation of Polyclonal Antibodies to PE_PGRS11
The polyclonal antibodies against PE_PGRS11 were generated in rabbits by subcutaneous injection of 1 mg of purified protein emulsified with an equal volume of Freund's adjuvant (Sigma-Aldrich). The first immunization was followed by two booster immunizations with Freund's incomplete adjuvant at 21-day intervals. The antibody titers in the serum were determined by ELISA 2 weeks after final immunization.
Sequence Comparisons and Model Building
Sequence similarity searches were carried out using BLAST, PSI-BLAST, and FASTA against the NCBI non-redundant database and Protein Data Bank. The PE_PGRS11 sequence was scanned against Conserved Domains Database to identify any known domains. The structure of the domain thus identified was built by homology modeling using Modeler. To understand the importance of different regions in the template structure, structural homologues of the chosen template (Protein Data Bank code 1C81) were identified through Dali searches. Structurally conserved regions among the homologues were identified and used as restraints on the target, as implemented in Modeler, by expressing them as probability density functions. The initial models that satisfied these along with the stereochemical restraints were optimized by a combination of conjugate gradients and molecular dynamics with simulated annealing. The stereochemical quality of the model was validated using PROCHECK and WHATIF.
Ligand Docking
Conserved active site residues in the target sequence were identified by aligning it with homologous sequences with known structures using the sequence annotated by structure tool. Probable active site pockets were identified independently by various geometric methods using the modeled structure with the help of CASTp, Molecular Operating Environment, and the Site Finder module in Insight II. The chosen ligand 3-phosphoglyceric acid was docked into the target structure using AutoDock to test the possibility of binding and the precise location of binding sites. The possible binding conformations and orientations were also analyzed by clustering methods embedded in AutoDock. To address the issue of protein flexibility and any conformational fine tuning of the active site residues, upon ligand binding, a postdocking energy minimization was carried out using the Discover suite by allowing full freedom to all the atoms of the protein and ligand. The protein-drug interactions such as hydrogen bonding, hydrophobic interactions, and aromatic stacking were evaluated using Ligand-Protein Contacts software. As a control exercise, the same ligand was stripped out of the original Protein Data Bank file and docked back into the template structure using an identical protocol and parameter set. The Accelrys software package (Insight II modules, Accelrys Inc.) was used to visualize, analyze, and manipulate the structures.
Estimation of Enzymatic Activity of Phosphoglycerate Mutase Domain of PE_PGRS11
Phosphoglycerate mutase activity of PE_PGRS11 was assayed by coupling the reaction to enolase as described previously (14, 15). Briefly, the conversion of 3-phosphoglyceric acid to 2-phosphoglyceric acid was determined by estimating NADH oxidation at 340 nm in an assay mixture containing 79 mm triethanolamine, pH 7.6, 6.6 mm d-(−)-3-phosphoglyceric acid, 0.70 mm adenosine 5′-diphosphate, 1.3 mm 2,3-diphospho-d-glyceric acid, 0.15 mm β-NADH, 2.5 mm MgSO4, 99 mm KCl, 14 units of pyruvate kinase, 20 units of l-lactic acid dehydrogenase, 3 units of enolase, and 0.03–0.06 unit of PE_PGRS11 purified protein.
Establishment of M. tuberculosis H37Ra Dormant Cultures Using Wayne's Model for Dormancy
M. tuberculosis H37Ra dormant cultures were established as described previously (16). In brief, 40 ml of albumin, dextrose, and catalase supplement was added to 360 ml of sterile Dubos broth base containing 500 μg/ml methylene blue as an oxygen depletion indicator (Difco). The medium was inoculated with a preinoculum having an A600 of 0.6, and the culture was incubated at 37 °C with agitation at 130 rpm. On the 12th day, the culture was harvested after reaching the stage of non-replicating persistence, washed with Tween-saline (0.8% (w/v) NaCl and 0.05% (v/v) Tween 80) and processed for RNA isolation.
Establishment of Nutrient-depleted Stationary Phase Growth Condition for M. tuberculosis H37Ra
For generating the nutrient-starved stationary phase culture, M. tuberculosis H37Ra cells were grown in a rotatory shaker at 37 °C until A600 reached 2.5 (∼15 days) followed by standing, non-shaking condition at 37 °C for another 30 days for gradual depletion of nutrients and microaerophilic submerged growth. After 30 days, bacteria were harvested, washed with Tween-saline, and processed for the isolation of total RNA.
Isolation of RNA from M. tuberculosis H37Ra and Quantitative Real Time PCR for PE_PGRS11
Total RNA was extracted from M. tuberculosis H37Ra cells using a well established hot phenol method (17) with slight modifications. In brief, harvested cells were lysed in 30 mm Tris-HCl, pH 7.4 containing 100 mm NaCl, 5 mm EDTA, 1% SDS, and 5 mm vanadyl ribonucleoside complex by sonication. The sonication conditions included four pulses of 10-s duration at 50% duty cycle. Lysates were incubated on ice for 10 min and then extracted with 65 °C preheated phenol (saturated with 100 mm sodium acetate buffer, pH 5.2, containing 10 mm EDTA), phenol chloroform, and chloroform, twice each. Total RNA was precipitated using 0.3 m sodium acetate and an equal volume of isopropanol and was finally dissolved in RNase-free water. 1 μg of total RNA was reverse transcribed to cDNA using a first strand cDNA synthesis kit (MBI Fermentas) according to the manufacturer's instructions. A real time PCR amplification (Applied Biosystems) using SYBR Green PCR mixture (Finnzymes) was performed for quantification of PE_PGRS11, narG, and HspX. Amplification of 16 S rRNA was used as an internal control. The forward and reverse primer pairs used were as follows: PE_PGRS11 forward, 5′-ATCGTCATCGACTTCGTGCG-3′; PE_PGRS11 reverse, 5′-GGTCTGCTGCGTTCTGATCAAC-3′; HspX forward, 5′-ATGGCCACCACCCTTCCCGTTC-3′; HspX reverse, 5′-GACCATCGCGGACCATAATGTCGAC-3′; narG forward, 5′-ACTACGCCGACAACACCAAGTTCGCCGACG-3′; narG reverse, 5′-AGCGGCGCACATAGTCGACAAAGAACGGAA-3′; 16 S rRNA forward, 5′-GCACCGGCCAACTACGTG-3′, and 16 S rRNA reverse, 5′-GAACAACGCGACAAACCACC-3′.
Construction of Recombinant Adenovirus Expressing PE_PGRS11
Replication-deficient recombinant adenovirus expressing PE_PGRS11 was generated according to the protocol as described (18). Briefly, PE_PGRS11 was subcloned into pAdTrack-CMV vector after releasing the gene insert from pGEMT Easy-PE_PGRS11. PmeI-linearized pAdTrack-CMV-PE_PGRS11 was co-electroporated with pAdEasy-1 vector, which contains the adenoviral genome backbone except E1 and E3 proteins, into E. coli BJ5183 strain. The clones were screened for recombinants by confirmative restriction digestions. Recombinant adenoviral DNA was digested with PacI and transfected into HEK-293 cells. HEK-293 cells stably express E1and E3 proteins and help in generation and replication of recombinant adenovirus. The appearance of cells expressing green fluorescent protein (GFP) and clearing zones indicates the generation of recombinant adenovirus. Expression of PE_PGRS11 was established by immunoblotting as well as immunocytochemistry. After 7 days of transfection, cell lysate was prepared by repeated freezing and thawing followed by fresh infection of HEK-293 cells with the lysate to get high titer virus. The resultant viruses were purified by CsCl banding as described previously (19). Briefly, cell-free viral supernatants were prepared by centrifugation at 3000 × g for 10 min. The concentrated viral supernatant was subjected to CsCl ultracentrifugation to separate the viruses from the cellular proteins and medium components. Following ultracentrifugation, CsCl was removed by dialysis, and viral titers were calculated. For infection of A549 cells, 2 × 103 cells/well were seeded in 96-well culture plates followed by infection with recombinant adenovirus at a multiplicity of infection of 10 at 37 °C in normal growth medium.
Generation of Recombinant M. smegmatis Expressing PE_PGRS11
For generation of recombinant M. smegmatis overexpressing PE_PGRS11, PE_PGRS11 was subcloned into pJAM2 vector, which is an M. smegmatis and E. coli shuttle vector with an acetamide-inducible expression system. The PE_PGRS11 gene insert was released from pGEMT Easy-PE_PGRS11 with BamHI and XbaI restriction endonucleases and mobilized into pJAM2. M. smegmatis underwent electroporation with pJAM2-PE_PGRS11 to generate recombinant M. smegmatis followed by induction with acetamide to induce the expression of PE_PGRS11, which was further confirmed by immunoblotting. Similarly, M. smegmatis was transformed with pJAM2 vector to generate control transformants.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
Assessment of cellular proliferation by MTT assay was carried out as described previously (20). Briefly, 1.5 × 103 A549 cells/well were seeded in a 96-well plate. After 12 h of plating, the cells were treated as indicated followed by addition of MTT (20 μl of 5 mg/ml) 3 h prior to completion of the experiment. MTT is a tetrazolium salt that is converted by living cells into blue formazan crystals. The cell-free supernatant was removed from the wells 3 h after MTT addition, 200 μl of DMSO was added to dissolve the formazan crystal, and the absorbance was measured at 550 nm in an ELISA reader (Molecular Devices).
Treatment of A549 Cells with Pharmacological Inhibitors of Signaling Pathways
All the pharmacological inhibitors were obtained from Calbiochem. They were reconstituted in sterile, cell culture grade DMSO (Sigma-Aldrich) and used at the following concentrations after determining the viability of A549 cells in titration experiments using the MTT assay: U0126, 10 μm, LY294002, 50 μm, AKT inhibitor II, 10 μm, and NS-398, 20 μm. DMSO at 0.1% concentration was used as the vehicle control.
Immunoblotting Analysis
For immunoblot analysis, A549 cells were grown in 60-mm culture dishes and treated as indicated. Cells were washed twice with phosphate-buffered saline (PBS); lysed in modified radioimmune precipitation assay buffer (Upstate Biotechnology) consisting of 50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 1 mm PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mm Na3VO4, and 1 mm NaF; and incubated at 4 °C for 30 min. Cell lysates were collected after centrifugation at 13,000 × g for 10 min. The immunoblotting for the intended protein was carried out according to the protocol described elsewhere (21). Briefly, protein lysates were subjected to SDS-PAGE and transferred onto PVDF membranes (Millipore) by a semidry Western blotting (Bio-Rad) apparatus. Nonspecific binding was blocked with 5% nonfat dry milk powder in TBST (20 mm Tris-HCl, pH 7.4, 137 mm NaCl, and 0.1% Tween 20) for 60 min. The blots were incubated overnight at 4 °C or for 2 h at room temperature with primary antibodies diluted in TBST with 5% BSA. The primary antibody dilutions were made according to the manufacturer's instructions. After washing with TBST, blots were incubated with goat anti-rabbit or anti-mouse IgG secondary antibody conjugated to HRP (Jackson ImmunoResearch Laboratories) diluted in TBST for 2 h. After further rinsing in TBST, the immunoblots were developed with the ECL system (PerkinElmer Life Sciences) following the manufacturer's instructions.
Nuclear and Cytosolic Subcellular Fractionation
A549 cells were washed with ice-cold PBS and gently resuspended in ice-cold Buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, and 0.5 mm PMSF). After incubation on ice for 15 min, cell membranes were disrupted with 10% Nonidet P-40, and the nuclear pellets were recovered by centrifugation at 13,000 × g for 15 min at 4 °C. The supernatants from this step were used as cytosolic extracts. Nuclear pellets were lysed with ice-cold Buffer C (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, and 1 mm PMSF), and nuclear extracts were collected after centrifugation at 13,000 × g for 20 min at 4 °C.
Transient Transfections and Immunofluorescence
HEK-293 cells were transiently transfected with TLR2 overexpression and NF-κB-luciferase reporter constructs using ESCORT III (Sigma-Aldrich). For immunofluorescence studies, vector or TLR2 cDNA construct-transfected HEK-293 cells were seeded in a 35-mm dish on coverslips and treated with 2 μg/ml purified PE_PGRS11 protein for 1 h. Cells were fixed with cold methanol for 15 min and stained with anti-PE_PGRS11 antibodies for 1 h followed by incubation with secondary antibody (Cy5-conjugated anti-rabbit IgG) in the dark for 1 h at room temperature. Coverslips with cells were mounted on a slide with Fluoromount-G, and immunofluorescence images were acquired by a Zeiss LSM 510 Meta confocal laser scanning microscope. The images were analyzed for the integrated density of the fluorescence and for the area of the cells using Zeiss LSM image browser software.
Luciferase Assay
HEK-293 cells transfected with TLR2 cDNA and NF-κB-luciferase reporter constructs were treated with purified PE_PGRS11 for 12 h. Cells were lysed in reporter lysis buffer (Promega), and luciferase activity was assayed using luciferase assay reagent (Promega). The results were normalized for transfection efficiencies by assay of β-galactosidase activity.
Preparation of Subcellular Fractions of Mycobacterium
The subcellular fractionation was carried out according to the protocol described previously (22, 23). Briefly, the cells were lysed by sonication in breaking buffer (PBS, pH 7.4 with 1 mm EDTA, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 200 μm PMSF, and 100 mm Tris-HCl, pH 8.0). Cell debris were removed by centrifugation at 3000 × g for 5 min, and the supernatant was subjected to ultracentrifugation at 27,000 × g for 1 h at 4 °C. The pellet from this centrifugation step was considered the cell wall fraction and was resuspended in breaking buffer. The supernatant containing the membrane and cytosol fractions was again subjected to centrifugation at 100,000 × g for 4 h at 4 °C, and this step was repeated with supernatant obtained in the earlier step. The supernatant fraction from both steps was considered as the cytosol fraction, whereas the pellets were considered as the membrane fraction. All the fractions were quantified for the amount of protein by the Bradford method and subjected to immunoblotting for analysis of PE_PGRS11 expression.
Proteinase K and Trypsin Sensitivity Assays
M. smegmatis harboring vector or pJAM2-PE_PGRS11 were induced with 2% acetamide for 6 h and harvested. Cells were washed once in TBS buffer (20 nm Tris-HCl, pH 7.5, 150 mm NaCl, and 3 mm KCl) and resuspended in 1 ml of the same buffer. Each sample was divided into two identical aliquots; one was mixed with proteinase K (MBI Fermentas) and one was mixed proteinase K (MBI Fermentas), with trypsin (Amresco) up to a concentration of 100 μg/ml. Both aliquots were incubated for the indicated time points at 37 °C. The reaction was stopped by adding 200 nm PMSF and 1 μg/ml aprotinin. Samples were centrifuged, and the pellet was washed three times in TBS and dissolved in SDS sample buffer (30% (v/v) glycerol, 10% (w/v) SDS, 312 mm Tris-HCl, pH 6.8, 0.25% (w/v) bromphenol blue, 0.25% (w/v) xylene cyanol, and 5% (v/v) β-mercaptoethanol) followed by analysis of PE_PGRS11 integrity by immunoblotting.
Patients and Control Subjects
The study population for serological characterization (n = 211) was comprised of tuberculosis (TB) bacillus-infected patients reporting to the National Jalma Institute of Leprosy and Other Mycobacterial Diseases in Agra, India. The patient population was categorized into different clinical groups as follows. Group 1 (n = 94) patients were diagnosed with pulmonary TB for the first time and had no history of chemotherapeutic treatment. Group 1 included 69 adults and 25 children. Group 2 (n = 30) patients exhibited relapsed infection, and all enlisted patients were adults. Group 3 (n = 31) patients, diagnosed with extrapulmonary TB infections, included nine adults and 22 children. The patients were categorized according to guidelines of the National TB Control Program, Central TB Division, Government of India. The diagnosis of pulmonary tuberculosis in group 1 and 2 patients was established based on clinical and radiological data together with the identification of acid-fast bacilli in sputum. In the case of group 2 patients with relapsed infection, the patients were diagnosed with pulmonary TB, but after a full course of antitubercular chemotherapeutic treatment, these patients had a recurrence of the infection and disease symptoms. Group 3 patients were primarily diagnosed with abdominal TB infection or tuberculosis meningitis (TBM). The diagnosis of extrapulmonary TB was confirmed by histological examination as well as with culture positivity of the bacillus in specimens obtained from extrapulmonary sites. The age ranges of adults and children were 18–60 and 2–15 years, respectively. The healthy donors (n = 56) were negative for active tuberculosis disease. Additionally, healthy donors included in the study were age- and gender-matched to the different clinical groups. The samples from HIV-positive subjects were excluded from the study.
Cerebrospinal fluid (CSF) samples (n = 73) were collected from patients reporting to the National Institute of Mental Health and Neuro Sciences, Bangalore, India. The diagnosis of TBM was established by detection of acid-fast bacilli in CSF of patients. Other disease control samples (n = 18) included other meningitis diseases like pyogenic meningitis, viral meningitis, etc. Healthy controls (n = 12) included cases of death by accident or meningitis-suspected cases but proven to be clinically healthy. The samples from HIV-positive subjects were not included in the study. The samples were collected without gender or age bias. All the included subjects for both serum and CSF characterization had given written consent, and the current study was carried out after approval from the institutional bioethics committee.
Serological Characterization of PE_PGRS11
ELISAs were carried out in 96-well microtiter plates (Nunc). ELISA plates were incubated with purified recombinant PE_PGRS11 protein overnight at 4 °C followed by three washes with PBS-Tween 20 (0.05%) buffer. After blocking with 3% BSA in PBS for 1 h at 37 °C, wells were incubated with human serum (1:400 dilution in blocking buffer) or CSF samples (1:10) for 1 h at 37 °C followed by washing with PBS-Tween 20 buffer. The plates were further incubated with HRP-labeled anti-human IgG or IgM secondary antibody for 1 h at 37 °C followed by development with o-phenylenediamine tetrahydrochloride. The absorbance values were measured at 492 nm using an ELISA reader (Molecular Devices).
Zone of Inhibition Assay
The zone of inhibition or disc diffusion assay was performed as described previously (24). In brief, culture plates were inoculated with M. smegmatis transformed with pJAM2 and pJAM2-PE_PGRS11 by wiping with sterile cotton swabs dipped in the respective cultures. Then sterile filter disks dipped in H2O2 (30%) were placed on culture plates followed by incubation of the plates at 37 °C for 24 h. After 24 h, the diameter of the zone of inhibition around each disk was measured.
Statistical Analysis
Levels of significance for comparison between samples were determined by Student's t test distribution. The data in the graphs are expressed as the mean ± S.D. or S.E. GraphPad Prism 3.0 software (GraphPad software) was used for all statistical analysis. Comparisons of immunoreactivities within clinical groups were carried out using one-way analysis of variance followed by Bonferroni's multiple comparison test.
RESULTS
Hypothetical PE_PGRS ORF Encodes a Functional Phosphoglycerate Mutase
Studies have advocated that the members of PE_PGRS family proteins including hypothetical PE_PGRS11 protein could have critical roles in the pathogenesis of tuberculosis as well as in modulating host innate and adaptive immune responses to pathogenic mycobacteria (8, 9). In view of these observations, initial bioinformatics analysis of PE_PGRS11 was performed as described to identify domains of importance, and in this regard, a search against the domain database showed the presence of the phosphoglycerate mutase domain. The phosphoglycerate mutase domain of PE_PGRS11 exhibited significant sequence similarity to phosphoglycerate mutases characterized across diverse organisms with the core phosphoglycerate mutase domain containing critical catalytic residues required for enzymatic activity (Fig. 1A). Secondary structure prediction, homology domain modeling, and ligand docking studies were carried out to ascertain the phosphoglycerate mutase domain of PE_PGRS11 (Fig. 1A). Because this is the first report of a PE_PGRS antigen possessing a functional phosphoglycerate mutase domain, the current study characterized PE_PGRS11 in terms of its biochemical and functional properties in greater detail.
FIGURE 1.
In silico analyses and functional characterization of multifunctional protein PE_PGRS11. A, i, the presence of the phosphoglycerate mutase domain toward the C terminus of PE_PGRS11 protein upon the search against the domain database. Phosphoglycerate mutase matched with region 287–374 of PE_PGRS11 upon comparison with four Protein Data Bank structures, 1K6M, 1C81, 1TIP, and 1V7Q. All the active site residues of PE_PGRS11 (by active site prediction methods) were identical with these templates. ii, a three-dimensional model of the PE_PGRS11 phosphoglycerate mutase domain constructed on the basis of template 1K6M. iii, representation of the active site residues of phosphoglycerate mutase with 3-phosphoglyceric acid as a ligand upon automated docking simulations with the AutoDock 3.0 software suite using Silicon Graphics station Octane. B, a typical hyperbolic substrate saturation plot for the phosphoglycerate mutase enzyme activity of PE_PGRS11. The inset shows a Lineweaver-Burk plot to derive Km and the Vmax values for the phosphoglycerate mutase enzyme activity of PE_PGRS11. C, quantitative real time RT-PCR analysis of PE_PGRS11 or HspX transcripts assessed in the RNA isolated from M. tuberculosis grown under the indicated growth conditions. Error bars represent mean ± S.E. (n = 3). The data represent three independent experiments.
Catalytic Properties of Phosphoglycerate Mutase Domain of PE_PGRS11
The gene encoding PE_PGRS11 was PCR-amplified, cloned, and heterologously overexpressed in E. coli strain BL21(DE3) pLysS as a His-tagged protein. The recombinant PE_PGRS11 protein with mobility corresponding to 57 kDa, which is the same as the calculated theoretical molecular mass, was induced with isopropyl 1-thio-β-d-galactopyranoside. The PE_PGRS11 protein was purified to homogeneity (supplemental Fig. S2A) and was utilized to characterize the phosphoglycerate mutase enzymatic activity as well as to generate polyclonal antibodies. Upon analysis of kinetic properties, recombinant PE_PGRS11 protein exhibited Michaelis-Menten kinetics as Km and Vmax for the enzyme were calculated as 1.82 mm and 2.645 μm/min, respectively (Fig. 1B). The substrate saturation curves were hyperbolic for the phosphoglycerate mutase enzyme, and the effects of pH or metal ions on enzyme kinetics were also studied. The pH plays a critical role in determining the enzymatic activity of phosphoglycerate mutase. Phosphoglycerate mutase of PE_PGRS11 was found to be broadly active between pH 6 and 9 with a pH optimum of 7.5 (supplemental Fig. S2B). The pH optimum is similar to that of diverse eukaryotic as well as bacterial phosphoglycerate mutases (25, 26). For example, phosphoglycerate mutase from rabbit muscle demonstrated a pH optimum of 7, whereas purified enzyme from Hyphomicrobium exhibited maximum catalytic activity at pH 7.3. Similarly, phosphoglycerate mutase from Pseudomonas exhibited maximum activity over the pH range 7.0–7.6. Metal ions like Mg2+ can act as essential cofactors for most of the enzymes that catalyze phosphoglycerate mutase enzymatic activity and may have significant structural or catalytic roles (25, 27). In this context, we investigated phosphoglycerate mutase activity in the absence or presence of increasing concentrations of Mg2+, Na+, Li+, and K+, and as shown, phosphoglycerate mutase activity was severely abrogated in the absence of Mg2+. However, Na+, K+, and Li+ did not significantly modulate the enzymatic activity (supplemental Fig. S2C and data not shown). Importantly, these metal ion properties of the phosphoglycerate mutase domain of PE_PGRS11 are shared in common with phosphoglycerate mutases reported from nematodes like Caenorhabditis elegans and Brugia malayi and bacteria like Pseudomonas and Hyphomicrobium (25, 27). In addition, we cloned and expressed the recombinant phosphoglycerate mutase domain of PE_PGRS11 to homogeneity. The purified phosphoglycerate mutase domain of PE_PGRS11 also exhibited robust enzymatic properties similar to those of full-length recombinant PE_PGRS11 protein (data not shown).
Mutational Analysis of Phosphoglycerate Mutase Domain of PE_PGRS11
A sequence comparison between the phosphoglycerate mutase domain of PE_PGRS11 with four Protein Data Bank structures including that of the crystal structure of phosphoglycerate mutase from Thermus thermophilus revealed candidate residues His, Arg, and Ser for the catalytic triad of the phosphoglycerate mutase domain of PE_PGRS11 (Fig. 1A). Extensive published reports have utilized alanine as the substitution residue of choice for mutational analysis because of its small and mostly inert methyl functional group, which eliminates the side chain beyond the β carbon but does not amend the main chain conformation (28, 29). Importantly, amino acid substitution with alanine does not impose extreme electrostatic or steric effects, and alanine forms the basis for alanine scanning, a commonly used technique to analyze the contribution of specific residues to a given protein (30). In regard to our study, critical catalytic residues at the active site of the phosphoglycerate mutase domain of PE_PGRS11 are hydrophilic amino acids. As predicted by secondary structure prediction, homology domain modeling, and ligand docking studies, critical catalytic residues were mutated to alanine, thus adversely affecting the enzyme function without altering the overall conformation of the protein. In this perspective, different single mutants of the phosphoglycerate mutase domain of PE_PGRS11, R289A, H290A, and S348A; double mutant R289A,H290A; and triple mutant R289A,H290A,G291A were generated. The recombinant mutant proteins were purified to homogeneity and had the same molecular weight as the wild-type protein as evaluated by similar mobilities on SDS-PAGE or immunoblot analysis (data not shown). Furthermore, circular dichroism (CD) spectra of mutant proteins did not demonstrate any significant conformational change (data not shown). As shown in supplemental Fig. S2D, all mutants failed to demonstrate any significant phosphoglycerate mutase activity, and inclusion of a high molar excess of any of the mutant proteins failed to demonstrate any significant activity as compared with wild-type PE_PGRS11 (data not shown).
Transcriptional Analysis Reveals That PE_PGRS11 Is a Hypoxia-responsive Gene
Many reported observations suggest that several members of the PE_PGRS family of antigens respond to different environmental cues including hypoxia (31). During latency, M. tuberculosis bacilli undergo a non-replicating persistence state without compromising viability and retain their ability to resume growth upon favorable conditions (32). Similarly, M. tuberculosis, under an in vitro nutrient-depleted stationary phase growth condition, exhibits a drastic reduction of respiration rate, indicating a very low metabolic rate or even metabolic inactivity. In addition, nutrient-depleted stationary phase tubercle bacilli are reported to maintain full viability, further suggesting functional similarity of the nutrient-depleted growth environment with in vivo latent infection conditions (33, 34). In this perspective, RNA was isolated from mycobacteria grown to mid-log or stationary phase as well as grown under hypoxic conditions, and PE_PGRS11 transcript levels were assessed. For the hypoxia-related expression, the M. tuberculosis H37Ra culture was subjected to Wayne's slow oxygen depletion model, which is believed to mimic the conditions of granulomas (16, 32). The analysis of PE_PGRS11 transcript levels under different growth conditions was carried out by quantitative RT-PCR as well as semiquantitative RT-PCR. The data presented in Fig. 1C demonstrate that the expression level of PE_PGRS11 was up-regulated in M. tuberculosis grown under hypoxic conditions when compared with the RNA levels in mycobacteria grown until mid-log phase. HspX was used as a positive control; it is known to be up-regulated in mycobacteria under conditions like hypoxia as well as during infection (Fig. 1C). In addition, narG, known to have unaltered expression during hypoxia and stationary phase, was utilized as a negative control (supplemental Fig. S2E) (35). The differential expression at the transcript level in response to differential environmental cues suggests that PE_PGRS11 might be regulated differentially at the level of transcription under the above mentioned conditions. In this perspective, primer extension analysis of RNA of mycobacteria grown under mid-log phase, hypoxia, or stationary phase was carried out. We observed that the PE_PGRS11-specific primer extension product could be detected only in the RNA isolated from M. tuberculosis subjected to a hypoxic environment compared with stationary phase, implicating PE_PGRS11 as a hypoxia-responsive gene (data not shown).
Enforced Expression of PE_PGRS11 Imparts Resistance against Oxidative Stress
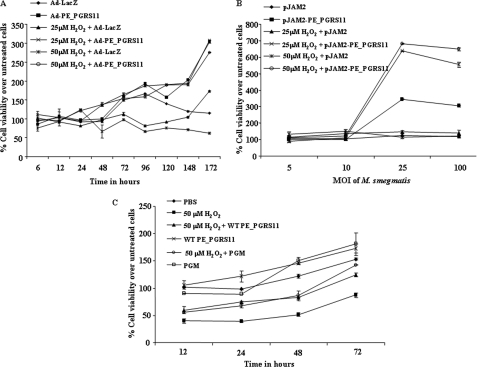
Glycolytic enzymes are reported to modulate cellular life spans by imparting resistance against oxidative stress. Significantly, although phosphoglycerate mutase overexpression resulted in an augmented rate of glycolysis and escape from senescence, its depletion was found to significantly shorten the cellular lifespan (36, 37). In this perspective, reports have suggested that the Warburg effect could contribute to evasion mechanisms of cellular life span limitations imposed by oxidative stress (38). Furthermore, deletion of phosphoglycerate mutase in Bordetella bronchiseptica culminated in enhanced susceptibility to the superoxide radical-generating chemical paraquat (39). Interestingly, the phosphoglycerate mutase mutant exhibited susceptibility to antimicrobial peptides and demonstrated decreased fitness in the mouse typhoid model (40). To study the relative role of the phosphoglycerate mutase domain of PE_PGRS11, we examined whether alveolar epithelial A549 cells, ectopically expressing PE_PGRS11 antigen, were resistant to cellular life span restrictions imposed by oxidative stress. For enforced expression, we generated recombinant replication-deficient adenovirus expressing PE_PGRS11 (Ad-PE_PGRS11) or recombinant M. smegmatis expressing PE_PGRS11 antigen. In all our experiments where recombinant adenoviruses were utilized, a control adenovirus expressing β-galactosidase (Ad-LacZ) was also used. Infection of alveolar epithelial cells with Ad-PE_PGRS11 but not with Ad-LacZ protected them against hydrogen peroxide (H2O2)-induced cell death as analyzed by MTT viability assay (Fig. 2A). The percent viability increased up to 4.5-fold in Ad-PE_PGRS11-infected alveolar epithelial cells in comparison with respective Ad-LacZ-infected cells under oxidative stress (Fig. 2A). However, enforced expression of another PE_PGRS antigen, PE_PGRS17, by Ad-PE_PGRS17 failed to rescue cells under oxidative stress, implicating a significant role for the phosphoglycerate mutase domain of PE_PGRS11 (supplemental Fig. S3A). For further validation, infection of alveolar epithelial cells with recombinant M. smegmatis expressing PE_PGRS11 demonstrated increased viability from 4-fold (25 μm H2O2-treated cells) to 5.4-fold (50 μm H2O2-treated cells) compared with cells infected with M. smegmatis harboring vector control (Fig. 2B). Importantly, the recombinant phosphoglycerate mutase domain of PE_PGRS11 also exhibited protective efficacy against H2O2-induced oxidative stress, quite comparable with full-length recombinant PE_PGRS11 protein (Fig. 2C). In addition, a triple mutant (Arg, His, and Ser) of the catalytic triad of the phosphoglycerate mutase domain failed to rescue the cells from H2O2-induced oxidative stress (supplemental Fig. S3B). These observations clearly validate a critical role for the phosphoglycerate mutase domain of PE_PGRS11 in restricting the cellular lifespan forced by oxidative stress.
FIGURE 2.
PE_PGRS11 imparts resistance to lung epithelial cells against oxidative stress. A, MTT assay to determine cell viability of A549 cells infected with Ad-PE_PGRS11 or Ad-LacZ upon oxidative stress (25 μm H2O2 and 50 μm H2O2). B, determination of cell viability of A549 cells infected with recombinant M. smegmatis pJAM2-PE_PGRS11 or M. smegmatis pJAM2 vector alone after 48 h of oxidative stress (25 μm H2O2 and 50 μm H2O2). C, percent cell viability for A549 cells under oxidative stress (50 μm H2O2) upon treatment with recombinant wild-type PE_PGRS11 (WT PE_PGRS11) or the phosphoglycerate mutase domain of PE_PGRS11 (PGM). Error bars represent mean ± S.D. The data are representative of three independent experiments. MOI, multiplicity of infection.
PE_PGRS11-induced Activation of PI3K-ERK1/2-NF-κB Signaling Axis Requires TLR2 Triggering at Cell Surface
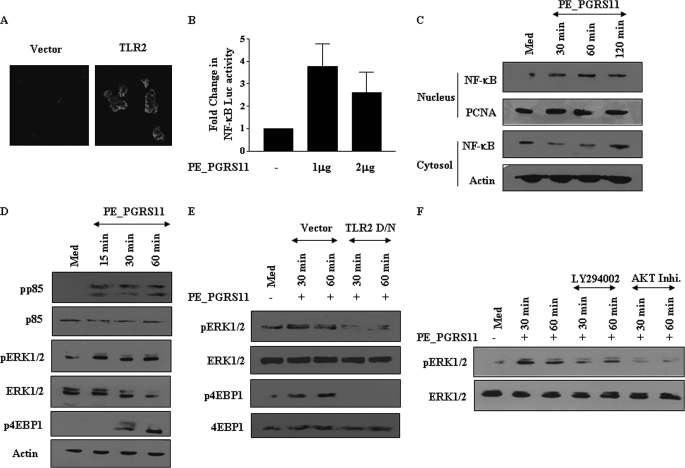
Pattern recognition receptors play a critical role in innate immunity, and TLR, a known pattern recognition receptor, is involved in recognition of pathogen-associated molecular patterns of pathogenic mycobacteria (41). Among TLRs, TLR2 triggering by mycobacteria often culminates in secretion of immunomodulatory cytokines and initiation of immune responses to Mycobacterium bacilli by activating cascades of intracellular signaling pathways (42–45). In the context of a significant role for TLRs, we investigated the ability of PE_PGRS11 to bind TLR2 expressed on the cell surface of transfected HEK-293 cells, and as shown in Fig. 3A, confocal microscopic studies suggest that PE_PGRS11 distinctively recognizes TLR2. These results are in agreement with our recent report wherein PE_PGRS11 specifically binds TLR2 and induces maturation and activation of human dendritic cells (46). TLR2 signaling in a variety of cellular systems often involves critical participation of the NF-κB signaling pathway. In this context, PE_PGRS11 binding to TLR2 triggered significant NF-κB promoter activity in TLR2-transfected HEK-293 cells compared with vector DNA-transfected cells (Fig. 3B). This finding was further evidenced by significant translocation of p65 NF-κB from the cytosol to the nucleus within 30 min of stimulation with PE_PGRS11 (Fig. 3C). Interestingly, the ability of PE_PGRS11 to trigger the NF-κB activation was not compromised in macrophages derived from TLR4-defective C3H/HeJ mice (data not shown), clearly implicating the selective role for TLR2 in PE_PGRS-mediated signaling events.
FIGURE 3.
PE_PGRS11 triggers TLR2-dependent activation of PI3K-ERK1/2-NF-κB signaling axis. A, TLR2- or vector-transfected HEK-293 cells were treated with PE_PGRS11. The cells were further stained with antibody specific to PE_PGRS11 followed by Cy5-labeled secondary antibody. The immunofluorescence staining of cells was analyzed by confocal microscopy. B, HEK-293 cells, transiently transfected with vector or TLR2 cDNA construct along with NF-κB reporter construct, were treated with different doses of PE_PGRS11 protein followed by analysis of NF-κB reporter activity (mean ± S.E., n = 3). C, A549 cells were treated with PE_PGRS11 protein for the indicated time points followed by immunoblotting of nuclear and cytosolic fractions of cells for NF-κB. D, activation of p85 (PI3K), ERK1/2, and 4EBP1 by PE_PGRS11. E, transfection of cells with TLR2 dominant-negative (D/N) cDNA construct abrogates PE_PGRS11-induced activation of ERK1/2 and 4EBP1. F, A549 cells were pretreated with LY294002 or AKT inhibitor (Inhi.) followed by analysis of PE_PGRS11-induced activation of ERK1/2. The results are representative of two independent experiments. PCNA, proliferating cell nuclear antigen; p, phospho; Med, medium.
The activation of the NF-κB pathway is often carefully titrated by activation levels of PI3K or mitogen-activated protein kinase signaling in diverse cell types including alveolar epithelial cells (47, 48). In mammalian cells, PI3K and ERK1/2 MAPK signaling pathways are widely acknowledged as key mediators of cell survival (49, 50). These pathways play decisive roles in controlling survival or apoptosis through the phosphorylation of numerous cellular proteins as well as by articulating the expression of an assortment of antiapoptotic genes including Bcl2 (51). In this context, we investigated the potential role of PE_PGRS11 antigen-TLR2-triggered activation of PI3K and ERK1/2 during PE_PGRS11-induced survival of A549 cells. Accordingly, PE_PGRS11 triggered activation of the PI3K or ERK1/2 pathway as analyzed by phosphorylation of the p85 subunit of PI3K, 4EBP1, or ERK1/2 (Fig. 3D). These activation events required the participation of TLR2 as the TLR2 dominant-negative construct significantly reduced PE_PGRS11-triggered activation of 4EBP1 and ERK1/2 (Fig. 3E). Furthermore, pharmacological intervention by specific inhibitor LY294002 (PI3K) or AKT inhibitor I (AKT) abrogated PE_PGRS11-induced activation of ERK1/2, alluding to a significant role for the TLR2-PI3K signaling axis in ERK1/2 activation (Fig. 3F). These results suggest that the NF-κB, PI3K, and ERK1/2 pathways characterize important key molecular links during PE_PGRS11-induced signaling cascades in host cells.
PE_PGRS11-induced Resistance to Oxidative Stress Necessitates Modulation of Specific Antiapoptotic Molecular Signatures
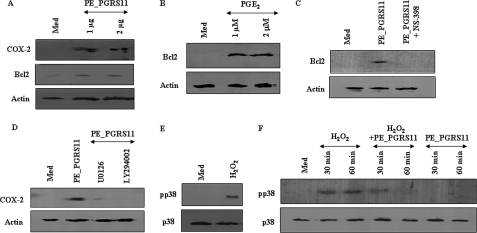
Crucial cell fate decisions adapt to diverse environmental perturbations like oxidative stress by reorganizing their signaling networks with concomitant expression of antiapoptotic genes, all of which could represent a significant host response. In this regard, expression of COX-2 and Bcl2 is often correlated with the survival of cells during oxidative stress (52, 53). COX-2 regulates p53 activation and inhibits p53- or genotoxic stress-induced apoptosis directly through close interaction between two molecules in a variety of cell types (52). In addition, PGE2, a product of COX-2 enzymatic activity, has been shown to modulate apoptosis and induce expression of Bcl2 in various cell types, suggesting a crucial role for COX-2 in the regulation of apoptosis (54). Bcl2, an important member of the Bcl family, is critical for regulation of apoptosis across diverse cell types. Bcl2 acts along the intrinsic mitochondrial apoptosis pathway that is activated in response to a number of stress stimuli including oxidative stress (53). In view of the above mentioned observations, we assessed the ability of PE_PGRS11 to trigger TLR2-mediated COX-2 or Bcl2 expression. As demonstrated, PE_PGRS11 induced the expression of COX-2, its activity product PGE2, or Bcl2 in a dose-dependent manner (Fig. 4A and data not shown). Interestingly, although PGE2 augmented the Bcl2 expression, inhibition of COX-2 activity markedly reduced PE_PGRS11-induced Bcl2 expression, evidently implicating the COX-2-driven signaling events in Bcl2 expression (Fig. 4, B and C). The expression of Bcl2 or COX-2 is reported to involve activation as well as extensive cross-talk between PI3K and mitogen-activated protein kinase pathways. Accordingly, signaling perturbation of PI3K or ERK1/2 by pharmacological inhibitors abrogated PE_PGRS11-driven COX-2 and Bcl2 expression (Fig. 4D and data not shown).
FIGURE 4.
PE_PGRS11 induced rescue from oxidative stress: activation of antiapoptotic and inhibition of apoptotic signaling axis. A, induced expression of COX-2 and Bcl2 upon PE_PGRS11 treatment as analyzed by immunoblotting. B, dose-dependent increase in Bcl2 expression upon PGE2 treatment. C, A549 cells were pretreated with NS-398, and PE_PGRS11-induced Bcl2 expression was analyzed by immunoblotting. D, pretreatment of A549 cells with U0126 and LY294002 leads to abrogation of PE_PGRS11-induced COX-2 expression. E, activation of p38 MAPK upon exposure of cells to H2O2. F, PE_PGRS11 abolishes H2O2-mediated activation of p38 MAPK. The blots are representative of three independent experiments. p, phospho; Med, medium.
As described, COX-2 and Bcl2 act as important effector molecules for cell survival under oxidative stress, and in this context, a member of the mitogen-activated protein kinase family, p38, is suggested to be preferentially activated by oxidative stress. p38 is the archetypal member of the second MAPK-related pathway, and a number of findings indicate that the activation of p38 may play decisive roles in the control of cell death. In the case of PC-12 neuronal cells or NIH-3T3 fibroblasts, interference with p38 kinase activation significantly abrogated apoptosis by nerve growth factor deprivation or UV irradiation (49, 55, 56). Critically, oxidative stress-induced apoptosis in a variety of cellular systems is suggested to involve activation of p38 MAPK (57). As shown, enforced expression of PE_PGRS11 in alveolar epithelial A549 cells imparted resistance to H2O2-induced oxidative stress (Fig. 2). In this context, exposure of alveolar epithelial cells to H2O2 resulted in the activation of p38 MAPK (Fig. 4E). Importantly, PE_PGRS11-triggered TLR2 signaling events abrogated oxidative stress-induced activation of p38 MAPK (Fig. 4F).
Furthermore, we evaluated the role of the phosphoglycerate mutase domain in PE_PGRS11-induced signaling events in alveolar epithelial A549 cells. Recombinant phosphoglycerate mutase of PE_PGRS11 effectively triggered the activation of the PI3K, ERK1/2, or NF-κB signaling pathway as analyzed by phosphorylation of the p85 subunit of PI3K, 4EBP1, or ERK1/2 and translocation of p65 NF-κB from the cytosol to the nucleus (supplemental Fig. S4, A and B). These observations are quite comparable with PI3K-ERK1/2-NF-κB signaling activation observed with full-length recombinant PE_PGRS11 protein. In concordance to results obtained with full-length recombinant PE_PGRS11 protein, the phosphoglycerate mutase domain alone induced the expression of COX-2 and Bcl2 as well as abrogated the oxidative stress-induced activation of p38 MAPK (supplemental Fig. S4, C and D). Taken together, TLR2-dependent expression of COX-2 and Bcl2 and abrogation of H2O2-induced p38 kinase activation by PE_PGRS11 clearly provide a mechanistic basis for the rescue of lung epithelial cells from oxidative stress.
Critical Role of PE_PGRS11 in Survival of Mycobacteria under Oxidative Stress
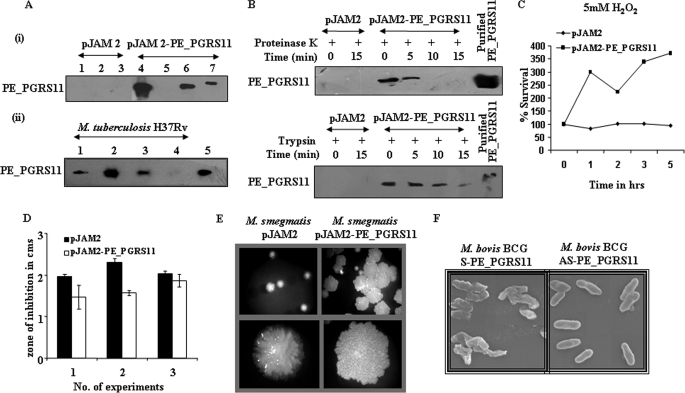
Many reports have suggested that PE_PGRS antigens localize to the cell wall by the PE domain anchoring the protein to the cell wall, thus allowing the antigenically variable PGRS domain to gain access to extracellular compartments from infected macrophages (10, 12, 13). To examine the subcellular localization of the PE_PGRS11, a recombinant M. smegmatis strain expressing PE_PGRS11 was subjected to cell fractionation studies and analyzed by immunoblotting using anti-PE_PGRS11 antibodies. Analysis of the cellular fractions of recombinant M. smegmatis demonstrated that the majority of PE_PGRS11 protein could be detected in the insoluble fraction of the cell preparation, and little if any PE_PGRS11 protein was detected in the soluble fraction (Fig. 5A). Furthermore, PE_PGRS11 could be detected only in the cell wall fraction but not in cytosol fractions derived from M. tuberculosis H37Rv (Fig. 5A). Assessment of the purity of cell wall fractions in terms of cytosolic contamination was carried out by probing the cellular fractions with monoclonal antibodies reactive to cytosolic catalase-peroxidase KatG of M. tuberculosis. Accordingly, although the cell wall fraction did not display any presence of KatG, cytosolic fractions exhibited reactivities to antibodies against KatG, thereby validating the purity of the cell wall preparations (data not shown). To investigate whether PE_PGRS11 is exposed on the mycobacterial surface, M. smegmatis overexpressing PE_PGRS11 was subjected to proteinase K and trypsin sensitivity assays. Proteinase K and trypsin sensitivity assays have been widely used to study the cell surface localization of bacterial proteins (58, 59). Data shown in Fig. 5B demonstrate that PE_PGRS11 was efficiently digested by the proteinase K treatment with complete digestion after 5min. A comparable pattern was obtained when M. smegmatis expressing PE_PGRS11 was subjected to trypsin digestion and analyzed by immunoblotting (Fig. 5B). These observations corroborate that PE_PGRS11 protein is exposed at the mycobacterial cell surface and might gain access to the extracellular environment.
FIGURE 5.
Subcellular localization and role of PE_PGRS11 in protection of mycobacteria against oxidative stress. A, i, recombinant M. smegmatis expressing PE_PGRS11 under an acetamide-inducible promoter was constructed, and different cellular fractions of M. smegmatis transformed with either pJAM2 or pJAM2-PE_PGRS11 were immunoblotted with antiserum to PE_PGRS11. Lane 1, cell wall fraction of induced M. smegmatis (pJAM2); lane 2, cytosol fraction of induced M. smegmatis (pJAM2); lane 3, cell membrane fractions of induced M. smegmatis (pJAM2); lane 4, cell wall fraction of induced M. smegmatis (pJAM2-PE_PGRS11); lane 5, cytosol fraction of induced M. smegmatis (pJAM2-PE_PGRS11); lane 6, cell membrane fraction of induced M. smegmatis (pJAM2-PE_PGRS11); lane 7, purified PE_PGRS11 protein. ii, expression of PE_PGRS11 was analyzed by immunoblotting in various cellular fractions of M. tuberculosis H37Rv cells. Lane 1, whole cell lysate of M. tuberculosis; lane 2, cell wall fraction of M. tuberculosis; lane 3, culture filtrate proteins of M. tuberculosis; lane 4, cytosol fraction of M. tuberculosis; lane 5, purified PE_PGRS11 protein. B, M. smegmatis transformed with pJAM2 or pJAM2-PE_PGRS11 were subjected to partial digestion by either proteinase K or trypsin for the indicated time points, and cell lysates were analyzed for integrity of PE_PGRS11 by immunoblotting. C, growth and survival of M. smegmatis transformed with pJAM2 or pJAM2-PE_PGRS11 in the presence of oxidative stress (5 mm hydrogen peroxide). D, analysis of zone of inhibition for M. smegmatis transformed with pJAM2 or pJAM2-PE_PGRS11 upon oxidative stress (30% H2O2) (mean ± S.E., n = 3). E, micrographs representing colony morphology of M. smegmatis transformed with pJAM2 or pJAM2-PE_PGRS11. F, electron micrographs of M. bovis BCG transformed with pMV361 sense PE_PGRS11 (M. bovis BCG S-PE_PGRS11) or pMV361 antisense PE_PGRS11 (M. bovis BCG AS-PE_PGRS11). The data represent two independent experiments.
The “respiratory burst” of invading host cells caused by reactive oxygen species represents one of the significant host effector functions in response to an infection. For example, H2O2 reacts with reduced copper or iron ions in the cytoplasm of bacteria, leading to generation of free hydroxyl radicals, which in turn culminates in single strand nicks in DNA and oxidation of proteins as well as biological membranes (60, 61). Interestingly, many bacteria including M. tuberculosis detoxify reactive oxygen species, thus providing the predominant challenge to its eradication by robust host immunity (62, 63). In this perspective, observations shown in Fig. 2 clearly suggest a protective role for PE_PGRS11 in providing resistance to epithelial cells against H2O2-triggered oxidative stress. Because a number of microbial cell surface proteins have been suggested to have a moonlighting property of protecting the bacteria against host-induced stress responses, we decided to investigate the potential role, if any, for PE_PGRS11 in executing resistance to mycobacteria against oxidative stress. In this regard, sensitivity assays with H2O2 were carried, and as shown, recombinant M. smegmatis overexpressing PE_PGRS11 exhibited significant proliferation under oxidative stress compared with vector-transformed M. smegmatis (Fig. 5C). For further validation, a well validated and more conclusive “zone of inhibition assay” was carried out wherein bacteria are plated uniformly on a culture plate followed by placement of a filter paper disk permeated with the compound to be tested (64). Accordingly, when tested against oxidative stress inducer H2O2, the size of the zone of clearance was significantly smaller in the case of recombinant M. smegmatis expressing PE_PGRS11 compared with M. smegmatis harboring vector, thus ascertaining a potential role for PE_PGRS11 in providing resistance to bacteria against oxidative stress (Fig. 5D).
Overexpression of PE_PGRS11 Contributes to Alterations in Colony Morphology
The characteristic lipid-rich cell wall is a critical attribute of Mycobacterium species, and each cell wall component plays a significant role in phenotypic features including colony morphology, resistance to various environmental stresses, virulence, etc. Furthermore, changes in colony morphology within a given species of Mycobacterium can have differential effects on the signaling events initiated in infected cells, resulting in a degree of difference in virulence as well as in mycobacterial survival (65–67). For example, among an isogenic pair of Mycobacterium avium morphotypes, whereas the virulent smooth transparent (SmT) morphotype proliferated in murine macrophages in a p38 mitogen-activated protein kinase activation-dependent manner, proliferation of the less virulent smooth opaque (SmO) morphotype to some extent was dependent upon ERK1/2 activation (68, 69). These studies clearly recognized the roles for phenotypic changes of the mycobacteria in virulence. In view of the above, we assessed the effects of overexpression of PE_PGRS11 on the colony morphology of recombinant M. smegmatis, and as demonstrated in Fig. 5E, ectopic expression of PE_PGRS11 resulted in the rough, glossy, bigger, strongly cohesive phenotype of colony morphology variants in comparison with the small, smooth, translucent phenotype of the vector-transformed counterpart. This observation in our opinion suggests a role for variation in colony morphology in adaptation of bacteria to resist stress conditions.
For further validation, M. bovis BCG expressing reduced levels of PE_PGRS11 protein was constructed by antisense RNA, and the expression levels of PE_PGRS11 were determined by immunoblotting. The sense and control transformants exhibited stronger and weaker reactivity, respectively, whereas no corresponding distinct band was observed in the antisense transformant (data not shown). To ascertain the expression of PE_PGRS11 with altered colony morphology, we analyzed the morphology of sense or antisense transformants by scanning electron microscopy. Data represented in Fig. 5F clearly show that antisense PE_PGRS11 transformants of M. bovis BCG exhibited a smooth, translucent phenotype in accordance with results obtained with M. smegmatis overexpressing PE_PGRS11. Taken together, these observations suggest a strong correlation of expression of PE_PGRS11 to the phenotypic changes in mycobacteria, which could result in the augmented ability to resist the hostile oxidative stress environment of the host cells.
Recombinant PE_PGRS11 Displays Differential B Cell Responses during Tuberculosis Infection
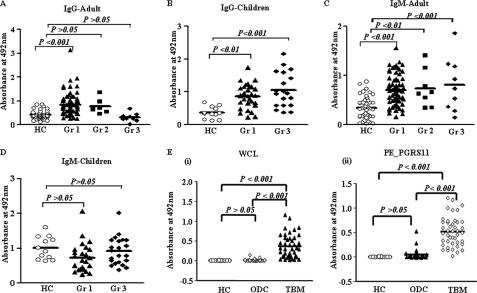
Our results have substantiated that PE_PGRS11 protein is exposed at the mycobacterial cell surface and might gain access to the extracellular compartments from infected host cells, suggesting accessibility of PE_PGRS11 to the immune system of the infected host. As described, PE_PGRS11 is reported to play a critical role in the pathogenesis of tuberculosis and in the host innate and adaptive immune responses to bacilli (8, 9). Furthermore, we have recently demonstrated that PE_PGRS11 recognizes TLR2, induces maturation and activation of human dendritic cells, and enhances the ability of dendritic cells to stimulate CD4+ T cells (46). In this regard, we evaluated the role of PE_PGRS11 as an antigen in a clinical setting. Experiments were designed to evaluate the potential humoral immune responses in sera from different clinical categories of tuberculosis patients to purified PE_PGRS11 protein. Recombinant protein was utilized to screen TB patient sera or CSF by ELISA, utilizing anti-human IgG-HRP and anti-human IgM-HRP antibodies as conjugates. As shown in Fig. 6A, sera from group 1 adult patients diagnosed with pulmonary TB mounted significantly higher IgG antibody responses against PE_PGRS11 than sera from healthy controls. Furthermore, similar results were observed in the case of group 1 child patients (Fig. 6B). However, PE_PGRS11 did not demonstrate statistically significant reactivity to IgG in sera from adult patients with relapsed infection in group 2 (Fig. 6A). The sera from group 2 child patients were not available for our studies. In the case of group 3 patients reporting extrapulmonary infections, only children but not adults elicited significant IgG immunoreactivity (Fig. 6, A and B). When analyzed for IgM antibody responses against PE_PGRS11, sera from adult patients of groups 1, 2, and 3 demonstrated stronger immunoreactivity in comparison with healthy controls (Fig. 6C). In contrast, PE_PGRS11 did not demonstrate any significant reactivity to IgM in sera from child patients from groups 1 and 3 (Fig. 6D). As mentioned, sera from child group 2 patients were not available for study.
FIGURE 6.
Differential immunoreactivity of PE_PGRS11 protein with sera or CSF from different categories of TB patients. A and B, PE_PGRS11 protein showed significant reactivity in all the groups in child TB patients for IgG antibody (B), whereas adult patients showed no reactivity except group 1 patients (A). C and D, when assessed for IgM antibody, PE_PGRS11 showed significant reactivity in all the categories of adult patients (C) but not in the case of any group of child TB patients (D). E, the IgG antibody reactivity to M. tuberculosis H37Rv whole cell lysate (WCL) and PE_PGRS11 in CSF from TBM patients in comparison with healthy controls and other meningitis disease controls. Gr 1, pulmonary infection; Gr 2, relapsed infection; Gr 3, extrasputum infection; HC, healthy control; ODC, other disease control.
We have shown previously that two PE_PGRS antigens demonstrated differential reactivities of the humoral antibodies, clearly suggesting a degree of difference in recognition by patient sera even though they share a similar N- and C-terminal region (7). In this regard, when analyzed, individual patients (group 1 adult and child) demonstrated differential humoral reactivity to PE_PGRS11 and PE_PGRS33, suggesting specificity of the humoral immune response against PE_PGRS11 (supplemental Fig. S5).
As a step further, we investigated the immunoreactivity potential of PE_PGRS11 in CSF of patients with TBM, a clinical case of extrapulmonary tuberculosis infection. As demonstrated in Fig. 6E, PE_PGRS11 exhibited statistically significant IgG antibody reactivities in CSF samples derived from TBM patients. Interestingly, the reactivity of PE_PGRS11 was similar to that of M. tuberculosis H37Rv whole cell lysate (Fig. 6E). However, neither PE_PGRS11 nor H37Rv whole cell lysate showed any significant reactivity to CSF samples from healthy controls or other disease controls. Furthermore, PE_PGRS11 exhibited a very high antigenic index for the potential antigenic determinants (data not shown), and our recent study has suggested that patients with pulmonary tuberculosis disease have a high frequency of T cells specific for PE_PGRS11 that secrete IFN-γ and IL-5 in response to PE_PGRS11 antigen stimulation (46). Additionally, DNA immunization experiments suggested that PE_PGRS11 could elicit robust T cell responses in vivo in mice (data not shown). Taken together, these results demonstrate that PE_PGRS11could be an immunodominant antigen expressed in vivo during infection and possesses probable serodiagnostic potential.
DISCUSSION
Diverse biological systems frequently modulate and reorganize intracellular physiological and metabolic pathways, representing important adaptive responses. Different stresses such as oxidative stress often result in both conserved and specific responses from the host cells, and in this context, pathogenic microbes have utilized these adaptive responses for their own survival amid robust host immune responses (62, 63). In the current study, we describe the functional attributes of a novel, unannotated cell wall-associated PE_PGRS antigen, PE_PGRS11, encoded by the M. tuberculosis genome. We demonstrate here that hypothetical PE_PGRS11 ORF encodes a functional phosphoglycerate mutase, and bioinformatics prediction of the phosphoglycerate mutase domain identified His, Arg, and Ser as the critical residues of the catalytic triad. In this regard, the catalytic properties of the phosphoglycerate mutase domain of PE_PGRS11 were quite similar to those of phosphoglycerate mutase from various biological systems (25–27). Importantly, mutation of any of the three residues of the catalytic triad abrogated the phosphoglycerate mutase activity of PE_PGRS11. As described, the PE_PGRS subfamily of PE genes is suggested to act as virulence factors because of their unique restriction as genes with a high probability of being essential in M. tuberculosis survival in the host (5, 10, 11, 70). Studies have reported that several PE_PGRS family antigens respond to different environmental cues including hypoxia, and in vivo selective expression of specific PE_PGRS antigens could be the contributory reason for the various pathological attributes of M. tuberculosis (8–11). The expression profiling investigations have revealed that PE_PGRS11 is selectively expressed in M. tuberculosis bacilli upon infection of macrophages and could be detected in lung tissues of infected mice (8, 9). Furthermore, our transcriptional analysis demonstrated that PE_PGRS11 is a hypoxia-responsive gene similar to HspX.
Across many cellular systems, a strong correlation between glycolytic enzymes, reactive oxygen species, and cell proliferation has been documented. Glycolytic enzymes are reported to modulate cellular life spans by eliciting protection from oxidative stress (36–38). For example, the highly active glycolytic metabolic pathway in embryonic stem cells contributes to immortality despite a strong p53 expression induced upon DNA damage (38). In this context, phosphoglycerate mutase has been demonstrated to increase the rate of glycolysis, decrease the accumulation of reactive oxygen species, and bypass senescence in mouse embryo fibroblasts (38). In addition, exposure of lung fibroblasts to hypoxia resulted in the induction of phosphoglycerate mutase enzymatic activity with concomitant elevations in phosphoglycerate mutase mRNA and protein levels. Induction of phosphoglycerate mutase contributed to the regulation of the glycolytic flux under hypoxic conditions and demonstrated its role in the adaptation of cells to hypoxia (71). Furthermore, a phosphoglycerate mutase-null mutant of B. bronchiseptica showed enhanced susceptibility to the superoxide radical as well as to antimicrobial peptides with concomitant decreased fitness in the mouse typhoid model (39, 40). In this context, we explored whether enforced expression of PE_PGRS11 imparts resistance against oxidative stress in alveolar epithelial cells. Alveolar epithelial cells in addition to alveolar macrophages act as first responders in a diverse set of infections including that of pathogenic mycobacteria and play a significant role in initiation as well as control of the magnitude of host immunity (72–74). As demonstrated, the ectopic expression of PE_PGRS11 in alveolar epithelial cells via recombinant adenovirus or recombinant M. smegmatis imparted resistance to H2O2-triggered oxidative stress. This observation is thus attributed to the phosphoglycerate mutase domain of PE_PGRS11, which is in accordance with reported studies describing a role for phosphoglycerate mutase in evading cellular life span restrictions mediated by oxidative stress.
We have recently demonstrated that PE_PGRS11 antigen triggers maturation of dendritic cells as exemplified by up-regulation of costimulatory and antigen-presenting molecules and increased production of proinflammatory cytokines. Furthermore, PE_PGRS11 induced CD4 T cell proliferation and secretion of cytokines in vitro and in vivo from T cells in patients with pulmonary tuberculosis (46). Here we show that PE_PGRS11 protein recognized TLR2 on alveolar epithelial cells and induced activation of PI3K, ERK1/2, and NF-κB pathways, thus representing important signaling partnership integration in host cells. This signaling cross-talk, induced by PE_PGRS11 antigen, culminated in the induction of the expression of COX-2, its activity product PGE2, and Bcl2 in a dose-dependent manner. Furthermore, PE_PGRS11 antigen treatment abolished H2O2-triggered activation of p38 MAPK, which is a major kinase involved in stress-induced apoptosis in various cell types. Importantly, PE_PGRS11 protein was exposed at the mycobacterial cell surface and executed resistance to mycobacteria against oxidative stress. Interestingly, overexpression of PE_PGRS11 contributed to alterations in colony morphology, and in our opinion, this variation in colony morphology could be an adaptation of bacteria to resist stress conditions. Furthermore, PE_PGRS11 exhibited immunoreactivity to sera from adult patients with pulmonary tuberculosis and child patients with pulmonary or extrapulmonary infection. The observed humoral immune response against a hypoxia-responsive gene (PE_PGRS11) in patients with active tuberculosis disease is interesting. As reported earlier, many hypoxia-specific mycobacterial antigens like Rv1168c, Rv1169c, Rv2430c, and others have been demonstrated to elicit robust humoral/antibody reactivities in patients with active pulmonary tuberculosis disease (with active lesions) (7, 75, 76). Interestingly, PE_PGRS62 elicited strong antibody responses in patients with latent infection as well as in patients with active pulmonary tuberculosis disease (77). These observations suggest that the expression levels of these antigens including PE_PGRS11 upon reaching a certain threshold level during hypoxic or other stages of infection could trigger a multitude of host immune responses, which could be sustained in terms of strength as well as magnitude during the life span of the infected individuals. Overall, these observations suggest that PE_PGRS11 is expressed in vivo during disease pathogenesis and could significantly contribute to immune evasion strategies of pathogenic mycobacteria against oxidative stress.
Supplementary Material
Acknowledgments
We are thankful to Dr. Douglas Golenbock, University of Massachusetts Medical School, Worcester, MA, for the kind gift of reagents. We thank Dr. D. N. Rao and his laboratory members, Department of Biochemistry, Indian Institute of Science, Bangalore, India, for kind help and advice during the course of the current investigation. We acknowledge the kind help of Sneha, Department of Biotechnology Confocal Facility, for assistance in confocal microscopy studies.
This work was supported by funds from the Department of Biotechnology (DBT); a collaborative grant (to the Indian Institute of Science and Karolinska Institute) from VINNOVA (The Swedish Governmental Agency for Innovation Systems) and DBT, India; the Department of Science and Technology (DST); and the Council for Scientific and Industrial Research and by infrastructure support from the Indian Council of Medical Research (Center for Advanced Study in Molecular Medicine), DST (Fund for Improvement of Science and Technology Infrastructure in Universities and Higher Educational Institutions), and University Grants Commission (special assistance).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PE
- proline-glutamic acid
- PPE
- proline-proline-glutamic acid
- PE_PGRS
- proline-glutamic acid-polymorphic GC-rich sequences
- COX-2
- cyclooxygenase-2
- TBM
- tuberculosis meningitis
- CSF
- cerebrospinal fluid
- BCG
- bacillus Calmette-Guérin
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- TB
- tuberculosis
- TLR
- Toll-like receptor
- PGE2
- prostaglandin E2.
REFERENCES
- 1.Józefowski S., Sobota A., Kwiatkowska K. (2008) BioEssays 30, 943–954 [DOI] [PubMed] [Google Scholar]
- 2.Andersen P., Doherty T. M. (2005) Nat. Rev. Microbiol. 3, 656–662 [DOI] [PubMed] [Google Scholar]
- 3.Brandt L., Feino Cunha J., Weinreich Olsen A., Chilima B., Hirsch P., Appelberg R., Andersen P. (2002) Infect. Immun. 70, 672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adindla S., Guruprasad L. (2003) J. Biosci. 28, 169–179 [DOI] [PubMed] [Google Scholar]
- 5.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 6.Koh K. W., Lehming N., Seah G. T. (2009) Mol. Immunol. 46, 1312–1318 [DOI] [PubMed] [Google Scholar]
- 7.Narayana Y., Joshi B., Katoch V. M., Mishra K. C., Balaji K. N. (2007) Clin. Vaccine Immunol. 14, 1334–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. (2003) J. Exp. Med. 198, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talaat A. M., Lyons R., Howard S. T., Johnston S. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4602–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan M. J., Delogu G., Chen Y., Bardarov S., Kriakov J., Alavi M., Jacobs W. R., Jr. (2001) Infect. Immun. 69, 7326–7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dheenadhayalan V., Delogu G., Brennan M. J. (2006) Microbes Infect. 8, 262–272 [DOI] [PubMed] [Google Scholar]
- 12.Beatty W. L., Russell D. G. (2000) Infect. Immun. 68, 6997–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beatty W. L., Ullrich H. J., Russell D. G. (2001) Eur. J. Cell Biol. 80, 31–40 [DOI] [PubMed] [Google Scholar]
- 14.Grisolia S., Tecson J. (1967) Biochim. Biophys. Acta 132, 56–67 [DOI] [PubMed] [Google Scholar]
- 15.Grisolia S., Wallace R., Mendelson J. (1975) Physiol. Chem. Phys. 7, 219–223 [PubMed] [Google Scholar]
- 16.Wayne L. G., Hayes L. G. (1996) Infect. Immun. 64, 2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson M. (1988) Nucleic Acids Res. 16, 10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker T. C., Noel R. J., Coats W. S., Gómez-Foix A. M., Alam T., Gerard R. D., Newgard C. B. (1994) Methods Cell Biol. 43, 161–189 [DOI] [PubMed] [Google Scholar]
- 20.Wajapeyee N., Somasundaram K. (2003) J. Biol. Chem. 278, 52093–52101 [DOI] [PubMed] [Google Scholar]
- 21.Narayana Y., Balaji K. N. (2008) J. Biol. Chem. 283, 12501–12511 [DOI] [PubMed] [Google Scholar]
- 22.Hirschfield G. R., McNeil M., Brennan P. J. (1990) J. Bacteriol. 172, 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee B. Y., Hefta S. A., Brennan P. J. (1992) Infect. Immun. 60, 2066–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao S., Young G., Oberg C., Nakaoka K. (2008) Flavour Fragrance J. 23, 444–449 [Google Scholar]
- 25.Hill B., Attwood M. M. (1976) J. Gen. Microbiol. 96, 185–193 [DOI] [PubMed] [Google Scholar]
- 26.Stankiewicz P. J., Hass L. F. (1986) J. Biol. Chem. 261, 12715–12721 [PubMed] [Google Scholar]
- 27.Raverdy S., Zhang Y., Foster J., Carlow C. K. (2007) Mol. Biochem. Parasitol. 156, 210–216 [DOI] [PubMed] [Google Scholar]
- 28.Lefèvre F., Rémy M. H., Masson J. M. (1997) Nucleic Acids Res. 25, 447–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W., Williams C. H., Jr., Massey V. (1996) J. Biol. Chem. 271, 17226–17233 [DOI] [PubMed] [Google Scholar]
- 30.Van Petegem F., Duderstadt K. E., Clark K. A., Wang M., Minor D. L., Jr. (2008) Structure 16, 280–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voskuil M. I., Schnappinger D., Rutherford R., Liu Y., Schoolnik G. K. (2004) Tuberculosis 84, 256–262 [DOI] [PubMed] [Google Scholar]
- 32.Wayne L. G., Sohaskey C. D. (2001) Annu. Rev. Microbiol. 55, 139–163 [DOI] [PubMed] [Google Scholar]
- 33.Betts J. C., Lukey P. T., Robb L. C., McAdam R. A., Duncan K. (2002) Mol. Microbiol. 43, 717–731 [DOI] [PubMed] [Google Scholar]
- 34.Gengenbacher M., Rao S. P., Pethe K., Dick T. (2010) Microbiology 156, 81–87 [DOI] [PubMed] [Google Scholar]
- 35.Sohaskey C. D., Wayne L. G. (2003) J. Bacteriol. 185, 7247–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondoh H., Lleonart M. E., Bernard D., Gil J. (2007) Histol. Histopathol. 22, 85–90 [DOI] [PubMed] [Google Scholar]
- 37.Kondoh H., Lleonart M. E., Gil J., Wang J., Degan P., Peters G., Martinez D., Carnero A., Beach D. (2005) Cancer Res. 65, 177–185 [PubMed] [Google Scholar]
- 38.Kondoh H., Lleonart M. E., Gil J., Beach D., Peters G. (2005) Drug Discov. Today Dis. Mech. 2, 263–267 [Google Scholar]
- 39.West N. P., Jungnitz H., Fitter J. T., McArthur J. D., Guzmán C. A., Walker M. J. (2000) Infect. Immun. 68, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paterson G. K., Cone D. B., Peters S. E., Maskell D. J. (2009) Microbiology 155, 3403–3410 [DOI] [PubMed] [Google Scholar]
- 41.Trinchieri G., Sher A. (2007) Nat. Rev. Immunol. 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 42.Almeida P. E., Silva A. R., Maya-Monteiro C. M., Töröcsik D., D'Avila H., Dezsö B., Magalhães K. G., Castro-Faria-Neto H. C., Nagy L., Bozza P. T. (2009) J. Immunol. 183, 1337–1345 [DOI] [PubMed] [Google Scholar]
- 43.Bansal K., Kapoor N., Narayana Y., Puzo G., Gilleron M., Balaji K. N. (2009) PLoS One 4, e4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bansal K., Narayana Y., Patil S. A., Balaji K. N. (2009) J. Leukoc. Biol. 85, 804–816 [DOI] [PubMed] [Google Scholar]
- 45.Pecora N. D., Gehring A. J., Canaday D. H., Boom W. H., Harding C. V. (2006) J. Immunol. 177, 422–429 [DOI] [PubMed] [Google Scholar]
- 46.Bansal K., Elluru S. R., Narayana Y., Chaturvedi R., Patil S. A., Kaveri S. V., Bayry J., Balaji K. N. (2010) J. Immunol. 184, 3495–3504 [DOI] [PubMed] [Google Scholar]
- 47.Chang M. S., Chen B. C., Yu M. T., Sheu J. R., Chen T. F., Lin C. H. (2005) Cell. Signal. 17, 299–310 [DOI] [PubMed] [Google Scholar]
- 48.Méndez-Samperio P., Pérez A., Rivera L. (2009) Cell Immunol. 256, 12–18 [DOI] [PubMed] [Google Scholar]
- 49.Kyriakis J. M., Avruch J. (2001) Physiol. Rev. 81, 807–869 [DOI] [PubMed] [Google Scholar]
- 50.Marte B. M., Downward J. (1997) Trends Biochem. Sci. 22, 355–358 [DOI] [PubMed] [Google Scholar]
- 51.Creson T. K., Yuan P., Manji H. K., Chen G. (2009) J. Mol. Neurosci. 37, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi E. M., Heo J. I., Oh J. Y., Kim Y. M., Ha K. S., Kim J. I., Han J. A. (2005) Biochem. Biophys. Res. Commun. 328, 1107–1112 [DOI] [PubMed] [Google Scholar]
- 53.Frenzel A., Grespi F., Chmelewskij W., Villunger A. (2009) Apoptosis 14, 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Souto E. O., Miyoshi H., Dubois R. N., Gores G. J. (2001) Am. J. Physiol. Gastrointest. Liver Physiol. 280, G805–G811 [DOI] [PubMed] [Google Scholar]
- 55.De Zutter G. S., Davis R. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6168–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kralova J., Dvorak M., Koc M., Kral V. (2008) Oncogene 27, 3010–3020 [DOI] [PubMed] [Google Scholar]
- 57.Yamagishi N., Saito Y., Hatayama T. (2008) FEBS J. 275, 4558–4570 [DOI] [PubMed] [Google Scholar]
- 58.Cascioferro A., Delogu G., Colone M., Sali M., Stringaro A., Arancia G., Fadda G., Palù G., Manganelli R. (2007) Mol. Microbiol. 66, 1536–1547 [DOI] [PubMed] [Google Scholar]
- 59.Delogu G., Pusceddu C., Bua A., Fadda G., Brennan M. J., Zanetti S. (2004) Mol. Microbiol. 52, 725–733 [DOI] [PubMed] [Google Scholar]
- 60.Higuchi Y. (2003) Biochem. Pharmacol. 66, 1527–1535 [DOI] [PubMed] [Google Scholar]
- 61.Payne C. M., Bernstein C., Bernstein H. (1995) Leuk. Lymphoma 19, 43–93 [DOI] [PubMed] [Google Scholar]
- 62.Chan J., Fan X. D., Hunter S. W., Brennan P. J., Bloom B. R. (1991) Infect. Immun. 59, 1755–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan J., Fujiwara T., Brennan P., McNeil M., Turco S. J., Sibille J. C., Snapper M., Aisen P., Bloom B. R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 2453–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barker L. P., Lien B. A., Brun O. S., Schaak D. D., McDonough K. A., Chang L. C. (2007) Planta Med. 73, 559–563 [DOI] [PubMed] [Google Scholar]
- 65.Chowdhury R. P., Saraswathi R., Chatterji D. (2010) IUBMB Life 62, 67–77 [DOI] [PubMed] [Google Scholar]
- 66.Schorey J. S., Cooper A. M. (2003) Cell Microbiol. 5, 133–142 [DOI] [PubMed] [Google Scholar]
- 67.Smeulders M. J., Keer J., Speight R. A., Williams H. D. (1999) J. Bacteriol. 181, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blumenthal A., Ehlers S., Ernst M., Flad H. D., Reiling N. (2002) Infect. Immun. 70, 4961–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tse H. M., Josephy S. I., Chan E. D., Fouts D., Cooper A. M. (2002) J. Immunol. 168, 825–833 [DOI] [PubMed] [Google Scholar]
- 70.Brennan M. J., Delogu G. (2002) Trends Microbiol. 10, 246–249 [DOI] [PubMed] [Google Scholar]
- 71.Takahashi Y., Takahashi S., Yoshimi T., Miura T. (1998) Eur. J. Biochem. 254, 497–504 [DOI] [PubMed] [Google Scholar]
- 72.Bansal K., Narayana Y., Balaji K. N. (2009) Scand. J. Immunol. 69, 11–19 [DOI] [PubMed] [Google Scholar]
- 73.Bermudez L. E., Goodman J. (1996) Infect. Immun. 64, 1400–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin Y., Zhang M., Barnes P. F. (1998) Infect. Immun. 66, 1121–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choudhary R. K., Mukhopadhyay S., Chakhaiyar P., Sharma N., Murthy K. J., Katoch V. M., Hasnain S. E. (2003) Infect. Immun. 71, 6338–6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khan N., Alam K., Nair S., Valluri V. L., Murthy K. J., Mukhopadhyay S. (2008) Clin. Vaccine Immunol. 15, 974–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koh K. W., Soh S. E., Seah G. T. (2009) Infect. Immun. 77, 3337–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.