Abstract
Hepatic stellate cells (HSCs), vitamin A-storing liver pericytes, undergo myofibroblastic trans-differentiation or “activation” to participate in liver wound healing. This cellular process involves loss of regulation by adipogenic transcription factors such as peroxisome proliferator-activated receptor γ (PPARγ). Necdin, a melanoma antigen family protein, promotes neuronal and myogenic differentiation while inhibiting adipogenesis. The present study demonstrates that necdin is selectively expressed in HSCs among different liver cell types and induced during their activation in vitro and in vivo. Silencing of necdin with adenovirally expressed shRNA, reverses activated HSCs to quiescent cells in a manner dependent on PPARγ and suppressed canonical Wnt signaling. Promoter analysis, site-directed mutagenesis, and chromatin immunoprecipitation demonstrate that Wnt10b, a canonical Wnt induced in activated HSCs, is a direct target of necdin. Necdin silencing abrogates three epigenetic signatures implicated in repression of PPARγ: increased MeCP2 (methyl CpG binding protein 2) and HP-1α co-repressor recruitments to Pparγ promoter and enhanced H3K27 dimethylation at the exon 5 locus, again in a manner dependent on suppressed canonical Wnt. These epigenetic effects are reproduced by antagonism of canonical Wnt signaling with Dikkopf-1. Our results demonstrate a novel necdin-Wnt pathway, which serves to mediate antiadipogenic HSC trans-differentiation via epigenetic repression of PPARγ.
Keywords: Adipocyte, Cell Differentiation, Epigenetics, Fibrinogenesis, PPAR, H3K27-Dimethylation, MeCP2, PPAR-γ, Wnt10b
Introduction
Hepatic stellate cells (HSCs)2 are desmin-positive mesenchymal cells located in the subendothelial (perisinusoidal) space of hepatic sinusoids. They store vitamin A, function as hepatic pericytes, communicate with hepatocytes via a gap junction and the release of soluble factors, and produce the components of the normal matrix milieu of the perisinusoidal space. HSCs also actively participate in matrix remodeling and wound healing through their production of matrix metalloproteinases and extracellular matrix proteins and are considered as one of the principal cell types responsible for the genesis of liver fibrosis (1). The most unique feature of the manner in which HSCs participate in liver fibrogenesis is their myofibroblastic trans-differentiation (activation) characterized by depletion of vitamin A storage, cell proliferation, induction of matrix genes, and acquisition of the myofibroblastic phenotype via induction of α-smooth muscle actin. Although major classes of mediators are identified for HSC trans-differentiation, including soluble factors (cytokines, hormones, and lipid mediators), reactive oxygen species, and altered extracellular matrix milieu (1), the fundamental understanding of cell lineage and cell fate regulation of HSCs is elusive. In this regard, Asahina et al. (2) has demonstrated recently the mesenchymal origin of fetal HSCs and “submesothelial cell” as a potential precursor for HSCs. Intriguingly, HSCs express markers for cell types derived from multipotent mesenchymal progenitor cells such as neural cells, chondrocytes, osteoblasts, smooth muscle cells, and adipocytes (3), suggesting the HSC lineage may indeed be located somewhere within these mesenchymal lineages.
In 1999, we proposed that HSC-myofibroblastic trans-differentiation may be similar to adipocyte-preadipocyte de-differentiation (8). Arguments for this notion included: 1) quiescent HSC store substantial amounts of triglycerides besides vitamin A-like adipocytes (4); 2) both quiescent HSC and adipocytes express type IV collagen, whereas activated HSC and preadipocytes express interstitial collagens; 3) cytokines (TNF-α and leptin) or growth factors (EGF/TGFα and TGFβ), which are known to inhibit adipocyte differentiation, also activate HSCs (5); and 4) adipocyte-specific genes such as leptin are expressed by HSCs (6). Indeed, the expression of PPARγ, a master regulator for adipocyte differentiation (7), is reduced in activated HSCs (8), and the ligands for this nuclear receptor inhibit HSC activation in vitro (9, 10). Furthermore, ectopic expression of PPARγ or SREBP-1c (sterol regulatory element binding protein-1), another adipogenic transcription factor, or treatment with the adipocyte differentiation mixture, reverses activated HSCs to quiescent cells with the restored capacity to store vitamin A (9, 10). In further support of our notion, activated HSCs show induced expression of canonical Wnt proteins such as Wnt10b and Wnt3a (11), which are known to suppress adipocyte differentiation via inhibition of the adipogenic transcription factors C/EBPα and PPARγ (12, 13). Forced expression of the Wnt co-receptor antagonist Dikkopf-1 (Dkk-1) reduces increased nuclear β-catenin, restores expression of PPARγ, and converts culture-activated HSCs to quiescent lipid-storing cells (11).
Necdin is a maternally imprinted gene, which encodes a 325-amino acid protein expressed in neurons, skeletal and smooth muscle cells, chondrocytes, adipocytes, and skin fibroblasts (14–19). Necdin belongs to the melanoma antigen family of proteins, which has a conserved central region termed the melanoma antigen homology domain. Necdin knock-out mice exhibit a phenotype similar to the neurobehavioral disorder Prader Willi syndrome, suggesting necdin deficiency impairs neuronal development (20). Indeed, necdin enhances post-mitotic differentiation of neurons (21). Necdin also promotes differentiation of skeletal (17) and smooth muscle (18) cells while inhibiting adipocyte differentiation (19). Some of these cell fate regulations are mediated at least in part via the interaction of necdin with Wnt1 promoter through homeodomain proteins such as Msx2 and Dlx2 (17, 21).
Based on the tissue expression pattern and antiadipogenic action of necdin, and its cell fate regulation based on canonical Wnt signaling, we hypothesized that necdin is expressed in HSCs to regulate their trans-differentiation. Indeed, our study demonstrates that necdin is expressed selectively in HSCs among different liver cell types and induced in HSC trans-differentiation. Its causal role in trans-differentiation is supported by morphologic and biochemical reversal of culture-activated HSCs to quiescent cells by necdin silencing, and this effect is shown dependent on PPARγ and inhibition of canonical Wnt expression. Furthermore, the necdin-Wnt pathway causes epigenetic repression of PPARγ, which underlies HSC trans-differentiation.
MATERIALS AND METHODS
Cell Isolation and Culture
HSCs were isolated from normal male Wistar rats as described previously (22) by the Non-Parenchymal Liver Cell Core of the Southern California Research Center for Alcoholic Liver and Pancreatic Diseases and Cirrhosis. HSCs also were isolated from rats with liver fibrosis resulting from 10-day cholestasis due to ligation of the common bile duct or with hepatotoxic liver fibrosis induced by provision of phenobarbital in drinking water (500 mg/liter) plus subcutaneous injection of CCl4 (0.1 ml/100 g of body weight) given as a 2-fold dilution with mineral oil twice a week for 3 weeks. The use of animals for this study was approved by the Institutional Animal Care and Use Committee of the University of Southern California and Department of Veterans Affairs Greater Los Angeles Healthcare System. HSCs isolated from normal rats were cultured on plastic with low glucose DMEM supplemented with 10% FBS and antibiotics for 1 day or 7 days for analysis of quiescent or activated HSCs. HSCs were isolated from the liver fibrosis models, cultured on plastic in the medium containing 2% FBS, and analyzed immediately after overnight culture. Cell morphology was assessed by phase contrast microscopy, intracellular vitamin A content by UV-excited autofluorescence, and intracellular lipid content by oil red O staining. For promoter analysis via transient transfection, the spontaneously immortalized cell line (BSC) established from experimental cholestatic liver fibrosis (23) or the Huh7 hepatoma cell line, which does not express necdin, was used. Kupffer cells were isolated by an essentially identical procedure except for the use of the cells at the arabinogalactan gradient interface of 1.043/1.058 and 1.058/1.075 and subsequent adherence method as described previously (24). Hepatocytes were isolated by the standard method of in situ collagenase digestion of the liver and low speed centrifugation (50 × g for 1 min). Sinusoidal endothelial cells were isolated by magnetic cell sorting using SE-1 antibody as described previously (25). Purity of the cells isolated exceeded 95% for all cell types.
RNA Extraction and Real-time PCR
Total RNA was extracted from the cells using the RNeasy mini kit (Qiagen). One microgram of RNA was reverse-transcribed to cDNA by using SuperScript III First-Strand Synthesis System (Invitrogen) and amplified by 40 cycles using primers listed below and the SYBR Green PCR Master mix reagent (Applied Biosystems). Each Ct value was first normalized to 36B4 Ct value and compared between the treatment and control samples. Primer sequences used were as follows: PPARγ, 5′-CTGAAGCTCCAAGAATACCAAA; and 5′-AGAGTTGGGTTTTTTCAGAATAATAAGG; LXRα, 5′-GGCCCTGCATGCCTATGT and 5′-CATTAGCATCCGTGGGAACA; α1(I) collagen, 5′-TCGATTCACCTACAGCACGC and 5′-GACTGTCTTGCCCCAAGTTCC; C/EBPβ, 5′-AAGAAGTCGGTGGATAAGAACAG and 5′-GTTGCGCTGTTTGGCTTTATCTC; ACC, 5′-CCCAACAGAATAAAGCTACTCTGG and 5′-TCCTTTTGTGCAACTAGGAACGT; fatty acid synthase, 5′-CCTGGACAGCATTCCAAACCT and 5′-AGCACATCTCGAAGGCTACACA; 36B4, 5′-TTCCCACTGGCTGAAAAGGT and 5′-CGCAGCCGCAAATGC.
Immunoblot Analysis
HSCs were cultured in a 10-cm dish for 7 days followed by infection with Ad.LacZ.shRNA or Ad.Necdin.shRNA described below at 100 multiplicity of infection (MOI) for an additional 3 days. The cells were then washed with PBS once, and nuclear and cytosolic proteins were isolated as described previously (26). An equal amount of the nuclear or cytosolic extract (20 μg) was separated by SDS-PAGE and electroblotted to nitrocellulose membranes. Necdin or β-catenin were detected by incubating with rabbit polyclonal anti-necdin antibody (Abcam) or rabbit polyclonal anti-β-catenin antibody (Santa Cruz Biotechnology) at a concentration of 0.2–2 μg/10 ml in TBS (100 mm Tris-HCl, 1.5 m NaCl, pH 7.4) with 5% nonfat milk followed by incubation with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) at 1 μg/10 ml. Proteins were detected by a chemiluminescent method using the Pierce ECL kit (Amersham Biosciences).
Construction and Use of Recombinant Adenovirus Vectors
Replication-incompetent recombinant adenovirus expressing necdin shRNA was constructed using BLOCK-iT Adenoviral RNAi expression system (Invitrogen) according to the manufacturer's instructions. To select the best shRNA against the rat necdin gene, we first designed four shRNA oligonucleotides by using the Invitrogen shRNA designer. Of these, one starting at +916 (5′-GGCCAGAGTCTTTAAGAAGGA-3′) was shown to be most effective. An additional sequence of CACC was added at the 5′ end, and AAAA was added to the 5′ end of the complementary sequence. These two DNA oligonucleotides were annealed to generate dsDNA, which was subsequently cloned into the pENTR/U6 vector using the BLOCK-iT U6 RNAi Entry vector kit. The U6 RNAi cassette in the pENTR/U6 necdin shRNA vector was transferred to the adenoviral expression plasmid by LR recombination reaction using the Gateway LR Clonase II enzyme mix and pAd/BLOCK-iT-DEST Gateway Vector kit. Isolated adenoviral expression clones were then digested with PacI to expose the inverted terminal repeats and transfected into 293A cells using Targefect F-2 (Advanced Targeting Systems, San Diego, CA) for production of a crude adenoviral stock. Large-scale amplification of adenoviral vector was performed in 293A cells as described previously (26, 27). The titer of the purified virus was determined by the standard plaque-forming assay with 293A cells. An adenovirus expressing β-galactosidase shRNA (Ad.LacZ.shRNA) was constructed as a control shRNA vector. Necdin-silencing efficiency was tested in day 6 culture-activated rat HSCs with an MOI of 50, 100, and 200. Adenovirus expressing GFP (26), a dominant-negative mutant of PPARγ (a gift from Dr. Krishna K. Chatterjee of the University of Cambridge) (28), or Dkk-1 (a gift from Dr. Calvin Kao, Stanford University), were similarly amplified and purified, and their titers were determined for the use at an MOI of 50 or 100.
Transient Transfection and Reporter Gene Assay
To determine whether necdin silencing affects Wnt10b promoter activity, the rat HSC cell line BSC was transiently transfected with the Wnt10b promoter (−705/+33)-luciferase construct (a gift from Dr. Dwight J. Klemm, University of Michigan) by using Targefect F-2 (Advanced Targeting Systems) followed 6 h later by infection with Ad.LacZ.shRNA or Ad.Necdin.shRNA at 100 MOI for 3 days. The cell lysate was collected for determination of both firefly and Renilla luciferase activities using the Dual-Luciferase reporter assay system (Promega), and the results were normalized by Renilla luciferase activity. Site-directed mutagenesis was performed at the most proximal GN box at −108/−96 within the Wnt10b proximal promoter using the QuikChange site-directed mutagenesis kit (Strategene, La Jolla, CA), and these mutants were tested for necdin-induced promoter activity in Huh7 cells, which do not express necdin.
ChIP
For assessing necdin recruitment to Wnt10b promoter, carrier ChIP was performed using Raj cells as the source of carrier chromatin. For this analysis, Raji cells (1.4 × 107) were added to cultured HSCs (0.2 × 106 cells) with or without necdin silencing or Wnt3 treatment and were fixed briefly with 1% formaldehyde on the rotating platform for 5–10 min at room temperature followed by addition of glycine to a final concentration of 0.125 m. Scraped HSCs and Raji cells in the media were spun down and washed with cold PBS with protease inhibitors. After lysis of the cells with SDS buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1) with protease inhibitors, the lysate was sonicated and snap-frozen in aliquots. For chromatin IP, diluted samples were first precleared with protein G-agarose and then incubated with antibody against necdin, MeCP2, HP-1α, or H3K27me2 (Abcam) at 1 μg/μl at 4 °C for overnight followed by precipitation with protein G-agarose. After elution of immunoprecipitated complex, cross-linking was reversed with 5 n NaCl, and proteins were digested with protease K. Extracted chromatin was subjected to real-time PCR using the primers flanking a segment within Pparγ promoter or Pparγ exon 5 as described recently (29), or five overlapping segments of the proximal Wnt10b promoter (−143/+42, −268/−119, −402/−248, −597/−383, −703/−574). Ct values of the samples with non-immune IgG were subtracted and compared with their respective input Ct values.
RESULTS
Necdin Is Selectively Expressed in HSCs
We first examined the expression of necdin in four liver cell types isolated from the rat, HSCs, Kupffer cells, sinusoidal endothelial cells, and hepatocytes, which collectively constitute 90∼95% of cells in the liver. Real-time PCR analysis revealed necdin mRNA was expressed at the highest level in HSCs among these cell types. The levels in Kupffer cells, sinusoidal endothelial cells, and hepatocytes were 3∼12% of that detected in HSCs (Fig. 1A). This selective expression is confirmed by immunoblot analysis, which detected a strong ∼38 kD band in HSCs but a very faint band in Kupffer and sinusoidal endothelial cells (Fig. 1B).
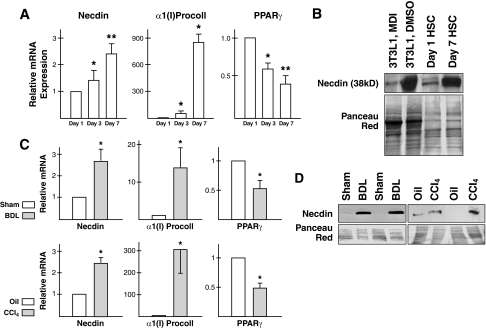
FIGURE 1.
Necdin mRNA (A) and protein (B) are selectively expressed in HSCs among different liver cell types. KC, Kupffer cells; SEC, sinusoidal endothelial cells; Hep, hepatocytes.
Necdin Is Induced in Activated HSCs
Next, we examined the expression of necdin in culture-activated HSCs. When primary HSCs are cultured on plastic, the cells undergo spontaneous activation on days 3 and 7 after plating as characterized by induction of the bona fide HSC activation markers, α1(I) procollagen and down-regulation of Pparγ, the gene that confers HSC quiescence (Fig. 2A) (26, 27). The necdin mRNA level is induced as rat primary HSCs become activated on days 3 and 7. Necdin protein level also is conspicuously up-regulated in day 7 culture-activated HSCs as compared with day 1 quiescent HSCs (Fig. 2B). This pattern of expression is merely identical to that in preadipocytic fibroblasts (3T3L1 cells treated with the vehicle DMSO) versus 3T3L1 cells that fully differentiated to adipocytes by the adipocyte differentiation mixture containing (isobutylmethyl xanthine, dexamethasone, insulin) (Fig. 2B). We then examined whether necdin induction also can be demonstrated in activated HSCs isolated from experimental models of liver fibrosis. For this analysis, HSCs were isolated from rats with cholestatic liver fibrosis induced by bile duct ligation (BDL) or those with hepatotoxic liver fibrosis resulting from repetitive injections of carbon tetrachloride (CCl4), as well as their controls: rats with sham-operation and those injected with oil, the vehicle for CCl4. Real-time PCR analysis of RNA extracted from these cells showed a 2.5-fold increase in necdin mRNA in HSCs from BDL or CCl4 rats as compared with the respective controls (Fig. 2C). Concomitantly, the α1(I) procollagen mRNA level was increased and PPARγ mRNA decreased much like what we observed in culture-activated HSCs (Fig. 2C). The necdin protein level was also increased in BDL and CCl4 HSCs (Fig. 2D). These results demonstrate that HSC activation in vitro or in vivo is associated with induction of necdin.
FIGURE 2.
Necdin expression is induced in activated HSCs. A, note that a time-dependent increase in necdin mRNA in cultured HSCs on plastic concomitant with induction of the HSC activation marker α1(I) procollagen (α1(I)Procoll) and suppression of the HSC quiescence marker PPARγ. B, necdin protein level is conspicuously increased in HSCs cultured for 7 days as compared with quiescent day 1 cells in a manner identical to a change in adipocytes (3T3L1 cells treated with the adipocyte differentiation mixture MDI) versus preadipicytes (3T3L1 treated with the solvent DMSO). C, necdin induction also is confirmed in “in vivo activated” HSCs isolated from fibrotic rat livers induced by BDL or repetitive carbon tetrachloride injection (CCl4), again concomitant with induction of α1(I) procollagen and down-regulation of PPARγ. D, necdin protein level is increased in activated HSCs from BDL- or CCl4-induced liver fibrosis as compared with respective controls. *, p < 0.05; **, p < 0.01 compared to Day 1 or controls. Error bars are standard deviations.
Necdin Silencing Reverses Activated HSCs to Quiescent Cells in a Manner Dependent on PPARγ
To test the role of necdin in HSC trans-differentiation, necdin expression was silenced with shRNA expressed by adenovirus (Ad.Necdin.shRNA). Adenovirus expressing shRNA against LacZ (Ad.LacZ.shRNA) was used as a control. As shown in Fig. 3A, infection of culture-activated HSCs with Ad.Necdin.shRNA at an MOI of 100 or 200 for 3 days, clearly reduced nuclear and cytosolic levels of necdin. This silencing resulted in a drastic morphologic reversal of HSCs to quiescent cells (Fig. 3B, upper panel), increased intracellular vitamin A content as assessed by UV-excited autofluorescence (middle panel), and increased lipid content stained by oil red O (lower panel). The reversal also was confirmed by reduced expression of α1(I) procollagen (Fig. 3C). We next determined the effects of necdin silencing on the expression of neuronal, myogenic, and adipogenic marker genes to gain insights into the mechanisms of the cell phenotype reversal. Necdin silencing did not regulate either neuronal or myogenic genes (Fig. 3D) but consistently up-regulated adipogenic transcription factors (Pparγ, C/ebpα, and Lxrα) and their target genes (acetyl-CoA carboxylase and fatty acid synthase) (Fig. 3E, left graph). As these genes are all down-regulated in day 7 culture-activated HSCs as compared with day 1 quiescent cells (Fig. 3E, right graph), the observed induction with necdin silencing should be interpreted as restoration of normal expression of the genes in quiescent HSCs.
FIGURE 3.
Necdin silencing reverses activated HSCs to quiescent cells in a manner dependent on PPARγ. A, nuclear and cytosolic necdin levels were effectively reduced by adenovirally expressed shRNA against necdin (Ad.Necdin.shRNA) in culture-activated HSCs as compared with the cells infected with adenovirus expressing shRNA against LacZ (Ad.LacZ.shRNA). B, necdin silencing with Ad.Necdin.shRNA at 100 MOI reverts activated HSCs to the cells with morphology of quiescent HSCs (Phase) and increases intracellular vitamin A content as assessed by UV-excited autofluorescence (UV) and intracellular lipid content as stained with oil red O (Oil Red). C, these effects are accompanied by reduced expression of α1(I) procollagen (α1(I)Coll). D, necdin silencing does not affect the expression of neuronal (left) or muscle cell (right) markers. E, necdin silencing in culture-activated HSC increases the expression of adipogenic transcription factors (Pparγ, C/ebpα, and Lxrα) and their target lipogenic genes, Acc and Fas (left). As these genes are all down-regulated in culture-activated cells (Day 7 HSC) compared with quiescent day 1 cells (right), the inductions seen after necdin silencing is considered as restoration of normal expressions. F, morphologic reversal of activated HSCs with Ad.Necdin.shRNA is completely abrogated by concomitant expression of dominant-negative PPARγ (+Ad.dnPPARγ) as assessed by phase contrast microscopy and UV-excited autofluorescence. Sma, smooth muscle actin. *, p < 0.05; **, p < 0.01; ***, p < 0.005 as compared to respective controls. Error bars are standard deviations.
As our previous study showed the pivotal importance of PPARγ in the maintenance of HSC quiescence (8, 26, 27) and the current study demonstrates necdin silencing restored PPARγ and HSC quiescence, we wondered whether the cell phenotype reversal by necdin silencing is mediated by PPARγ. To test this notion, culture-activated HSCs were co-infected with Ad.Necdin.shRNA and adenovirus that expresses a dominant-negative mutant of PPARγ (Ad.dnPPARγ) or GFP (Ad.GFP) as a control. This dnPPARγ carries the mutations at leucine and glutamic acid residues in the conserved AF-2 region, inhibiting co-activator recruitment and ligand-dependent release of co-repressors while promoting basal recruitment of co-repressors (28). Coexpression of this mutant indeed prevented the morphologic reversal achieved by necdin silencing: note the activated cell morphology by phase contrast microscopy and diminished vitamin A storage by UV-excited autofluorescence (Fig. 3F, bottom panel). Co-infection of Ad.dnPPARγ and Ad.LacZ.shRNA did not affect the cell morphology or UV-autofluorescence (Fig. 3F, second row).
Effects of Necdin Silencing Is Dependent on Inhibition of Canonical Wnt
Canonical Wnt signaling contributes to antiadipogenic effects (12, 13) and HSC activation (11) via suppression of PPARγ. Thus, we tested next the effects of necdin on Wnt expression. The expression of canonical (Wnt10b and Wnt3) and noncanonical (Wnt4) Wnt proteins by activated HSCs (11) were suppressed by necdin silencing (Fig. 4A). Nuclear level of β-catenin also was reduced (Fig. 4B), indicating a positive link between necdin and canonical Wnt. In fact, the phenotypic reversal of activated HSCs achieved by necdin silencing, was abrogated by concomitant treatment with Wnt3a (Fig. 4C, bottom panel) which also prevented restoration of PPARγ mRNA expression (Fig. 4D). Addition of Wnt3a to Ad.LacZ.shRNA-infected cells did not alter the activated cell morphology or minimal UV-autofluorescence (Fig. 4C, second panel). Here, we used Wnt3a as a representative canonical Wnt as it but not Wnt10b was readily available from a commercial source.
FIGURE 4.
Necdin silencing reduces the expression of canonical (Wnt10b and Wnt3a) and noncanonical (Wnt4) Wnts (A) and nuclear level of β-catenin (B) in culture-activated HSCs. C, morphologic reversal to the quiescent state achieved by necdin silencing is prevented by addition of recombinant Wnt3a (100 ng/ml). D, restored PPARγ mRNA expression by necdin silencing is abrogated by Wnt3a. *, p < 0.05 compared to LacZ.shRNA; †, p < 0.05 compared to Necdin.shRNA without Wnt3a.
Necdin Binds and Activates Proximal Wnt10b Promoter
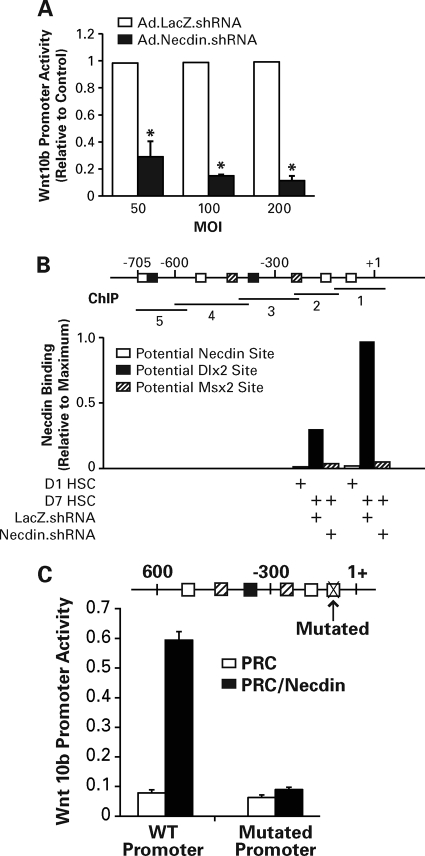
Regulation of canonical Wnts by necdin was demonstrated previously in neuronal and muscle cells (17, 21) and suggested by our results in HSCs. To address this link in HSCs, we have analyzed the activity of Wnt10b promoter (−705/+30) in the HSC line infected with Ad.Necdin.shRNA or Ad.LacZ.shRNA. As shown in Fig. 5A, necdin silencing at an MOI of 50∼200, suppressed the activity by 70%∼90%. We then examined necdin binding to the promoter by ChIP assay. We analyzed five overlapping 100∼150-bp segments covering the potential necdin sites, so-called GN boxes: multiple G clusters intervened with mono- or dinucleotides of A, T, and C (30), or potential Dlx3 or Mx2 sites to which necdin may bind via these homeodomain proteins (17, 21). Analysis was performed in day 1 quiescent HSCs, or day 7 culture-activated HSCs infected with Ad.Necdin.shRNA or Ad.LacZ.shRNA (MOI, 200). Necdin binding was detected in two most proximal segments (−143/+42 and −268/−119) but not in other regions of Wnt10b promoter in day 7 HSCs transduced with LacZ.shRNA. These bindings were abrogated by Ad.Necdin.shRNA (Fig. 5B). Day 1 HSCs showed minimal necdin binding in any areas within this promoter. Next, we mutated the most proximal necdin binding site by disrupting the three consecutive G clusters with T: −108/−96: GG(T)GTGG(T)GGTGG(T)GG; and compared the promoter activity between the wild type and the mutant with or without overexpression of necdin. As the BSC line overexpresses necdin and the Wnt10b promoter cannot be further stimulated with exogenous necdin, we used Huh7 cells that do not express necdin. Indeed, in this cell line, necdin expression increases WT Wnt10b promoter activity 6-fold but not with the mutation in the GN box (Fig. 5C), confirming positive regulation of Wnt10b promoter with necdin which requires the most proximal intact necdin binding GN box.
FIGURE 5.
A, necdin silencing with Ad.Necdin.shRNA at 50 ∼ 200 MOI inhibits Wnt10b proximal promoter (−705/+30) activity in the hepatic stellate cell line (BSC). B, ChIP analysis detects recruitment of necdin to two most proximal ChIP sites (−143/+42 and −268/−119) labeled 1 and 2 within the Wnt10b promoter in day 7 culture-activated HSC infected with Ad.LacZ.shRNA. These binding are abrogated by necdin silencing with Ad.Necdin.shRNA. Day 1 quiescent HSCs show minimal necdin binding to these sites. ChIP data are expressed as relative to maximal recruitment at the most proximal site seen in day 7 HSC tansduced with LacZ.shRNA. C, mutations of a potential necdin binding GN box at −108/−96: GG(T)GTGG(T)GGTGG(T)GG within the Wnt10b proximal promoter, abrogate necdin-inducible promoter activity in Huh7 cells. *, p < 0.05 compared to Ad.LacZ.shRNA.
Necdin Silencing Rescues HSCs from Epigenetic PPARγ Repression via Suppressed Canonical Wnt
We next examined the mechanisms by which necdin suppresses PPARγ expression. Epigenetic repression mediated by the methyl-CpG binding protein MeCP2, has been shown recently to underlie PPARγ silencing in activated HSCs (29). According to this finding, induced expression of MeCP2 in HSC activation, results in increased MeCP2 binding to the 5′ end of Pparγ, and consequent recruitment of the transcriptional repressor HP-1α. MeCP2 also positively regulates EZH2, causing H3K27 methylation for the formation of repressive chromatin in the exon 5 of Pparγ in activated HSCs (29). Thus, we tested the effects of necdin silencing with Ad.Necdin.shRNA on MeCP2 and HP-1α binding to Pparγ and H3K27 dimethylation (H3K27me2). As canonical Wnt is shown to be downstream of necdin and to negatively regulate PPARγ expression, we also have tested the effects of concomitant Wnt3a treatment of HSCs infected with Ad.Necdin.shRNA. Indeed, necdin silencing largely abrogated the increases in both MeCP2 and HP-1α binding to the Pparγ promoter and H3K27me2 (histone 3 lysine 27 dimethylation) at the 3′ Pparγ exon locus in culture-activated HSCs as the cells were reverted to quiescent cells, and these effects were prevented completely by addition of Wnt3a (Fig. 6, A–C). These results suggest necdin achieves epigenetic repression of Pparγ transcription via canonical Wnt signaling. This notion was further tested by determining the effect of adenovirally expressed Dkk-1, the Wnt co-receptor antagonist, on the identical epigenetic signatures shown for PPARγ repression. As shown in Fig. 6D, MeCP2 binding to the Pparγ promoter and H3K27me2 at the 3′ Pparγ exon locus, were attenuated significantly by Dkk-1 transduction. Furthermore, Dkk-1 restored PPARγ mRNA expression in culture-activated HSCs (Fig. 6D, right panel).
FIGURE 6.
Necdin silencing reduces epigenetic PPARγ repression. Necdin shRNA reduces MeCP2 (A) and HP-1α (B) binding to Pparγ promoter and H3K27me2 at the exon 5 locus (C), and these effects were abrogated by recombinant Wnt3a (A–C). Adenovirally expressed Dkk-1 largely attenuates MeCP2 enrichment in the Pparγ promoter and H3K27me2 enrichment at exon 5 (D) and restored PPARγ expression (E). *, p < 0.05 compared to LacZ.shRNA or Day 1 Ad.GFP; † compared to Necdin.shRNA without Wnt3a or Day 7 Ad.GFP.
DISCUSSION
Our present findings on HSC-specific expression of necdin and its anti-adipogenic role in HSC activation provide additional evidence in support of the concept that HSC-myofibroblastic trans-differentiation involves similar mechanisms to those responsible for adipocyte-preadipocyte dedifferentiation. Besides the role of necdin in neuronal and muscle cell differentiation, this melanoma antigen family protein is expressed at a high level in preadipocytes and suggested to inhibit adipogenesis via induction of Wnt (30). Indeed, we show Wnt10b, a canonical Wnt typically induced in HSC activation (11), is a direct target of necdin. Our ChIP analysis reveals the region containing the typical necdin binding GN boxes (−143/+42) has the highest level of necdin recruitment in activated HSCs, which is completely abrogated by necdin silencing. Furthermore, necdin silencing reduces the activity of this Wnt10b proximal promoter, Wnt10b mRNA expression, and the nuclear β-catenin level. Moreover, our site-directed mutagenesis analysis reveals that the GN box located at −108/−96: (GGGTGGGGTGGG) is largely responsible for necdin-induced Wnt10b proximal promoter activity. As predicted from our earlier observation of a reversal of culture-activated HSCs to quiescent cells by antagonism of canonical Wnt signaling with Dkk-1 (11), a similar phenotypic reversal is demonstrated by necdin silencing. This reversal is dependent on restored activity of PPARγ as suggested by complete prevention of the reversal with the dominant-negative mutant of PPARγ.
We also have obtained novel insights into how necdin via Wnt suppresses PPARγ expression. Necdin silencing abrogates two key epigenetic signatures, which recently have been demonstrated for repression of PPARγ in activated HSC: namely, MeCP2 recruitment to the promoter and H3K27me2 in the 3′ exon 5 (29). These rescue effects also are prevented by concomitant Wnt3a treatment, suggesting canonical Wnt signaling mediates these epigenetic modifications. Indeed, this notion is supported by our result that both MeCP2 recruitment and PPARγ repression are reversed by Dkk-1 transduction.
A recent study demonstrates that chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) is a direct target of β-catenin and mediates Wnt-induced recruitment of the COUP-TFII-SMRT (silencing mediator for retinoid and thyroid hormone receptors) corepressor complexes to the first introns of Pparγ1 and Pparγ2, resulting in hypoacetylation of the local chromatin and repression of Pparγ (31). Conversely, another study reports Wnt10b is a direct target of COUP-TFII-mediated negative regulation, and this regulation may participate in adipogenesis (32). In activated HSCs, COUP-TFII is induced.3 How necdin and Wnt10b cross-interact with COUP-TFII for PPARγ regulation in HSC trans-differentiation, is yet to be determined. More specifically, whether COUP-TFII/SMRT-mediated epigenetic regulation occurs in Wnt-induced PPARγ repression in HSCs, needs to be addressed. Another remaining question is how necdin is induced in HSC activation. Activated cAMP response element binding protein is a critical initiator of adipogenesis (33) partly via suppression of anti-adipogenic genes Wnt and Pref-1 (34). Activated cAMP response element binding protein also is required for HSC quiescence (35) and may be a negative regulator of the necdin-Wnt pathway identified by the present study. Although we identified necdin to be upstream of Wnt, how other preadipocyte-selective or anti-adipogenic genes regulate necdin expression in HSCs, is unknown at the present time. Of equal importance is epigenetic regulation of necdin via paternal allele-specific histone acetylation, which appears to contribute to tissue-specific activity (36).
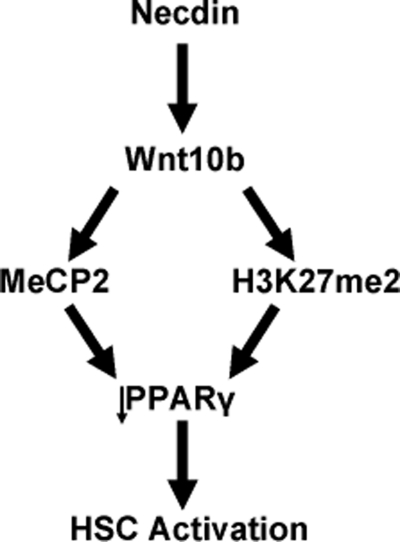
In summary, our study has identified necdin as a new regulator for the cell fate of HSCs. Induced necdin favors myofibroblastic transdifferentiation of HSCs via transcriptional activation of Wnt10b and Wnt-mediated epigenetic repression of PPARγ involving MeCP2 and H3K27me2 as schematically summarized in Fig. 7. The identified pathway may serve as a new therapeutic target for liver fibrosis.
FIGURE 7.
The necdin-Wnt10b pathway activates HSCs via epigenetic repression of PPARγ involving MeCP2 and H3K27me2. This schematic diagram summarizes our finding of necdin-mediated Wnt10b up-regulation causing increased MeCP2 recruitment to the Pparγ promoter and increased H3K27 dimethylation at the Pparγ locus with consequent PPARγ repression and HSC activation.
Acknowledgments
We thank Drs. Jelena Mann and Derek Mann (Newcastle University) for sharing and making helpful suggestions on carrier ChIP protocol.
This work was supported, in whole or in part, by National Institutes of Health Grants P50AA011999, R24AA012885, and R21AA016682. This work was also supported by the Medical Research Service of the Department of Veterans Affairs.
N.-L. Zhu, J. Wang, and H. Tsukamoto, unpublished data.
- HSC
- hepatic stellate cell
- PPARγ
- peroxisome proliferator-activated receptor γ
- MOI
- multiplicity of infection
- BDL
- bile duct ligation
- COUP-TFII
- chicken ovalbumin upstream promoter-transcription factor II.
REFERENCES
- 1.Friedman S. L. (2008) Physiol. Rev. 88, 125–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asahina K., Tsai S. Y., Li P., Ishii M., Maxson R. E., Jr., Sucov H. M., Tsukamoto H. (2009) Hepatology 49, 998–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geerts A. (2001) Semin. Liver Dis. 21, 311–335 [DOI] [PubMed] [Google Scholar]
- 4.Yamada M., Blaner W. S., Soprano D. R., Dixon J. L., Kjeldbye H. M., Goodman D. S. (1987) Hepatology 7, 1224–1229 [DOI] [PubMed] [Google Scholar]
- 5.Tsukamoto H. (1999) Alcohol Clin. Exp. Res. 23, 911–916 [PubMed] [Google Scholar]
- 6.Potter J. J., Womack L., Mezey E., Anania F. A. (1998) Biochem. Biophys. Res. Commun. 244, 178–182 [DOI] [PubMed] [Google Scholar]
- 7.Spiegelman B. M., Flier J. S. (1996) Cell 87, 377–389 [DOI] [PubMed] [Google Scholar]
- 8.Miyahara T., Schrum L., Rippe R., Xiong S., Yee H. F., Jr., Motomura K., Anania F. A., Willson T. M., Tsukamoto H. (2000) J. Biol. Chem. 275, 35715–35722 [DOI] [PubMed] [Google Scholar]
- 9.Galli A., Crabb D., Price D., Ceni E., Salzano R., Surrenti C., Casini A. (2000) Hepatology 31, 101–108 [DOI] [PubMed] [Google Scholar]
- 10.Marra F., Efsen E., Romanelli R. G., Caligiuri A., Pastacaldi S., Batignani G., Bonacchi A., Caporale R., Laffi G., Pinzani M., Gentilini P. (2000) Gastroenterology 119, 466–478 [DOI] [PubMed] [Google Scholar]
- 11.Cheng J. H., She H., Han Y. P., Wang J., Xiong S., Asahina K., Tsukamoto H. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 294, G39–49 [DOI] [PubMed] [Google Scholar]
- 12.Ross S. E., Hemati N., Longo K. A., Bennett C. N., Lucas P. C., Erickson R. L., MacDougald O. A. (2000) Science 289, 950–953 [DOI] [PubMed] [Google Scholar]
- 13.Bennett C. N., Ross S. E., Longo K. A., Bajnok L., Hemati N., Johnson K. W., Harrison S. D., MacDougald O. A. (2002) J. Biol. Chem. 277, 30998–31004 [DOI] [PubMed] [Google Scholar]
- 14.Taniguchi N., Taniura H., Niinobe M., Takayama C., Tominaga-Yoshino K., Ogura A., Yoshikawa K. (2000) J. Biol. Chem. 275, 31674–31681 [DOI] [PubMed] [Google Scholar]
- 15.Gérard M., Hernandez L., Wevrick R., Stewart C. L. (1999) Nat. Genet. 23, 199–202 [DOI] [PubMed] [Google Scholar]
- 16.Yoshikawa K. (2000) Neurosci. Res. 37, 1–14 [DOI] [PubMed] [Google Scholar]
- 17.Kuwajima T., Taniura H., Nishimura I., Yoshikawa K. (2004) J. Biol. Chem. 279, 40484–40493 [DOI] [PubMed] [Google Scholar]
- 18.Brunelli S., Tagliafico E., De Angelis F. G., Tonlorenzi R., Baesso S., Ferrari S., Niinobe M., Yoshikawa K., Schwartz R. J., Bozzoni I., Ferrari S., Cossu G. (2004) Circ. Res. 94, 1571–1578 [DOI] [PubMed] [Google Scholar]
- 19.Tseng Y. H., Butte A. J., Kokkotou E., Yechoor V. K., Taniguchi C. M., Kriauciunas K. M., Cypess A. M., Niinobe M., Yoshikawa K., Patti M. E., Kahn C. R. (2005) Nat. Cell Biol. 7, 601–611 [DOI] [PubMed] [Google Scholar]
- 20.Ren J., Lee S., Pagliardini S., Gérard M., Stewart C. L., Greer J. J., Wevrick R. (2003) J. Neurosci. 23, 1569–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwajima T., Nishimura I., Yoshikawa K. (2006) J. Neurosci. 26, 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukamoto H., Horne W., Kamimura S., Niemelä O., Parkkila S., Ylä-Herttuala S., Brittenham G. M. (1995) J. Clin. Invest. 96, 620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sung C. K., She H., Xiong S., Tsukamoto H. (2004) Am. J. Physiol. Gastrointest. Liver Physiol. 286, G722–G729 [DOI] [PubMed] [Google Scholar]
- 24.Xiong S., She H., Takeuchi H., Han B., Engelhardt J. F., Barton C. H., Zandi E., Giulivi C., Tsukamoto H. (2003) J. Biol. Chem. 278, 17646–17654 [DOI] [PubMed] [Google Scholar]
- 25.Tokairin T., Nishikawa Y., Doi Y., Watanabe H., Yoshioka T., Su M., Omori Y., Enomoto K. (2002) J Hepatol. 36, 725–733 [DOI] [PubMed] [Google Scholar]
- 26.Hazra S., Xiong S., Wang J., Rippe R. A., Krishna V., Chatterjee K., Tsukamoto H. (2004) J. Biol. Chem. 279, 11392–11401 [DOI] [PubMed] [Google Scholar]
- 27.She H., Xiong S., Hazra S., Tsukamoto H. (2005) J. Biol. Chem. 280, 4959–4967 [DOI] [PubMed] [Google Scholar]
- 28.Gurnell M., Wentworth J. M., Agostini M., Adams M., Collingwood T. N., Provenzano C., Browne P. O., Rajanayagam O., Burris T. P., Schwabe J. W., Lazar M. A., Chatterjee V. K. (2000) J. Biol. Chem. 275, 5754–5759 [DOI] [PubMed] [Google Scholar]
- 29.Mann J., Chu D. C., Maxwell A., Oakley F., Zhu N. L., Tsukamoto H., Mann D. A. (2010) Gastroenterology 138, 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto K., Taniura H., Uetsuki T., Yoshikawa K. (2001) Gene 272, 173–179 [DOI] [PubMed] [Google Scholar]
- 31.Okamura M., Kudo H., Wakabayashi K., Tanaka T., Nonaka A., Uchida A., Tsutsumi S., Sakakibara I., Naito M., Osborne T. F., Hamakubo T., Ito S., Aburatani H., Yanagisawa M., Kodama T., Sakai J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5819–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L., Xie X., Qin J., Jeha G. S., Saha P. K., Yan J., Haueter C. M., Chan L., Tsai S. Y., Tsai M. J. (2009) Cell Metab. 9, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reusch J. E., Colton L. A., Klemm D. J. (2000) Mol. Cell. Biol. 20, 1008–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacDougald O. A., Burant C. F. (2005) Nat. Cell Biol. 7, 543–545 [DOI] [PubMed] [Google Scholar]
- 35.Houglum K., Lee K. S., Chojkier M. (1997) J. Clin. Invest. 99, 1322–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau J. C., Hanel M. L., Wevrick R. (2004) Nucleic Acids Res. 32, 3376–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]