Abstract
Here, we demonstrate that p68 (DDX5) and p72 (DDX17), two homologous RNA helicases and transcriptional cofactors, are substrates for the acetyltransferase p300 in vitro and in vivo. Mutation of acetylation sites affected the binding of p68/p72 to histone deacetylases, but not to p300 or estrogen receptor. Acetylation additionally increased the stability of p68 and p72 RNA helicase and stimulated their ability to coactivate the estrogen receptor, thereby potentially contributing to its aberrant activation in breast tumors. Also, acetylation of p72, but not of p68 RNA helicase, enhanced p53-dependent activation of the MDM2 promoter, pointing at another mechanism of how p72 acetylation may facilitate carcinogenesis by boosting the negative p53-MDM2 feedback loop. Furthermore, blocking p72 acetylation caused cell cycle arrest and apoptosis, revealing an essential role for p72 acetylation. In conclusion, our report has identified for the first time that acetylation modulates RNA helicases and provides multiple mechanisms how acetylation of p68 and p72 may affect normal and tumor cells.
Keywords: Gene Transcription, p300, Protein Acylation, Protein Stability, RNA Helicase
Introduction
One prominent post-translational modification is the acetylation of lysine residues. In particular, histones are reversibly modified by acetylation, and this normally stimulates gene transcription. However, acetylation also occurs on a plethora of non-histone proteins and modulates their activity in various ways, for instance by affecting DNA binding, transactivation, protein stability, or intracellular localization (1, 2).
The acetyltransferase p300 is capable of modifying all four core histones and is essential for development (3, 4). Moreover, chromosomal translocations involving the p300 gene have been observed in hematologic malignancies and mutations in several solid tumors, suggesting an important role for p300 in human carcinogenesis (5, 6). Among the known interactants of p300 are the homologous RNA helicases p68 (also called DEAD-box protein 5, DDX5)2 and p72 (DDX17) that synergize with p300 to activate gene transcription (7, 8). Several transcription factors, including estrogen receptor α (ERα), androgen receptor, Smad3, MyoD, and the tumor suppressor p53, not only bind to p300 but also to p68/p72 (9–14). Accordingly, p68/p72 and p300 are co-recruited into and jointly modulate the activity of various multiprotein complexes involved in transcriptional control (15).
Knocking out p68 or p72 causes embryonic or neonatal lethality, indicating that p68 as well as p72 are crucial during development (16). Additionally, these two RNA helicases are overexpressed in colorectal tumors and thought to promote tumorigenesis by stimulating β-catenin, whose aberrant activation is responsible for neoplastic transformation in the vast majority of colon tumors (17–19). Similarly, p68 is overexpressed in and may contribute to the formation of prostate tumors by coactivating the androgen receptor (12). In addition, both p68 and p72 are overexpressed in breast tumors (20, 21) and stimulate ERα, whose abnormal activation is an underlying cause in the development of most human breast tumors (9, 14). Thus, p68 and p72 play important roles in the physiology of normal and diseased cells, warranting investigation into how these two homologous RNA helicases are regulated. Here, we examined whether p68 and p72 are acetylated by p300 and whether this is a means to modulate their function.
EXPERIMENTAL PROCEDURES
Luciferase Assays
CV-1 cells were grown in 6-cm dishes and transiently transfected by the calcium phosphate coprecipitation method (22). The TORU luciferase reporter construct (23) was cotransfected with a p68 expression vector as described before (7). Where indicated, cells were treated with 15 mm sodium butyrate 12 h prior to lysis. Cells were lysed 36 h after transfection (24) and luciferase activity was measured in a Berthold Lumat (25, 26).
MDA-MB-231 cells were grown in 12-well plates and transiently transfected using the calcium phosphate coprecipitation method (27). As reporter plasmid, 500 ng of ERE-luc (28) or 200 ng of MDM2-luc (29) were employed. Where indicated, 1600 ng of HA-tagged p68 or p72 or empty vector pEV3S (30), 30 ng of pSG5-ERα or empty vector pSG5, and 10 ng of pcDNA3-p53 or empty vector pcDNA3 were cotransfected. In case of stimulation with 1 nm estradiol or respective control, 5% charcoal-stripped serum was employed after transfection. To determine levels of p68 or p72 protein expression, cells were lysed in 10 mm Tris-HCl, 30 mm Na4P2O7 (pH 7.1), 50 mm NaCl, 50 mm NaF, 0.5 mm Na3VO4, 0.2 mm DTT, 1% sodium deoxycholate, 1% Triton X-100, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μg/ml pepstatin A, 1 mm phenylmethylsulfonyl fluoride, 0.2 mm DTT. Immunoprecipitations with anti-HA 12CA5 monoclonal antibodies were then performed essentially as described (31) and followed by anti-HA Western blotting (32).
Detection of Acetylated Lysine in Vivo
To probe for in vivo acetylation, immunoprecipitations with two different acetyllysine antibodies (AcK1: 06–933, Upstate; AcK2: PAN-AC1, Abcam) were performed as described before (33). Acetylated proteins were then detected by Western blotting (34). Alternatively, immunoprecipitations were performed with Myc 9E10 monoclonal antibody followed by Western blotting with acetyllysine antibodies (9441, New England Biolabs). Where indicated, 293T cells were transfected with expression vectors for p300-HA (35), Ras-G12V (36), Raf-BXB (37), or HER2/Neu-V664E (38) as described before (33).
In Vitro Acetylation
Glutathione S-transferase (GST) fusion proteins were produced and purified according to standard procedures (39). These proteins served as substrates for in vitro acetylation assays that were performed for 60–90 min at 30 °C in 50 mm Hepes (pH 7.4), 10 mm sodium butyrate, 1 mm DTT, 0.1 mm phenylmethylsulfonyl fluoride in the presence of [14C]acetyl coenzyme A. Included were also the catalytic domains of p300, P/CAF, or ACTR fused to GST or p300-HA that was immunoprecipitated with HA antibodies from transfected 293T cells (33, 40).
Mass Spectrometry
293T cells were plated in 10-cm dishes and transiently transfected with 18 μg of 6Myc-p68 or 6Myc-p72 with or without 9 μg of p300-HA expression plasmid (41). Cells were lysed as described above in the presence of 10 mm sodium butyrate and immunoprecipitations performed with anti-Myc 9E10 monoclonal antibody. After SDS-PAGE on a 8% polyacrylamide gel, colloidal Coomassie Brilliant Blue G-250 was employed for staining, the appropriate bands cut out, digested with either trypsin or chymotrypsin, and subjected to liquid chromatography-tandem mass spectrometry.
Pulse-chase Experiments
HeLa cells (Tet-On cell line, Clontech) grown in 6-cm dishes were transiently transfected by the calcium phosphate coprecipitation method (42) with 8 μg of 6Myc-p68 or 6Myc-p72 expression vectors. 36 h after transfection, cells were pulsed for 2 h with 100 μCi of [35S]methionine followed by a chase with non-radioactive methionine (33). At various time points, cells were harvested, and immunoprecipitations with anti-Myc 9E10 monoclonal antibody performed as described (43). Immunoprecipitates were subjected to SDS-PAGE, and radioactivity incorporated into 6Myc-p68 and 6Myc-p72 was quantitated with the help of a PhosphorImager.
Coimmunoprecipitations
To study complex formation between p68/p72 and ERα, 293T cells were grown in 6-cm dishes and transiently transfected with 8 μg of 6Myc-p68 or 6Myc-p72 and 1 μg of Flag-ERα expression vector (44). Cells were lysed 8 h after transfection in 2.5 mm Tris-HCl, 7.5 mm Na4P2O7 (pH 7.1), 12.5 mm NaCl, 12.5 mm NaF, 0.25% Triton X-100, 0.5 mm Na3VO4, 0.2 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μg/ml pepstatin A. Immunoprecipitations with anti-Myc 9E10 antibody were performed essentially as described before (40). Coimmunoprecipitated ERα was detected by Western blotting utilizing anti-Flag M2 monoclonal antibody (45). To probe for complex formation with p300, 293T cells were transiently transfected with 3 μg p300-HA and 1 μg of 6Myc-p68 or 6Myc-p72 constructs (46), immunoprecipitations performed with anti-Myc 9E10 antibody as described (47) and coprecipitated p300 detected by anti-HA Western blotting. To explore complex formation with histone deacetylases, 293T cells were similarly transfected with plasmids encoding HA-p68 or HA-p72 (8 μg) and Flag-HDAC (1 μg), anti-Flag immunoprecipitations performed and followed by Western blotting employing anti-HA antibodies (48).
Flow Cytometry
293T cells were seeded at 25% confluency in 10-cm dishes (49). They were transiently transfected for 12 h with 24 μg of HA-p68 or HA-p72 and additionally with 1 μg of GFP expression vector employing the calcium phosphate coprecipitation method (50). Cells were fixed with ethanol 8 h or 24 h afterward, stained with propidium iodide overnight in the presence of RNase A, and then subjected to flow cytometry.
RESULTS
Acetylation of p68 RNA Helicase by p300
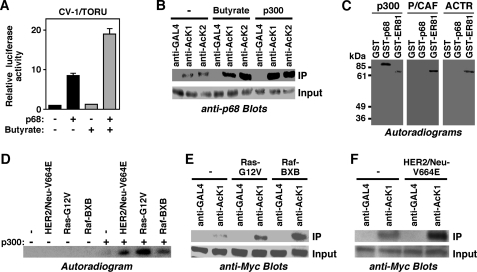
Previously, we showed that the RNA helicase p68 and the acetyltransferase p300 cooperate in activating transcription mediated by a TPA oncogene responsive unit (TORU) (7). We noted that treatment of cells with sodium butyrate, an inhibitor of deacetylating enzymes (51), stimulated the ability of p68 to induce a TORU luciferase reporter gene, whereas sodium butyrate had no effect on transcription in the absence of p68 (Fig. 1A). This suggested that p68 may be acetylated by p300 and that this acetylation enhances its coactivation potential. Indeed, p68 is acetylated in vivo, because endogenous p68 was immunoprecipitated with two different acetyllysine antibodies but not with GAL4 control antibodies (Fig. 1B). Moreover, treatment of cells with sodium butyrate as well as overexpression of p300 led to increased acetylation of endogenous p68 (Fig. 1B), indicating that p68 is a substrate for p300 in vivo.
FIGURE 1.
p68 is acetylated by p300. A, activation of the TORU luciferase reporter by p68. Where indicated, CV-1 cells were transfected with p68 expression vector and treated with sodium butyrate. B, 293T cells were untreated or either treated with sodium butyrate or transfected with a p300 expression vector. Immunoprecipitation (IP) was performed with two different acetyllysine antibodies (AcK1, AcK2) or control GAL4 antibodies. Immunoprecipitated p68 was then revealed by blotting with a p68 antibody. The bottom panel shows that comparable amounts of total p68 were present in each immunoprecipitation experiment. C, indicated GST fusion proteins were incubated with recombinant p300, P/CAF or ACTR in the presence of [14C]acetyl coenzyme A. Acetylation was revealed by determining the incorporation of radioactivity through autoradiography. D, p300-HA and HER2/Neu-V664E, Ras-G12V, or Raf-BXB were coexpressed as indicated in 293T cells. Anti-HA immunoprecipitates were then employed in an in vitro acetylation assay with [14C]acetyl coenzyme A. Shown is the incorporation of radioactivity into GST-p68. E, 293T cells were transfected with 6Myc-p68 and the indicated oncogenic mutants of Ras or Raf. Immunoprecipitations were preformed with control (GAL4) or acetyllysine (AcK1) antibodies. Shown are anti-Myc blots of the immunoprecipitates and the corresponding inputs. F, analogous, impact of HER2/Neu on the in vivo acetylation of p68.
We then performed in vitro acetylation experiments utilizing the catalytic domains of p300 and two other acetyltransferases, P/CAF and ACTR, which are phylogenetically unrelated to p300 (1). No acetylation was observed on the GST moiety, but GST-p68 was robustly acetylated by p300 in vitro (Fig. 1C). In contrast, P/CAF and ACTR were unable to acetylate p68. This was not due to catalytic inactivity of our recombinant P/CAF and ACTR proteins, because they readily acetylated a known substrate, the transcription factor ER81 (33, 40), similarly as p300. Thus, p68 can be acetylated by p300 both in vitro and in vivo.
Oncoproteins such as Raf, Ras, and HER2/Neu stimulate the enzymatic activity of p300, most likely by inducing the MAP kinase pathway resulting into the phosphorylation of p300 (33). Accordingly, the ability of p300 to acetylate p68 in vitro was drastically enhanced when p300 was immunoprecipitated from cells overexpressing oncogenic mutants of HER2/Neu, Ras, or Raf (Fig. 1D). In addition, overexpression of Raf, Ras, or HER2/Neu led to enhanced in vivo acetylation of p68 (Fig. 1, E and F). These results strongly suggest that p68 acetylation levels rise during the formation of HER2/Neu-, Ras-, or Raf-dependent neoplasias that account for >50% of all human tumors.
Mapping of Acetylation Sites
To determine which lysine residues become acetylated in p68, we split the molecule into six parts and studied which of those would be acetylated by p300 in vitro. Only the N-terminal 80 amino acids of p68 were a substrate for p300 (Fig. 2A). Further subdividing these 80 amino acids into three subfragments covering amino acids 1–35, 36–47, and 48–80 revealed that all three subfragments were acetylated by p300 in vitro (Fig. 2B). Thus, potentially all nine lysine residues in the N-terminal 80 amino acids of p68 (see Fig. 2B, top) may become acetylated.
FIGURE 2.
Acetylation of p68 by p300 in vitro. A, indicated amino acids of p68 were fused to GST and incubated with p300 and [14C]acetyl coenzyme A in vitro. Acetylation was revealed by autoradiography. The bottom panel (Coomassie Blue stain) shows the amounts of GST fusion proteins utilized. B, similar, in vitro acetylation of indicated GST-p68 fusion proteins and mutants thereof. The top shows amino acids 1–80 of p68 with all nine lysine residues pointed out.
Indeed, when we mutated either Lys-32 or Lys-33 in GST-p681–35 to arginine, acetylation was not abolished, indicating that both Lys-32 and Lys-33 were acetylated by p300 (Fig. 2B, left panels). Next, we mutated each of the four lysine residues in p68 amino acids 36–47 to arginine. None of these individual mutations abolished acetylation of respective GST-p6836–47 fusion proteins, but acetylation with the K40R and K43R mutants seemed to be reduced. In addition, the double mutant K40/43R was no longer acetylated, indicating that both Lys-40 and Lys-43 were acetylated by p300 (Fig. 2B, middle panels). Finally, we also observed that Lys-56 and Lys-80, but not Lys-53, were acetylated by p300 (Fig. 2B, right panels). Altogether, these in vitro acetylation experiments implied that Lys-32, Lys-33, Lys-40, Lys-43, Lys-56, and Lys-80 are targeted by p300.
To confirm that these in vitro acetylation sites are also utilized in vivo, we coexpressed p68 with and without p300 in 293T cells, immunopurified p68 and analyzed it after proteolytic cleavage by liquid chromatography-tandem mass spectrometry. No acetylated peptides derived from p68 were detectable in the absence of coexpressed p300, but several acetylated p68 peptides were observable in the presence of ectopic p300. One example is given in Fig. 3A, validating that both Lys-32 and Lys-33 are substrates for p300 in vivo; please note that also peptides without acetylation on Lys-32 and/or Lys-33 were observed (not shown), indicating that lysine acetylation occurs at a substoichiometric level. We also confirmed acetylation on Lys-40 and Lys-43 in vivo, but none of the tryptic and chymotryptic peptides encompassing Lys-56 or Lys-80 showed any in vivo acetylation. This discrepancy to our in vitro results may be due to the fact that Lys-56 and Lys-80 are not accessible for p300 in the full-length p68 protein, but are in the GST-p6848–80 molecule, pointing at one limitation of our in vitro acetylation experiments with short fragments of p68. Moreover, in contrast to our in vitro acetylation assays with GST-p6836–47, we observed acetylation of Lys-44 and Lys-45 by mass spectrometry, again pointing at the limited value of in vitro acetylation data with short p68 fragments. In conclusion, p300 acetylates the following six lysine residues in p68 in vivo: Lys-32, Lys-33, Lys-40, Lys-43, Lys-44, and Lys-45.
FIGURE 3.
Mapping of in vivo acetylation sites in p68/p72. A, 293T cells were cotransfected with p68 and p300 expression vectors. Immunopurified p68 was digested with trypsin and subjected to tandem mass spectrometry. Shown is the fragmentation of a peptide encompassing amino acids 26–40 as well as expected masses of b and y ions in case of acetylation of both Lys-32 and Lys-33. B, 6Myc-p68 or mutants thereof were coexpressed with or without p300 in 293T cells. After anti-Myc immunoprecipitation, acetylation was probed with acetyllysine antibodies (top). The bottom panel shows that comparable amounts of Myc-tagged proteins were expressed. C, in vitro acetylation of indicated p72 amino acids fused to GST by recombinant p300. D, alignment of acetylated amino acids in p68 and p72. E, as in panel B, in vivo acetylation of wild-type p72 or its K29/30/42R mutant was assessed.
Accordingly, we found that a mutant of p68, in which these six lysine residues were changed to arginine (K32/33/40/43/44/45R mutant), was no longer recognizable with an acetyllysine antibody upon p300 expression (Fig. 3B); please note that no acetylation was detectable in the absence of coexpressed p300. As a control, we also mutated a reported sumoylation site in p68, lysine 53 (21, 52), and detected no significant difference of the respective K53R mutant to become acetylated compared with wild-type p68, also suggesting that sumoylation does not interfere with acetylation. Interestingly, we observed in our mass spectrometric analyses that acetylation of Lys-43 and Lys-44 depended on Lys-40 acetylation. Consistently, mutation of Lys-32, Lys-33, Lys-40, and Lys-45 sufficed to abolish all p300-mediated acetylation in p68 as determined by mass spectrometry and by blotting with acetyllysine antibodies (not shown).
Next, we assessed acetylation of p72 RNA helicase. Like p68, p72 was acetylated in vitro by p300 only in its N-terminal region (Fig. 3C). Mass spectrometric analysis revealed that p72 becomes partially acetylated on three lysine residues in vivo (Lys-29, Lys-30, and Lys-42), which are homologous to Lys-32, Lys-33, and Lys-45 in p68 (see Fig. 3D); please note that no p72 acetylation was observable in the absence of p300 (compare lanes 1 and 2 in Fig. 3E). In contrast to Lys-43 and Lys-44 in p68, no acetylation was observed on the homologous lysine residues Lys-40 and Lys-41 in p72. This could be explained by the facts that p72 has an arginine at position 37 instead of the homologous lysine 40 in p68, and that acetylation on Lys-40 is a prerequisite for Lys-43 and Lys-44 acetylation in p68. Finally, the p72 triple mutant K29/30/42R was no longer acetylated by p300 in vivo as determined by mass spectrometry and immunoblotting (Fig. 3E). Altogether, these data demonstrate that multiple in vivo acetylation sites exist in the N termini of p68 and p72 that may regulate their function.
Impact of Acetylation on p68/p72 Stability
The stability of a protein can be affected by post-translational modifications. One conceivable mechanism is that acetylation prevents the ubiquitylation of the same lysine residue and thus proteasome-mediated degradation, as for instance shown for the Smad7 protein (53). To test whether acetylation of p68/p72 affects their stability, we determined the half-lives of wild-type p68/p72 and their acetylation site mutants by performing pulse-chase experiments with [35S]methionine in HeLa cells. The half-life of p68 decreased from 29.1 h to 13.8 h upon mutation of its acetylation sites (Fig. 4, A and B), indicating that acetylation stabilizes p68. Similarly, the stability of p72 decreased by half upon mutation of its acetylation sites (Fig. 4, A and B). Thus, acetylation significantly stabilizes both p68 and p72. However, this is not due to a competitive inhibition of ubiquitylation at the same lysine residues, because if this were the case, our K → R mutants, which can be neither acetylated nor ubiquitylated at the identified acetylation sites, should be at least as stable as the wild-type p68/p72 proteins. Rather, acetylation might prevent ubiquitylation at lysine residues that are different from the acetylation sites, possibly by precluding the binding of ubiquitin ligases.
FIGURE 4.
Acetylation increases protein stability. Wild-type p68 and p72 or their acetylation site mutants (K32/33/40/45R and K29/30/42R, respectively) were expressed in HeLa cells and protein half-lives determined by pulse-chase experiments. A, representative experiment showing the remaining amount of 35S-labeled p68/p72 proteins at indicated time-points of the chase with non-radioactive methionine. A PhosphorImager was employed to detect and measure the amount of 35S-labeled p68/p72. B, summary of three independent experiments.
Acetylation Selectively Affects Interaction with HDACs
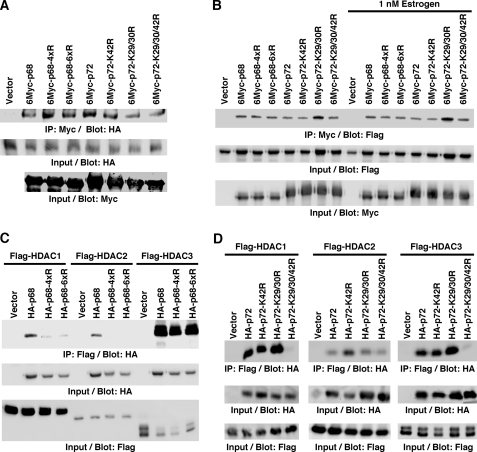
Post-translational modifications are known to influence the physical interaction between proteins. Both p68 and p72 bind to p300 (7, 8), and it is conceivable that acetylation of p68/p72 alters their affinity for p300. However, we observed no significant difference between wild-type p68/p72 and their respective acetylation site mutants in their ability to interact with p300 in coimmunoprecipitation experiments (Fig. 5A). Likewise, acetylation of p68/p72 did not interfere with the reported binding to ERα (9, 14) in the presence or absence of estrogen (Fig. 5B).
FIGURE 5.
Acetylation promotes interaction with selected HDACs. A, interaction with p300. Myc-tagged p68 and p72 were coexpressed with HA-tagged p300. After anti-Myc immunoprecipitation, coprecipitated p300 was detected by anti-HA blotting. The bottom two panels show input levels of HA-tagged p300 and Myc-tagged p68/p72. 4xR and 6xR are the K32/33/40/45R and K32/33/40/43/44/45R mutants of p68, respectively. B, similar, interaction of Myc-tagged p68/p72 with Flag-tagged ERα in the presence and absence of estrogen. C, coimmunoprecipitation of HA-tagged p68 (wild-type or 4xR or 6xR mutant) with Flag-tagged HDACs. D, analogous, analysis of the interaction of p72 with HDACs.
Histone deacetylase (HDAC) 1, 2, and 3 are further interaction partners of p68/p72 (21, 54). Thus, we also tested the impact of acetylation on the ability of p68 and p72 to interact with these HDACs in coimmunoprecipitation assays. Notably, mutation of the p68 acetylation sites diminished its interaction with HDAC1 and 2, but not HDAC3 (Fig. 5C). Acetylation of p72 was likewise important for its interaction with HDAC1, but had no significant effect on HDAC2 binding (Fig. 5D). However, HDAC3 binding was suppressed upon mutation of all p72 acetylation sites, pointing at differences in the molecular consequences of p68 and p72 acetylation. Altogether, these data indicate that acetylation promotes the interaction of p68 and p72 with selected HDACs.
Acetylation of p68/p72 Stimulates Coactivation Potential
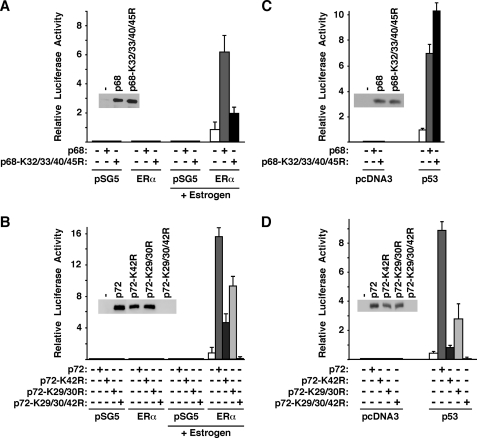
A well-known property of p68 and p72 is their ability to stimulate transcription mediated by ERα (9, 14), and therefore we tested whether this would be affected by acetylation of p68/p72. We employed MDA-MB-231 breast cancer cells that are ERα-negative and the ERE-luc luciferase reporter that is driven by an estrogen response element. Neither in the absence of transfected ERα nor in the absence of estrogen stimulation was luciferase activity measurable. However, when ERα was stimulated with estrogen, robust luciferase activity was observed (Fig. 6A). When wild-type p68 was coexpressed, it stimulated ERα activity ∼6-fold, whereas the K32/33/40/45R mutant raised ERα-dependent transcription only by ∼2-fold. A control Western blot indicated that this was not due to unequal expression of wild-type p68 and its K32/33/40/45R mutant (see inset in Fig. 6A). Thus, acetylation enhances the ability of p68 to coactivate ERα.
FIGURE 6.
Acetylation of p68 and p72 stimulates their coactivation potential. A, MDA-MB-231 cells were transfected with HA-tagged p68 (wild-type or K32/33/40/45R mutant) and ERα or empty expression vector pSG5. Cells were stimulated with estrogen as indicated. Resultant luciferase activities derived from the cotransfected ERE-luc reporter are depicted. The inset is an anti-HA blot showing that comparable amounts of wild-type and K32/33/40/45R p68 were expressed upon ERα cotransfection and estrogen stimulation. B, analogous, activation of the ERE-luc reporter by p72 or three mutants thereof. C, stimulation of the MDM2-luc reporter by p68 in MDA-MB-231 cells. As indicated, p53 or the parental pcDNA3 expression vector was cotransfected. The anti-HA blot in the inset shows the comparable expression of wild-type p68 and its K32/33/40/45R mutant upon p53 coexpression. D, similar, activation of the MDM2-luc reporter by p72.
Similarly, we observed that the ability of p72 to coactivate ERα was markedly reduced upon mutation of the acetylation site Lys-42 (Fig. 6B), whereas mutation of the acetylation sites Lys-29 and Lys-30 resulted in less reduction of p72-mediated coactivation. When we expressed the triple mutant K29/30/42R in MDA-MB-231 cells, massive cell death was induced and accordingly, we were unable to observe the K29/30/42R mutant protein in Western blots or any significant luciferase activity (Fig. 6B), thereby not allowing us to establish whether acetylation on Lys-29/ Lys-30 cooperates with Lys-42 acetylation to stimulate p72 function. Regardless, our data demonstrate that acetylation of p72 stimulates ERα-dependent transcription.
Another gene activated by p68 and p72 is the MDM2 gene, which is a target of the tumor suppressor p53 (8, 10). We observed that p68 synergized with p53 in the activation of the MDM2 promoter in the p53-negative MDA-MB-231 cells, as expected (Fig. 6C). However, mutation of the acetylation sites did not reduce the p68 coactivation potential in this case; if at all, it was slightly enhanced with the K32/33/40/45R mutant. In contrast, mutation of the p72 acetylation sites had the same effect as observed before with the ERE-luc reporter: the K42R mutant was severely impaired in its coactivation potential, and the K29/30R mutant less so (Fig. 6D). Altogether, these data demonstrate that acetylation of p68 and p72 can, but does not necessarily, raise their coactivation potential, pointing to promoter-specific effects of this post-translational modification.
Acetylation of p72 Is Required for Cell Survival
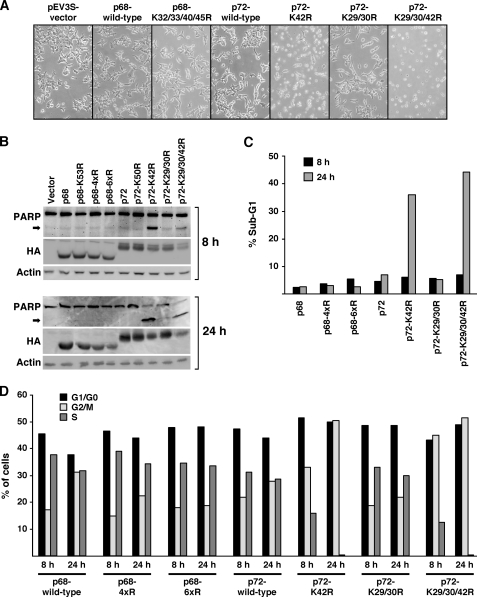
Finally, we asked the question how acetylation of p68 and p72 affects cell physiology. To this end, we transiently transfected 293T cells for 12 h and then imaged cells by phase-contrast microscopy 24 h thereafter. We did not observe any obvious changes in cell morphology when comparing cells transfected with vector control, p68 or its K32/33/40/45R acetylation site mutant (Fig. 7A). However, there was a marked difference between wild-type p72 and its K29/30R mutant on the one hand and the K42R and K29/30/42R mutants on the other hand: round cell bodies were observed with the K42R mutant and even more so with the K29/30/42R mutant, which is indicative of cell death. Indeed, Western blotting revealed that poly(ADP-ribose) polymerase (PARP), a prominent substrate of caspases and early marker of apoptosis, becomes cleaved already 8 h and even more so 24 h after transfection in the presence of the K42R and K29/30/42R mutants of p72 (Fig. 7B). Furthermore, a drastic increase in cells with a sub-G1 DNA content, another marker of apoptosis, was observed 24 h after transfection with the K42R and K29/30/42R mutants of p72 (Fig. 7C). Altogether, these data indicate that acetylation of p72 on K42 is required for 293T cell survival.
FIGURE 7.
Cell survival and cell cycling depend on p72 acetylation. A, 293T cells were transfected with indicated HA-tagged proteins and observed 24 h afterward by phase-contrast microscopy. B, corresponding Western blots showing the expression of PARP 8 h or 24 h after transfection. The arrow points to PARP cleaved by caspases. The bottom panels reveal expression levels of HA-tagged p68/p72 and of endogenous actin. 4xR and 6xR p68 mutants are K32/33/40/45R and K32/33/40/43/44/45R, respectively. Lys-53 in p68 and Lys-50 in p72 are reported sumoylation sites and respective K → R mutants served as a control. C, representative experiment showing the percentage of 293T cells with a sub-G1 DNA content (indicative of apoptosis) 8 h or 24 h after transfection with indicated expression vectors. D, corresponding cell cycle distribution.
Furthermore, we studied the cell cycle distribution of transfected 293T cells by flow cytometry. Whereas there was little, if any, impact of p68 acetylation or acetylation of Lys-29 and Lys-30 in p72 on the distribution of cells in G1/G0, G2/M, and S phase, the K42R and K29/30/42R mutants of p72 showed a significant decrease of cells in S phase already 8 h after transfection, and no S phase cells were observable 24 h after transfection (Fig. 7D). This suggests that acetylation of p72 on Lys-42 is essential for 293T cell cycle progression.
DISCUSSION
In this report, we have identified multiple lysine residues in the N termini of p68 and p72 RNA helicase that are acetylated by p300 in vitro and in vivo. This post-translational modification of p68/p72 has profound effects on their function and is the first demonstration that RNA helicases can be regulated by acetylation.
Post-translational Modifications in p68/p72
The helicase domain residing in the center of p68 and p72 is highly conserved between these two proteins and also among the greater family of RNA helicases (55). Amino acids flanking the helicase domain are much less conserved evolutionary and thought to selectively modulate the activity of RNA helicases (56, 57). One way of functionally regulating a protein is through post-translational modifications. Indeed, we mapped six and three acetylation sites in p68 and p72, respectively, that are all located N-terminal of the helicase domain. Similarly, attachment of SUMO, a small molecule sharing high structural homology with ubiquitin, has been reported to occur on Lys-53 in p68 and on Lys-50 in p72 (21, 52), further suggesting that the N terminus of p68 and p72 is critical for the modulation of their activity.
Blocking acetylation in p68 led to the suppression of its interaction with HDAC1 and HDAC2, but had no effect on the binding to HDAC3, p300 or ERα, whereas acetylation of p72 enhanced its interaction with HDAC1 and HDAC3. This reveals one mechanism of how acetylation may regulate the activity of p68/p72 by selectively modulating the binding to partner proteins. Because two out of five interactions tested in this report were influenced by acetylation of p68 or p72, it is highly likely that acetylation will affect the ability of p68 and p72 to form complexes with many more of the known and to-be-discovered interaction partners and thus constitutes a seminal post-translational modification of p68/p72. Like acetylation, sumoylation promoted the interaction of p68 and p72 with HDAC1, but not with HDAC2 or HDAC3 (21, 52), indicating that acetylation and sumoylation differentially affect p68/p72 function. Indeed, in contrast to acetylation (see Fig. 6B), sumoylation represses the ability of p72 to coactivate ERα (21).
Histone acetylation is generally associated with transcribed regions of the chromatin, suggesting that recruitment of HDACs leads to the repression of gene transcription. Following this argument, the enhanced recruitment of HDAC1 and HDAC2 upon acetylation of p68 and of HDAC1 and HDAC3 upon acetylation of p72 should lead to repression of transcription, but our data indicate that acetylation of p68 and p72 stimulates ERα-dependent transcription. This may argue that enhanced HDAC recruitment is not the prevailing acetylation-induced incident with regard to the coactivation potential of p68/p72. For instance, enhanced HDAC recruitment could be more than counterbalanced by increased coactivator binding upon acetylation of p68/p72. However, recent studies indicated that HDACs may actually activate selected transcriptional complexes (58, 59). If so in case of the ERα/p68/p72 complexes, this would provide a mechanism of how acetylation stimulates the coactivation potential of p68 and p72 through enhanced HDAC recruitment.
Tyrosine phosphorylation of p68 has been reported to be elevated in cancer cells (60). In particular, phosphorylation of Tyr-593 on p68 is induced by platelet-derived growth factor in colon cancer cells and promotes epithelial-mesenchymal-transition (19). Additionally, p68 that is phosphorylated on Tyr-593 and Tyr-595 protects glioblastoma cells from TRAIL-induced apoptosis (61). Thus, tyrosine phosphorylation C-terminal of the p68 helicase domain as well as N-terminal acetylation and sumoylation may jointly govern the activity of p68, and it remains to be determined whether these different post-translational modifications are interdependent.
Enhanced Coactivation Potential and Cell Survival Mediated by p68/p72 Acetylation
Both p68 and p72 physically interact with ERα and thereby stimulate estrogen-dependent transcription (9, 14). However, it has remained unknown how the interaction between p68/p72 and ERα is regulated. Our data indicate that acetylation of p68/p72 does not lead to obvious changes in association with ERα, but enhances their ability to coactivate ERα-mediated transcription. Thus, acetylation of p68/p72 may impact on the pleiotropic effects of estrogen during embryonal development and in adult tissues.
In addition, acetylation of p72 raised its ability to stimulate p53-dependent MDM2 transcription, but p68 acetylation had no significant impact in this regard, marking a clear difference between these two homologous proteins. The tumor suppressor p53 activates the MDM2 promoter, thereby establishing a negative feedback loop, because MDM2 is a ubiquitin ligase causing the destruction of p53 (62–64). Accordingly, p72 acetylation is predicted to strengthen this negative feedback loop leading to less intracellular p53. Therefore, one mechanism of how acetylation of p72 contributes to tumorigenesis may be by reducing levels of the p53 tumor suppressor.
Mutation of Lys-42 alone or more pronouncedly of all three acetylation sites in p72 resulted in cell death and cell cycle blockage in 293T cells, whereas acetylation of p68 had no significant effect. Thus, p72 acetylation is seminal for cell survival and proliferation, an unsuspected result showing, to our knowledge for the first time, an essential function for post-translational modification of an RNA helicase. At present, we do not know why p72 acetylation is required for cell survival, but this is independent of p53, because apoptosis was observed with the K29/30/42R mutant in MDA-MB-231 cells, which are p53-negative, as well as in 293T cells, in which endogenous p53 is incapacitated by the presence of the adenoviral E1A and E1B proteins and SV40 large T antigen. Interestingly, p300-mediated acetylation of p53 at Lys-373 was reported to promote cell death, whereas acetylation of the androgen receptor led to less apoptosis (65, 66). This indicates that p300-mediated acetylation of transcription factors has opposite effects on cell survival depending on which protein becomes acetylated.
Whereas mutation of Lys-42 in p72 was sufficient to induce massive apoptosis and stop cell proliferation in 293T cells, it did not do so in MDA-MB-231 cells. However, blocking acetylation of Lys-29, Lys-30, and Lys-42 together resulted also in cell death in MDA-MB-231 cells, and even in 293T cells it was slightly more detrimental than mutating Lys-42 alone. Thus, there are cell type-specific differences in which sites have to be acetylated to prevent p72 from inducing apoptosis. In the same vein, we did not observe increased apoptosis with any of the p72 acetylation site mutants in HeLa cells (this being the reason why we chose this cell line to determine half-lives of p68 and p72 by pulse-chase experiments), implicating that acetylation of the ubiquitously expressed p72 RNA helicase is not always essential for cell survival.
At present, it remains unknown why acetylation of p68 compared with p72 has different consequences with regard to MDM2 transcription and cell survival. Although p68 and p72 share 92% similarity within their helicase domains, their N- and C-terminal domains are much less conserved (15). Thus, it is likely that p68 and p72 are recruited into different protein complexes by virtue of these divergent domains, and therefore acetylation of these paralogous RNA helicases may be futile in one case (e.g. p68), but crucial in the other case (e.g. p72) to affect biological consequences. Future studies should comprehensively explore the different spectrum of interactants for p68/p72 and determine how their acetylation modulates the activity of p68/p72-containing protein complexes.
Role of p68/p72 Acetylation in Tumors
Raf, Ras, and HER2/Neu stimulate the enzymatic activity of p300, most likely by inducing its MAP kinase-dependent phosphorylation (33). Accordingly, we observed that p68 acetylation was enhanced upon overexpression of these oncoproteins, mutations of which are found in the majority of all human tumors. Thus, p68/p72 acetylation levels are predicted to be enhanced in and therefore potentially contribute to the genesis of many different tumors.
Estrogen plays a causal role in the development of ∼70% of all breast tumors and drugs targeting estrogen metabolism or ERα are a mainstay in breast cancer therapy (67, 68). ERα-positive breast tumors often display hyperactivation of MAP kinases, in part due to the non-genomic actions of estrogen (69, 70). Thus, MAP kinase-mediated stimulation of p300 enzymatic activity is expected to enhance p68/p72 acetylation in ERα-positive breast tumors, which will lead to increased activation of ERα, providing one mechanism how post-translational modification of p68/p72 fosters breast carcinogenesis. In addition, enhanced acetylation of p68 and p72, which significantly increases their stability, may (partially) account for the fact that p68 and p72 are overexpressed in human breast tumors (20, 21). Consistently, we observed that overexpression of Raf1 in MCF7 breast cancer cells up-regulated both p68 and p72 at the protein level (see supplemental Fig. S1).
Approximately 30% of all breast tumors display HER2/Neu overexpression, and targeting this receptor with monoclonal antibodies is now an established practice in the treatment of breast tumors (71, 72). Because HER2/Neu stimulates the acetyltransferase activity of p300 (33), acetylation of p68/p72 should also be enhanced in HER2/Neu-positive breast tumors. However, these breast tumors are normally ERα-negative, precluding that acetylation of p68/p72 affects HER2/Neu-positive breast tumors by stimulating ERα. But enhanced acetylation of p72 could still increase MDM2 transcription, thereby leading to MDM2-mediated destruction of the tumor suppressor p53 and thus promoting tumorigenesis. And indeed, reduced p53 levels are observed in many of the most malignant, HER2/Neu-positive breast tumors (73).
Recently, it was shown that p68 is a cofactor of androgen receptor and overexpressed in prostate tumors (12). Moreover, HER2/Neu overexpression is also common in prostate cancer and increases with the progression of the disease (74–76). Again, HER2/Neu-stimulated p300 acetyltransferase activity may raise the coactivation potential of p68 and thereby its tumor promoting activity by stimulating the androgen receptor, the key villain in prostate cancer.
Both p68 and p72 are overexpressed in colorectal tumors and their expression levels correlate with the progression of the disease (17, 18). Knocking down p68 and p72 together in colon cancer cells resulted in reduced cell proliferation and tumor formation, indicating that they are causally involved in colon carcinogenesis. Moreover, p68 mediates epithelial-mesenchymal-transition in colon cancer cells, which is particularly important during metastasis. All of these processes are most likely related to the p68/p72-mediated activation of β-catenin, the linchpin of colon carcinogenesis (18, 19, 61). Notably, mutations in K-Ras and B-Raf (77–79) as well as overexpression of the receptor tyrosine kinase HER2/Neu (80–82) are often found in colorectal tumors. Our data suggest that one mechanism of how K-Ras, B-Raf, and HER2/Neu drive colon carcinogenesis is through stimulating p300 enzymatic activity and the resultant acetylation of p68/p72. This is predicted to increase p68 and p72 protein stability and thus account, at least in part, for the observed overexpression of p68 and p72 in colorectal tumors. And overexpression of p68 or p72 alone is sufficient to aberrantly activate β-catenin (18), thereby contributing to colon cancer formation.
CONCLUSION
In this report, we have demonstrated for the first time that two members of the RNA helicase superfamily are regulated by acetylation of lysine residues. Acetylation stimulates the coactivation potential of p68 and p72, modulates their interaction with partner proteins, enhances their protein stability and prevents p72-mediated cell cycle arrest and apoptosis, all of which potentially promotes tumorigenesis. Because oncogenic HER2/Neu, Ras, or Raf as well as estrogen can enhance p300-mediated acetylation of p68 and p72, this provides a novel pathway of how these oncoproteins and estrogen may exert their deleterious effects in tumor cells.
Supplementary Material
Acknowledgments
We thank Benjamin Madden for performing mass spectrometry. We are also grateful for help provided by the Mayo Clinic Flow Cytometry Core.
This work was supported by Grant W81XWH-06-1-0492 from the Dept. of Defense Breast Cancer Research Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- DDX
- DEAD-box protein
- ERα
- estrogen receptor α
- HDAC
- histone deacetylase
- PARP
- poly(ADP-ribose) polymerase
- TORU
- TPA oncogene responsive unit.
REFERENCES
- 1.Lee K. K., Workman J. L. (2007) Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 2.Yang X. J., Seto E. (2008) Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janknecht R., Hunter T. (1996) Curr. Biol. 6, 951–954 [DOI] [PubMed] [Google Scholar]
- 4.Goodman R. H., Smolik S. (2000) Genes Dev. 14, 1553–1577 [PubMed] [Google Scholar]
- 5.Janknecht R. (2002) Histol. Histopathol. 17, 657–668 [DOI] [PubMed] [Google Scholar]
- 6.Iyer N. G., Ozdag H., Caldas C. (2004) Oncogene 23, 4225–4231 [DOI] [PubMed] [Google Scholar]
- 7.Rossow K. L., Janknecht R. (2003) Oncogene 22, 151–156 [DOI] [PubMed] [Google Scholar]
- 8.Shin S., Janknecht R. (2007) J. Cell. Biochem. 101, 1252–1265 [DOI] [PubMed] [Google Scholar]
- 9.Endoh H., Maruyama K., Masuhiro Y., Kobayashi Y., Goto M., Tai H., Yanagisawa J., Metzger D., Hashimoto S., Kato S. (1999) Mol. Cell. Biol. 19, 5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Bates G. J., Nicol S. M., Wilson B. J., Jacobs A. M., Bourdon J. C., Wardrop J., Gregory D. J., Lane D. P., Perkins N. D., Fuller-Pace F. V. (2005) EMBO J. 24, 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caretti G., Schiltz R. L., Dilworth F. J., Di Padova M., Zhao P., Ogryzko V., Fuller-Pace F. V., Hoffman E. P., Tapscott S. J., Sartorelli V. (2006) Dev. Cell 11, 547–560 [DOI] [PubMed] [Google Scholar]
- 12.Clark E. L., Coulson A., Dalgliesh C., Rajan P., Nicol S. M., Fleming S., Heer R., Gaughan L., Leung H. Y., Elliott D. J., Fuller-Pace F. V., Robson C. N. (2008) Cancer Res. 68, 7938–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warner D. R., Bhattacherjee V., Yin X., Singh S., Mukhopadhyay P., Pisano M. M., Greene R. M. (2004) Biochem. Biophys. Res. Commun. 324, 70–76 [DOI] [PubMed] [Google Scholar]
- 14.Watanabe M., Yanagisawa J., Kitagawa H., Takeyama K., Ogawa S., Arao Y., Suzawa M., Kobayashi Y., Yano T., Yoshikawa H., Masuhiro Y., Kato S. (2001) EMBO J. 20, 1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Janknecht R. (2010) Am. J. Transl. Res. 2, 223–234 [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda T., Yamagata K., Fujiyama S., Matsumoto T., Koshida I., Yoshimura K., Mihara M., Naitou M., Endoh H., Nakamura T., Akimoto C., Yamamoto Y., Katagiri T., Foulds C., Takezawa S., Kitagawa H., Takeyama K., O'Malley B. W., Kato S. (2007) Nat. Cell Biol. 9, 604–611 [DOI] [PubMed] [Google Scholar]
- 17.Causevic M., Hislop R. G., Kernohan N. M., Carey F. A., Kay R. A., Steele R. J., Fuller-Pace F. V. (2001) Oncogene 20, 7734–7743 [DOI] [PubMed] [Google Scholar]
- 18.Shin S., Rossow K. L., Grande J. P., Janknecht R. (2007) Cancer Res. 67, 7572–7578 [DOI] [PubMed] [Google Scholar]
- 19.Yang L., Lin C., Liu Z. R. (2006) Cell 127, 139–155 [DOI] [PubMed] [Google Scholar]
- 20.Wortham N. C., Ahamed E., Nicol S. M., Thomas R. S., Periyasamy M., Jiang J., Ochocka A. M., Shousha S., Huson L., Bray S. E., Coombes R. C., Ali S., Fuller-Pace F. V. (2009) Oncogene 28, 4053–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mooney S. M., Grande J. P., Salisbury J. L., Janknecht R. (2010) Biochemistry 49, 1–10 [DOI] [PubMed] [Google Scholar]
- 22.Shin S., Kim T. D., Jin F., van Deursen J. M., Dehm S. M., Tindall D. J., Grande J. P., Munz J. M., Vasmatzis G., Janknecht R. (2009) Cancer Res. 69, 8102–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monté D., Coutte L., Baert J. L., Angeli I., Stéhelin D., de Launoit Y. (1995) Oncogene 11, 771–779 [PubMed] [Google Scholar]
- 24.De Haro L., Janknecht R. (2005) Genomics 85, 493–502 [DOI] [PubMed] [Google Scholar]
- 25.Wu J., Janknecht R. (2002) J. Biol. Chem. 277, 42669–42679 [DOI] [PubMed] [Google Scholar]
- 26.Fuchs B., Inwards C. Y., Janknecht R. (2004) Clin. Cancer Res. 10, 1344–1353 [DOI] [PubMed] [Google Scholar]
- 27.Goueli B. S., Janknecht R. (2003) Oncogene 22, 8042–8047 [DOI] [PubMed] [Google Scholar]
- 28.Paech K., Webb P., Kuiper G. G., Nilsson S., Gustafsson J., Kushner P. J., Scanlan T. S. (1997) Science 277, 1508–1510 [DOI] [PubMed] [Google Scholar]
- 29.Ries S., Biederer C., Woods D., Shifman O., Shirasawa S., Sasazuki T., McMahon M., Oren M., McCormick F. (2000) Cell 103, 321–330 [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R. (2001) J. Biol. Chem. 276, 41856–41861 [DOI] [PubMed] [Google Scholar]
- 31.Janknecht R. (2003) Oncogene 22, 746–755 [DOI] [PubMed] [Google Scholar]
- 32.Goueli B. S., Janknecht R. (2004) Mol. Cell. Biol. 24, 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goel A., Janknecht R. (2003) Mol. Cell. Biol. 23, 6243–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janknecht R., Hunter T. (1997) EMBO J. 16, 1620–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janknecht R., Wells N. J., Hunter T. (1998) Genes Dev. 12, 2114–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Block C., Janknecht R., Herrmann C., Nassar N., Wittinghofer A. (1996) Nat. Struct. Biol. 3, 244–251 [DOI] [PubMed] [Google Scholar]
- 37.Bruder J. T., Heidecker G., Rapp U. R. (1992) Genes Dev. 6, 545–556 [DOI] [PubMed] [Google Scholar]
- 38.Ben-Levy R., Paterson H. F., Marshall C. J., Yarden Y. (1994) EMBO J. 13, 3302–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knebel J., De Haro L., Janknecht R. (2006) J. Cell. Biochem. 99, 319–329 [DOI] [PubMed] [Google Scholar]
- 40.Goel A., Janknecht R. (2004) J. Biol. Chem. 279, 14909–14916 [DOI] [PubMed] [Google Scholar]
- 41.Janknecht R. (1996) Mol. Cell. Biol. 16, 1550–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bosc D. G., Goueli B. S., Janknecht R. (2001) Oncogene 20, 6215–6224 [DOI] [PubMed] [Google Scholar]
- 43.Papoutsopoulou S., Janknecht R. (2000) Mol. Cell. Biol. 20, 7300–7310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim T. D., Shin S., Janknecht R. (2008) Biochem. Biophys. Res. Commun. 366, 563–567 [DOI] [PubMed] [Google Scholar]
- 45.Shin S., Bosc D. G., Ingle J. N., Spelsberg T. C., Janknecht R. (2008) J. Cell. Biochem. 105, 866–874 [DOI] [PubMed] [Google Scholar]
- 46.Shin S., Janknecht R. (2007) Biochem. Biophys. Res. Commun. 359, 742–746 [DOI] [PubMed] [Google Scholar]
- 47.Rossow K. L., Janknecht R. (2001) Cancer Res. 61, 2690–2695 [PubMed] [Google Scholar]
- 48.Shin S., Janknecht R. (2007) Biochem. Biophys. Res. Commun. 353, 973–977 [DOI] [PubMed] [Google Scholar]
- 49.Bosc D. G., Janknecht R. (2002) J. Cell. Biochem. 86, 174–183 [DOI] [PubMed] [Google Scholar]
- 50.Dowdy S. C., Mariani A., Janknecht R. (2003) J. Biol. Chem. 278, 44377–44384 [DOI] [PubMed] [Google Scholar]
- 51.Boffa L. C., Vidali G., Mann R. S., Allfrey V. G. (1978) J. Biol. Chem. 253, 3364–3366 [PubMed] [Google Scholar]
- 52.Jacobs A. M., Nicol S. M., Hislop R. G., Jaffray E. G., Hay R. T., Fuller-Pace F. V. (2007) Oncogene 26, 5866–5876 [DOI] [PubMed] [Google Scholar]
- 53.Grönroos E., Hellman U., Heldin C. H., Ericsson J. (2002) Mol. Cell 10, 483–493 [DOI] [PubMed] [Google Scholar]
- 54.Wilson B. J., Bates G. J., Nicol S. M., Gregory D. J., Perkins N. D., Fuller-Pace F. V. (2004) BMC Mol. Biol. 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamm G. M., Nicol S. M., Fuller-Pace F. V., Lamond A. I. (1996) Nucleic Acids Res. 24, 3739–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rocak S., Linder P. (2004) Nat. Rev. Mol. Cell Biol. 5, 232–241 [DOI] [PubMed] [Google Scholar]
- 57.Bleichert F., Baserga S. J. (2007) Mol. Cell 27, 339–352 [DOI] [PubMed] [Google Scholar]
- 58.Nusinzon I., Horvath C. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zupkovitz G., Tischler J., Posch M., Sadzak I., Ramsauer K., Egger G., Grausenburger R., Schweifer N., Chiocca S., Decker T., Seiser C. (2006) Mol. Cell. Biol. 26, 7913–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L., Lin C., Liu Z. R. (2005) Mol. Cancer Res. 3, 355–363 [DOI] [PubMed] [Google Scholar]
- 61.Yang L., Lin C., Sun S. Y., Zhao S., Liu Z. R. (2007) Oncogene 26, 6082–6092 [DOI] [PubMed] [Google Scholar]
- 62.Bond G. L., Hu W., Levine A. J. (2005) Curr. Cancer Drug Targets 5, 3–8 [DOI] [PubMed] [Google Scholar]
- 63.Brooks C. L., Gu W. (2006) Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vazquez A., Bond E. E., Levine A. J., Bond G. L. (2008) Nat. Rev. Drug Discov. 7, 979–987 [DOI] [PubMed] [Google Scholar]
- 65.Fu M., Rao M., Wang C., Sakamaki T., Wang J., Di Vizio D., Zhang X., Albanese C., Balk S., Chang C., Fan S., Rosen E., Palvimo J. J., Jänne O. A., Muratoglu S., Avantaggiati M. L., Pestell R. G. (2003) Mol. Cell. Biol. 23, 8563–8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knights C. D., Catania J., Di Giovanni S., Muratoglu S., Perez R., Swartzbeck A., Quong A. A., Zhang X., Beerman T., Pestell R. G., Avantaggiati M. L. (2006) J. Cell Biol. 173, 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yager J. D., Davidson N. E. (2006) N. Engl. J. Med. 354, 270–282 [DOI] [PubMed] [Google Scholar]
- 68.Normanno N., Di Maio M., De Maio E., De Luca A., de Matteis A., Giordano A., Perrone F. (2005) Endocr. Relat. Cancer 12, 721–747 [DOI] [PubMed] [Google Scholar]
- 69.Santen R. J., Song R. X., McPherson R., Kumar R., Adam L., Jeng M. H., Yue W. (2002) J. Steroid Biochem. Mol. Biol. 80, 239–256 [DOI] [PubMed] [Google Scholar]
- 70.Castoria G., Migliaccio A., D'Amato L., Di Stasio R., Ciociola A., Lombardi M., Bilancio A., Di Domenico M., de Falco A., Auricchio F. (2008) Front. Biosci. 13, 1318–1327 [DOI] [PubMed] [Google Scholar]
- 71.Hynes N. E., Lane H. A. (2005) Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 72.Hall P. S., Cameron D. A. (2009) Eur. J. Cancer 45, 12–18 [DOI] [PubMed] [Google Scholar]
- 73.Lacroix M., Toillon R. A., Leclercq G. (2006) Endocr. Relat. Cancer 13, 293–325 [DOI] [PubMed] [Google Scholar]
- 74.Signoretti S., Montironi R., Manola J., Altimari A., Tam C., Bubley G., Balk S., Thomas G., Kaplan I., Hlatky L., Hahnfeldt P., Kantoff P., Loda M. (2000) J. Natl. Cancer Inst. 92, 1918–1925 [DOI] [PubMed] [Google Scholar]
- 75.Osman I., Mikhail M., Shuch B., Clute M., Cheli C. D., Ghani F., Thiel R. P., Taneja S. S. (2005) J. Urol. 174, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 76.Nishio Y., Yamada Y., Kokubo H., Nakamura K., Aoki S., Taki T., Honda N., Nakagawa A., Saga S., Hara K. (2006) Urology 68, 110–115 [DOI] [PubMed] [Google Scholar]
- 77.Bos J. L., Fearon E. R., Hamilton S. R., Verlaan-de Vries M., van Boom J. H., van der Eb A. J., Vogelstein B. (1987) Nature 327, 293–297 [DOI] [PubMed] [Google Scholar]
- 78.Forrester K., Almoguera C., Han K., Grizzle W. E., Perucho M. (1987) Nature 327, 298–303 [DOI] [PubMed] [Google Scholar]
- 79.Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B. A., Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G. J., Bigner D. D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J. W., Leung S. Y., Yuen S. T., Weber B. L., Seigler H. F., Darrow T. L., Paterson H., Marais R., Marshall C. J., Wooster R., Stratton M. R., Futreal P. A. (2002) Nature 417, 949–954 [DOI] [PubMed] [Google Scholar]
- 80.Kapitanović S., Radosević S., Kapitanović M., Andelinović S., Ferencić Z., Tavassoli M., Primorać D., Sonicki Z., Spaventi S., Pavelic K., Spaventi R. (1997) Gastroenterology 112, 1103–1113 [DOI] [PubMed] [Google Scholar]
- 81.Osako T., Miyahara M., Uchino S., Inomata M., Kitano S., Kobayashi M. (1998) Oncology 55, 548–555 [DOI] [PubMed] [Google Scholar]
- 82.Park D. I., Kang M. S., Oh S. J., Kim H. J., Cho Y. K., Sohn C. I., Jeon W. K., Kim B. I., Han W. K., Kim H., Ryu S. H., Sepulveda A. R. (2007) Int. J. Colorectal Dis. 22, 491–497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.