Abstract
Cystic fibrosis (CF) is a life-shortening disease caused by a mutation in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. To gain an understanding of the epithelial dysfunction associated with CF mutations and discover biomarkers for therapeutics development, untargeted metabolomic analysis was performed on primary human airway epithelial cell cultures from three separate cohorts of CF patients and non-CF subjects. Statistical analysis revealed a set of reproducible and significant metabolic differences between the CF and non-CF cells. Aside from changes that were consistent with known CF effects, such as diminished cellular regulation against oxidative stress and osmotic stress, new observations on the cellular metabolism in the disease were generated. In the CF cells, the levels of various purine nucleotides, which may function to regulate cellular responses via purinergic signaling, were significantly decreased. Furthermore, CF cells exhibited reduced glucose metabolism in glycolysis, pentose phosphate pathway, and sorbitol pathway, which may further exacerbate oxidative stress and limit the epithelial cell response to environmental pressure. Taken together, these findings reveal novel metabolic abnormalities associated with the CF pathological process and identify a panel of potential biomarkers for therapeutic development using this model system.
Keywords: Cystic Fibrosis, Energy Metabolism, Metabolomics, Oxidative Stress, Purine
Introduction
Cystic fibrosis (CF)2 is a fatal autosomal recessive disease that affects ∼30,000 patients in the United States and >70,000 individuals world-wide (1). More than 1000 new CF cases are diagnosed each year. The condition was first recognized in the 1930s as an inheritable gene defect that causes scar tissue (fibrosis) and cysts in the lungs and loss of pancreatic function (2). The most common manifestations of cystic fibrosis include progressive respiratory dysfunction due to repeated cycles of infection and inflammation, and chronic digestive disorders, including fat malabsorption due to pancreatic insufficiency. These symptoms are caused by the presence of unusually thick mucus that clogs airways and obstructs the pancreatic ducts, impairing normal function. Over time, these conditions lead to tissue remodeling, scarring and fibrosis, frequently resulting in the need for organ transplantation.
The scientific advances in understanding the genetic defect and the effects of specific mutations have been dramatic. In recent years, access to new treatments and improvements in standard of care have dramatically improved the outlook for people with CF (3, 4). The median life expectancy of a child born with CF in 2007 was 37.5 years (Cystic Fibrosis Foundation Patient Registry 2007 Annual Report, Bethesda, MD), a significant increase from 25 years, which was the median survival in 1985. Nevertheless, there is significant need for additional new therapies to further increase survival for people with CF.
CF is caused by mutations in cystic fibrosis transmembrane conductance regulator (CFTR), which encodes an ATP-gated, anion-selective channel that conducts chloride ions across the epithelial cell membrane. Chloride transport is a critical requirement for maintenance of an appropriately hydrated liquid layer on the epithelial surface. Mutations in CFTR lead to defective chloride transport, a major contributor to the pathological thickening of mucus in the lung and blockage of the ducts of other affected organs.
The application of genetics and molecular biology have had a significant impact on our understanding of the effects of gene mutations on the CFTR protein (5). The most prevalent mutation in the allele population is an in-frame deletion of Phe at the 508 position (F508del). In the case of F508del CFTR, the protein maturation process is severely disrupted, and the inactive protein remains in the endoplasmic reticulum and then undergoes proteasome degradation (6, 7). The mutation also impairs the ability of the channel to transport chloride, which is attributed to a reduced open probability (8) as well as to a reduced half-life of residency at the cell surface (9).
Much of the recent knowledge of the effects of the F508del mutation on CFTR function has been delineated through the use of a model system in which human bronchial epithelial cells are isolated from the explant lung tissue of CF and non-CF subjects after lung transplantation (10). The CF human bronchial epithelial cell has recently been used as a preclinical model system for the development of investigational new drugs (11).
Interestingly, the effects of CFTR gene mutations cause a myriad of downstream biological changes, and the relationship between these changes and the resulting clinical symptoms remains poorly understood. For example, levels of NO and S-nitrosothiols are low in the CF airway (12, 13), and low micromolar concentrations of S-nitrosylating agents have been shown to promote F508del CFTR maturation in human bronchial epithelial cells (14). People with CF have been observed to have increased energy expenditure, presumably due to the extra exertion required for breathing and chronic infection (15).
Global metabolomics is a new and powerful technology that can provide a relatively complete picture of the metabolism in biological systems and has recently been applied to a wide variety of important problems (16–19). We decided to apply this approach to understanding the basis of the changes in cellular metabolism that result from CFTR mutations with the hope of better understanding the mechanism by which the CFTR gene induces its disease effects.
EXPERIMENTAL PROCEDURES
Primary Human Airway Epithelial Cell Culture
Primary human airway epithelial cell cultures from three separate cohorts of CF (F508del homozygous) and non-CF (wild-type) subjects were provided by University of Iowa in Vitro Models and Cell Culture Core (study 1), University of North Carolina CF Center Tissue Culture Core (study 2), and University of Pittsburgh Cystic Fibrosis Research Center (study 3). Following lung transplantation, the primary cells were harvested under the Institutional Review Board-approved protocols of each institution.
For the University of Iowa cohort, the airway epithelial cells were obtained from 13 CF and 12 non-CF donors as previously described (20). Cells were isolated by enzyme digestion, and freshly isolated cells were seeded onto collagen-coated Millpore cell polycarbonate filters (Millipore, Bedford, MA). The cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and air. Twenty-four hours after plating, the apical medium was removed, and the cells were maintained at the air-liquid interface. These cultures develop a ciliated surface within 14 days of seeding. The culture medium consisted of a 1:1 mix of Dulbecco's modified Eagle's medium-Ham's F-12, 2% Ultroser G (Biosepra SA, Cergy-Saint-Christophe, France), penicillin (100 units/ml), streptomycin (100 μg/ml), gentamicin (50 μg/ml), fluconazole (2 μg/ml), and amphotericin B (1.25 μg/ml).
For the University of North Carolina cohort, the airway epithelial cells were obtained from 7 CF and 8 non-CF donors. Briefly, primary cells derived from single patient sources were expanded on plastic to generate passage 1 cells, which were plated at a density of 250,000 cells/cm2 on permeable membrane support (Transwell-Col, 12- or 24-mm diameter, Corning; Millicells, 12-mm diameter, Millipore) and maintained in a specialty media (21). Differentiation was induced under air-liquid interface. The cultures were considered “matured” (in ∼4–8 weeks) when significantly ciliated (>50% ciliated cells). Only fully differentiated, ciliated cultures were used in the experiments.
For the University of Pittsburgh cohort, the airway epithelial cells were obtained from 8 CF and 8 non-CF donors. The cells were cultured on human placental collagen-coated Costar Transwell filters (0.33 cm2) as described previously (22) and used for experimentation following 4–6 weeks of culture at an air-liquid interface.
Metabolomic Profiling
The metabolomic platforms were described previously in detail (23, 24). Briefly, the platform consisted of three independent platforms: ultrahigh performance liquid chromatography/tandem mass spectrometry (UHLC/MS/MS) optimized for basic species, UHLC/MS/MS optimized for acidic species, and gas chromatography/mass spectrometry (GC/MS). The major components of the process are summarized as follows.
Sample Extraction and GC/MS Analysis
The samples were extracted using an automated MicroLab STAR® system (Hamilton Company, Salt Lake City, UT) in 400 μl of methanol, containing the recovery standards. The samples destined for GC/MS analysis were dried under vacuum desiccation for a minimum of 24 hours and then derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide. The GC column was 5% phenyl, and the temperature ramp was from 40 to 300 °C in a 16-min period. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization.
UHPLC/MS/MS Analysis
UHPLC/MS/MS was carried out using a Waters Acquity UHPLC (Waters Corporation, Milford, MA) coupled to an LTQ mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA) equipped with an electrospray ionization source. Two separate UHPLC/MS injections were performed on each sample: one optimized for positive ions and one for negative ions. Chromatographic separation followed by full scan mass spectra was carried out to record retention time, molecular weight (m/z), and MS/MS of all detectable ions presented in the samples.
Metabolites were identified by automated comparison of the ion features in the experimental samples with a reference library of chemical standard entries that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as their associated MS/MS spectra. This library allowed the rapid identification of metabolites in the experimental with high confidence.
Data Imputation and Statistical Analysis
After the data were corrected for minor variation resulting from instrument interday tuning differences (24), the missing values for a given metabolite were imputed with the observed minimum detection value on the assumption that they were below the limits of detection. For the convenience of data visualization, the raw area counts for each biochemical were rescaled by dividing the value for a specific biochemical in each sample by the median value for that specific biochemical.
Statistical analysis of the data were performed using JMP (SAS), a commercial software package, and “R,” which is a freely available open-source, software package. Welch's two-sample t tests were performed on the log-transformed data to compare the CF and non-CF groups for each of the three data sets. Multiple comparisons were accounted for with the false discovery (FDR) rate method, and each FDR was estimated using q values (25).
To assess which metabolites consistently distinguished the CF and non-CF groups across the three studies, meta-analysis was performed using Fisher's statistic [Fisher]: Q = −2*log(p1*p2, respectively. The statistic Q has a χ2 distribution with 2m degrees of freedom, where m is the number of independent p values. Thus, if the metabolite was found in each study, the statistic has six degrees of freedom, whereas if it was found in only one study, it has two degrees of freedom (26).
RESULTS
Study Design, Metabolomic Profiles, and Statistical Analysis
Clinical samples often have substantial biological variation. To assess biochemical differences with strong statistical significance between CF and non-CF cells, it would be ideal to analyze a sufficiently large number of samples per experimental group. However, it was challenging to obtain a large number of both CF and non-CF donors for airway epithelial cells at a single clinical site. Thus, we completed three independent metabolomic analyses: a cohort of 13 CF and 12 non-CF samples collected by the University of Iowa in Vitro Models and Cell Culture Core (study 1); a cohort of 7 CF and 8 non-CF samples collected by the University of North Carolina CF Center Tissue Culture Core (study 2); and a cohort of 8 CF and 8 non-CF samples collected by the University of Pittsburgh Cystic Fibrosis Research Center (study 3).
In samples from each of the three cohorts, ∼400 metabolites were detected. Among these metabolites, 137, 162, and 198 metabolites with known chemical structures were identified for study 1, 2, and 3 respectively. These metabolites are listed in the supplemental material. The increased number of metabolites with known structures in the studies was due to reference library and platform method enhancements over the time the study samples were processed.
For each cohort, the differences in the metabolite levels between CF and non-CF samples were calculated by the ratio of their group means. The statistical significance of the differences was analyzed by Welch's t test. Meta-analysis was then employed to identify metabolites that showed reproducible changes between CF and non-CF across the three studies, with p < 0.05 deemed to be significant. In addition, multiple comparisons were accounted for with the FDR method, and each FDR was estimated using the q value. The full statistical analysis table is included in the supplemental material. After mapping the metabolites into general biochemical pathways according to the Kyoto Encyclopedia of Genes and Genomes (KEGG), it was apparent that the most significant differences between the CF and non-CF cells were in nucleotide metabolism, tryptophan metabolism, glutathione, organic osmolytes, and energy metabolism.
Nucleotide Metabolism
One of the most significant differences between the CF and non-CF cells was in nucleotide metabolism, especially purine biosynthesis. As shown in a condensed scheme in Fig. 1, both de novo and salvage pathways contribute to the biosynthesis of purines. Several metabolites in purine metabolism, including adenosine, inosine, hypoxanthine, and guanosine, were significantly decreased in the CF cells. Xanthine also showed a trend of decreasing in CF cells, although its p value narrowly missed the 0.05 cutoff. Pyrimidine metabolism was also affected by CF; cytidine was significantly decreased in the CF cells (Fig. 1).
FIGURE 1.
Relative levels of nucleotide between the CF and non-CF groups. A, purine degradation pathway and the levels of adenosine, guanosine, inosine, xanthine, and hypoxanthine by box plots. For the box plots, the top and bottom of the boxes represent the 75th and 25th percentile, respectively. The top and bottom bars (whiskers) represent the entire spread of the data points for the subjects, excluding extreme points, which are indicated with circles (○). The hyphen indicates the median value. Metabolite names in bold font indicate significantly altered metabolites between CF and non-CF groups (p < 0.05). Metabolites in regular font indicate no significant differences between CF and non-CF groups. Metabolites in italic font indicate not measured in this study. B, level of cytidine by box plots. The y axis is the relative levels of the metabolites.
Tryptophan Metabolism
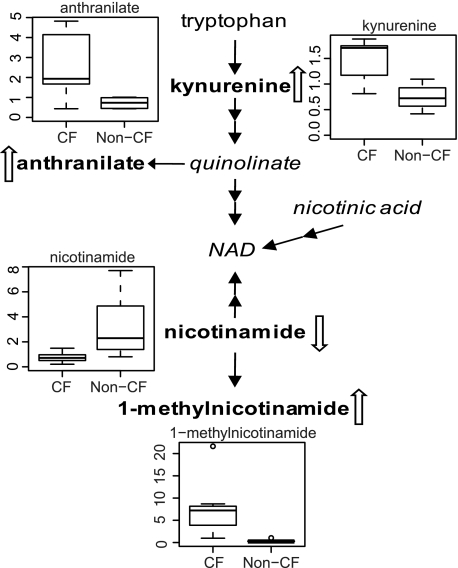
A clear difference in tryptophan metabolism was detected between the CF and non-CF cells. The main tryptophan catabolic route is via the kynurenine pathway. Tryptophan is converted to kynurenine and, ultimately, to precursors for NAD synthesis. As shown in Fig. 2, an ∼2-fold increase in the levels of kynurenine and anthranilate was observed in CF cells. Downstream of tryptophan catabolism, the metabolite 1-methylnicotinamide showed a dramatic 24-fold elevation, whereas the level of its precursor, nicotinamide, was significantly decreased.
FIGURE 2.
NAD biosynthesis and relative levels of anthranilate, kynurenine, nicotinamide, and 1-methylnicodinamide by box plots. The explanation for the box plots can be found in the legend of Fig. 1.
Glutathione Biosynthesis
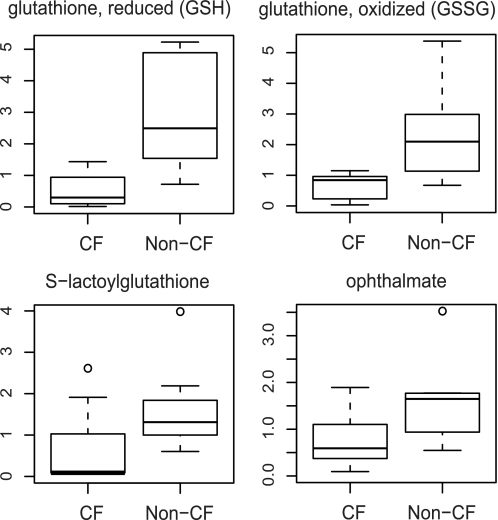
Glutathione and its associated metabolites showed significant differences between CF and non-CF cells (Fig. 3). The levels of both oxidized glutathione (GSSG) and reduced glutathione (GSH) in CF cells decreased to <30% of those of non-CF cells. In addition, ophthalmate (Glu-2-aminobutyrate-gly), a metabolite related to the synthesis of GSH, showed similar decreases (Fig. 3). Ophthalmate is an analog of glutathione and has been proposed as an indicator for GSH biosynthesis (27). Consistent with the reduced glutathione level, S-lactoylglutathione, a metabolite derived from glutathione detoxification, was significantly lower in the CF cells (Fig. 3).
FIGURE 3.
Relative levels of reduced glutathione, oxidized glutathione, S-lactoylglutathione, and ophthalmate between the CF and non-CF groups by box plots. The explanation for the box plots can be found in the legend of Fig. 1.
Osmolytes
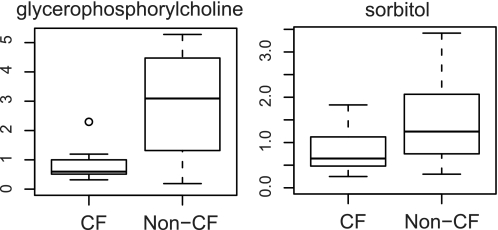
The levels of two major cellular osmolytes, sorbitol and glycerophosphorylcholine, were significantly reduced in the CF cells compared with the non-CF cells (Fig. 4). Organic osmolytes play an important function in maintaining cell volume and fluid balance, which is vital for proper cellular functions and cell survival (28).
FIGURE 4.
Relative levels of glycerophosphorylcholine and sorbitol between the CF and non-CF groups by box plots. The explanation for the box plots can be found in the legend of Fig. 1.
Glucose Metabolism
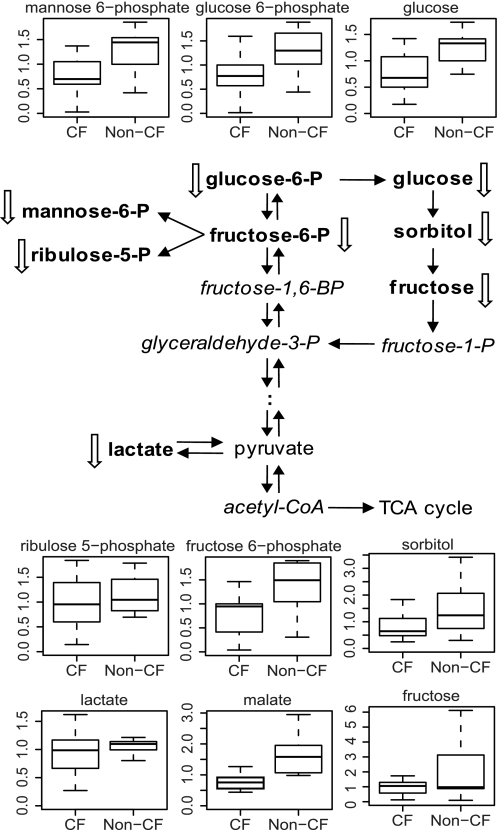
Glucose plays a central function in cellular metabolism to produce energy and biosynthetic precursors of nucleotides and fatty acids. The levels of glucose and various glycolytic intermediates, including glucose 6-phosphate, fructose 6-phoshpate, and lactate, were significantly reduced in CF cells. In addition, the levels of various metabolites in other branches of glucose metabolism were decreased in CF cells. Decreases were measured in the levels of ribulose-5-phosphate from the pentose phosphate pathway, malate from the tricarboxylic acid cycle, and sorbitol and fructose from the sorbitol pathway (Fig. 5). Collectively, these results may indicate that glucose metabolism was suppressed in the CF cells.
FIGURE 5.
Glucose metabolism and the levels of fructose, fructose 6-phosphate, glucose, glucose 6-phosphate, lactate, malate, mannose 6-phosphate, ribulose 5-phosphate, and sorbitol between the CF and non-CF groups by box plots. The explanation for the box plots can be found in the legend of Fig. 1.
DISCUSSION
Although it is known that CF is caused by mutations in the CFTR gene, the specific means by which defective CFTR signaling leads to the disease phenotypes such as alteration of airway surface liquid volume, mucus deposition, inflammation, and deficient anti-microbial properties, remains incompletely understood. Murine models of CF do not recapitulate human CF lung disease (29), and although there is significant promise that the CF pig model will prove useful for defining pathogenesis (30), the lack of high fidelity animal models of CF has hampered progress and propelled the use of primary human airway cells derived from CF patients.
Human airway epithelial cell cultures of cells isolated from lung tissue following lung transplantation have been widely used as a model system to study CF disease (10, 31) and have been utilized as a preclinical model for CF drug development programs (11). To gain insights into the molecular and biochemical mechanisms of the disease process, we used untargeted global metabolomic technology to compare the biochemical profiles of CF and wild-type cell cultures. Collectively, we profiled three independent cohorts of samples. After statistical analyses were performed on each study individually, we used meta-analysis to examine the metabolic differences consistently displayed in all three studies. This analytical approach has several advantages. First, the combination of the three datasets generated a larger number of samples for each experimental group, which facilitates the identification of CF-altered metabolism with high statistical significance. Second, analysis of the three independent cohorts of samples provided the ability to overcome experimental artifacts associated with sample collection and cell culture methods that are specific to an individual site. Third, the studies can serve as validations for one another.
One of the hallmarks of the CF disease phenotype is the depletion of airway surface liquid, leading to the formation of thickened mucus plaques and plugs in the airway surfaces. The underlying mechanism has been suggested to be accelerated Na+ absorption and defective Cl2− secretion in the airway epithelial cells (32). The CFTR mutation is known to affect the activities of other ion channels and transporters (33). The resulting accumulation of inorganic ions in the cytosol likely alters trans-epithelial potential. Thus, both cellular uptake and excretion processes may be significantly impacted in CF, which in turn, impacts metabolism. In fact, our analysis revealed biochemical perturbations that most likely result from the impact of aberrant inorganic ion accumulation on uptake/transport and regulation processes which contribute to the defects in airway surface liquid observed in CF patients.
Among the most significant metabolic perturbations were the decreases of various purine metabolites measured in CF cells. As shown in Fig. 1, the levels of adenosine, inosine, hypoxanthine, and guanosine were significantly lower in CF cells, suggesting that purine biosynthesis was reduced. In cell cultures purines are produced mainly from the salvage pathway which utilizes hypoxanthine or xanthine supplied in the medium as the precursor. It is likely that in CF cells the uptake of hypoxanthine or xanthine was compromised, thereby leading to the observed decrease in purine biosynthesis.
It is well established that purines can function as signal moleculars to regulation various physiological processes associated with health and disease. Especially under the context of CF, adenosine and other purine nucleotides are important regulators of airway surface liquid volume via activation of both CFTR-dependent and CFTR-independent purinergic receptors (34, 35). Results of using chamber studies of rat lung epthithelial cells suggest that adenosine can directly increase chloride ion effux through CFTR (36). Thus, decreased purine biosynthesis could exacerbate the depletion of airway surface liquid volume caused by the CFTR mutation. Interestingly, xanthine derivatives have been identified as compounds that can act as activators of CFTR (37, 38). The in vivo significance of this finding will need to be determined in future studies because epithelial airway cells produce purines via both de novo synthesis and the salvage pathway, which is not reliant on uptake of exogenous hypoxanthine or xanthine. However, whatever the in vivo case may be, these metabolites represent biomarker candidates with potential utility for CF drug discovery and development using primary human airway epithelial cell cultures.
To our knowledge this is the first study to measure metabolites of purine biosynthesis directly from lung epithelial cells. Our results are in contrast to the elevated purine levels that have been detected in exhaled breath condensate of CF patients, potentially as a result of neutrophil inflammation in the airway (39, 40).
The accumulation of cytosolic inorganic ions in the cell undoubtedly also interferes with cell volume regulation. In our metabolomic analysis, the levels of two major cellular organic osmolytes, sorbitol and glycerophosphorylcholine, were significantly diminished. Organic osmolytes play a central function in cell volume regulation. Unlike inorganic ions, they do not interfere with protein stability and ion gradient-driven transporters (41). The decreased osmolyte levels in CF cells observed in this study could reflect perturbations in the ability of CF cells to sense, regulate, synthesize, or transport these metabolites. Interestingly, the supplementation of kidney cells with organic osmolytes such as sorbitol, taurine, inositol, and glycerophosphorylcholine has been shown to promote proper CFTR protein maturation (28).
Several metabolite differences measured between CF and non-CF cells were in agreement with the current understanding of CF-associated metabolism. In our metabolomic analysis, both reduced and oxidized glutathione levels were significantly decreased in CF cells (Fig. 3). Consistent with the reduced level of glutathione were the observed decreases in S-lactoylglutathione, a metabolite derived from glutathione detoxification (42), S-nitrosoglutathione (12) and ophthalmate, a marker for glutathione production (27). Studies have indicated that S-nitrosoglutathione and S-nitrosylating compounds can promote CFTR cell surface expression and channel activation in CF epithelial cells (14, 43).
Mutations in CFTR lead to disruptions of GSH levels in both the intracellular and the extracellular milieu (44–47). CF patients have decreased GSH levels in the lung epithelial lining fluid and blood, but GSH levels in the lung itself appear to be unaffected (47). As CFTR is present on the apical side of lung epithelial cells, it has been proposed that CFTR may transport GSH into the epithelial lining where it may have multiple functions, including breaking disulfide bonds to reduce mucus viscosity, affecting mucus hydration and playing a role in the inflammatory response to infection (44, 45, 48).
Although CFTR has been shown to transport GSH, it is also possible that CFTR interacts with other GSH transporters and affects GSH transport through these interactions. CFTR has been shown to functionally and physically associate with MRP4, a cAMP transporter which is also a putative GSH transporter (49). GSH levels have been shown to be reduced in CFTR−/− mice compared with wild-type mice following Pseudomonas aeruginosa infection (50). GSH levels are also reduced in neutrophils of CF patients which may contribute to abnormal function and increased necrosis of neutrophils (48, 51). The poor response rate to infection may in part be due to an inability to increase GSH levels in response to infection (48).
Glutathione is the most abundant cellular redox molecule and is critical for maintaining the redox status in cells. Reduced glutathione levels may result in oxidative conditions, further contributing to the pathology of CF (52). Thus, increasing the cellular glutathione levels has been suggested as a therapeutic option for CF (51, 53). Inhaled GSH and N-acetylcysteine are under clinical evaluation (51, 54, 55). Oral N-acetylcysteine has been shown to increase, and whole blood and neutrophil concentrations of GSH (51) and an increase in extracellular glutathione in induced sputum (55) occur in CF patients.
In addition, we identified alterations in two biochemical pathways in CF cells that could further aggravate oxidative stress conditions. First, indications of suppressed glucose metabolism, including the pentose phosphate pathway, were observed in CF cells (Fig. 5). The pentose phosphate pathway is the major cellular source for the generation of NADPH. NADPH provides the reducing equivalents for the regeneration of reduced glutathione and thioredoxin, two principal antioxidants. Decreased metabolic flux through the pentose phosphate pathway may amplify cell sensitivity to oxidative stress. Second, our results show that tryptophan metabolism was altered in CF cells, resulting in the accumulation of kynurenine and anthranilate (Fig. 2). Kynurenine has been associated with oxidative stress induction and diseased conditions (56).
In the present study, significant differences in metabolism, including differences in pathways key to cellular function, between CF and non-CF cells were uncovered using metabolomic analysis. These results suggest that CF pathogenesis may be largely mediated at the level of metabolism. Furthermore, the biochemical profiles provide additional insights into CF disease and suggest metabolism-based disease mechanisms that can be developed into testable hypotheses. In addition, metabolites showing altered levels between CF and non-CF cells could be used as potential biomarkers for the discovery and development of novel therapeutics.
Supplementary Material
Acknowledgments
Joseph Pilewski, M.D., Joseph Zabner, M.D., and Scott Randell, Ph.D. kindly contributed to the primary bronchial epithelial cells used in this study.
This work was supported by Cystic Fibrosis Foundation Therapeutics, Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental material.
- CF
- cystic fibrosis
- CFTR
- CF transmembrane conductance regulator
- FDR
- false discovery rate
- MS/MS
- tandem mass spectrometry
- UHLC
- ultrahigh performance liquid chromatography.
REFERENCES
- 1.Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L., Drumm M. L., Iannuzzi M. C., Collins F. S., Tsui L. C. (1989) Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 2.Davis P. B. (2006) Am. J. Respir. Crit. Care Med. 173, 475–482 [DOI] [PubMed] [Google Scholar]
- 3.Flume P. A., Mogayzel P. J., Jr., Robinson K. A., Goss C. H., Rosenblatt R. L., Kuhn R. J., Marshall B. C. (2009) Am. J. Respir. Crit. Care Med. 180, 802–808 [DOI] [PubMed] [Google Scholar]
- 4.Stallings V. A., Stark L. J., Robinson K. A., Feranchak A. P., Quinton H. (2008) J. Am. Diet. Assoc. 108, 832–839 [DOI] [PubMed] [Google Scholar]
- 5.Guggino W. B., Stanton B. A. (2006) Nat. Rev. Mol. Cell Biol. 7, 426–436 [DOI] [PubMed] [Google Scholar]
- 6.Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. (1990) Cell 63, 827–834 [DOI] [PubMed] [Google Scholar]
- 7.Drumm M. L., Wilkinson D. J., Smit L. S., Worrell R. T., Strong T. V., Frizzell R. A., Dawson D. C., Collins F. S. (1991) Science 254, 1797–1799 [DOI] [PubMed] [Google Scholar]
- 8.Welsh M. J., Smith A. E. (1993) Cell 73, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 9.Swiatecka-Urban A., Brown A., Moreau-Marquis S., Renuka J., Coutermarsh B., Barnaby R., Karlson K. H., Flotte T. R., Fukuda M., Langford G. M., Stanton B. A. (2005) J. Biol. Chem. 280, 36762–36772 [DOI] [PubMed] [Google Scholar]
- 10.Fulcher M. L., Gabriel S. E., Olsen J. C., Tatreau J. R., Gentzsch M., Livanos E., Saavedra M. T., Salmon P., Randell S. H. (2009) Am. J. Physiol. Lung Cell. Mol. Physiol. 296, L82–L91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Goor F., Hadida S., Grootenhuis P. D., Burton B., Cao D., Neuberger T., Turnbull A., Singh A., Joubran J., Hazlewood A., Zhou J., McCartney J., Arumugam V., Decker C., Yang J., Young C., Olson E. R., Wine J. J., Frizzell R. A., Ashlock M., Negulescu P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18825–18830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasemann H., Gaston B., Fang K., Paul K., Ratjen F. (1999) J. Pediatr. 135, 770–772 [DOI] [PubMed] [Google Scholar]
- 13.Snyder A. H., McPherson M. E., Hunt J. F., Johnson M., Stamler J. S., Gaston B. (2002) Am. J. Respir. Crit. Care Med. 165, 922–926 [DOI] [PubMed] [Google Scholar]
- 14.Zaman K., Carraro S., Doherty J., Henderson E. M., Lendermon E., Liu L., Verghese G., Zigler M., Ross M., Park E., Palmer L. A., Doctor A., Stamler J. S., Gaston B. (2006) Mol. Pharmacol. 70, 1435–1442 [DOI] [PubMed] [Google Scholar]
- 15.Tomezsko J. L., Stallings V. A., Kawchak D. A., Goin J. E., Diamond G., Scanlin T. F. (1994) Pediatr. Res. 35, 451–460 [DOI] [PubMed] [Google Scholar]
- 16.Schnackenberg L. K., Beger R. D. (2006) Pharmacogenomics 7, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 17.Lawton K. A., Berger A., Mitchell M., Milgram K. E., Evans A. M., Guo L., Hanson R. W., Kalhan S. C., Ryals J. A., Milburn M. V. (2008) Pharmacogenomics 9, 383–397 [DOI] [PubMed] [Google Scholar]
- 18.Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 19.Boudonck K. J., Rose D. J., Karoly E. D., Lee D. P., Lawton K. A., Lapinskas P. J. (2009) Bioanalysis 1, 1645–1663 [DOI] [PubMed] [Google Scholar]
- 20.Rich D. P., Couture L. A., Cardoza L. M., Guiggio V. M., Armentano D., Espino P. C., Hehir K., Welsh M. J., Smith A. E., Gregory R. J. (1993) Hum. Gene Ther. 4, 461–476 [DOI] [PubMed] [Google Scholar]
- 21.Fulcher M. L., Gabriel S., Burns K. A., Yankaskas J. R., Randell S. H. (2005) Methods Mol. Med. 107, 183–206 [DOI] [PubMed] [Google Scholar]
- 22.Devor D. C., Bridges R. J., Pilewski J. M. (2000) Am. J. Physiol. Cell Physiol. 279, C461–C479 [DOI] [PubMed] [Google Scholar]
- 23.Ohta T., Masutomi N., Tsutsui N., Sakairi T., Mitchell M., Milburn M. V., Ryals J. A., Beebe K. D., Guo L. (2009) Toxicol. Pathol. 37, 521–535 [DOI] [PubMed] [Google Scholar]
- 24.Evans A. M., DeHaven C. D., Barrett T., Mitchell M., Milgram E. (2009) Anal. Chem. 81, 6656–6667 [DOI] [PubMed] [Google Scholar]
- 25.Storey J. D., Tibshirani R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher R. A. (1938) Statistical Methods for Research Workers, 7th Ed., Oliver and Boyd, Edinburgh, pp. 104–106 [Google Scholar]
- 27.Soga T., Baran R., Suematsu M., Ueno Y., Ikeda S., Sakurakawa T., Kakazu Y., Ishikawa T., Robert M., Nishioka T., Tomita M. (2006) J. Biol. Chem. 281, 16768–16776 [DOI] [PubMed] [Google Scholar]
- 28.Lang F. (2007) J. Am. Coll. Nutr. 26, Suppl. 5, 613S–623S [DOI] [PubMed] [Google Scholar]
- 29.Grubb B. R. (1999) Am. J. Physiol. Gastrointest. Liver Physiol. 277, G167–G174 [DOI] [PubMed] [Google Scholar]
- 30.Rogers C. S., Stoltz D. A., Meyerholz D. K., Ostedgaard L. S., Rokhlina T., Taft P. J., Rogan M. P., Pezzulo A. A., Karp P. H., Itani O. A., Kabel A. C., Wohlford-Lenane C. L., Davis G. J., Hanfland R. A., Smith T. L., Samuel M., Wax D., Murphy C. N., Rieke A., Whitworth K., Uc A., Starner T. D., Brogden K. A., Shilyansky J., McCray P. B., Jr., Zabner J., Prather R. S., Welsh M. J. (2008) Science 321, 1837–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruenert D. C., Finkbeiner W. E., Widdicombe J. H. (1995) Am. J. Physiol. Lung Cell. Mol. Physiol. 268, L347–L360 [Google Scholar]
- 32.Yancey P. H. (2005) J. Exp. Biol. 208, 2819–2830 [DOI] [PubMed] [Google Scholar]
- 33.Gabriel S. E., Clarke L. L., Boucher R. C., Stutts M. J. (1993) Nature 363, 263–268 [DOI] [PubMed] [Google Scholar]
- 34.Com G., Clancy J. P. (2009) Handb. Exp. Pharmacol. 193, 363–381 [DOI] [PubMed] [Google Scholar]
- 35.Kreda S. M., Okada S. F., van Heusden C. A., O'Neal W., Gabriel S., Abdullah L., Davis C. W., Boucher R. C., Lazarowski E. R. (2007) J. Physiol. 584, 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Factor P., Mutlu G. M., Chen L., Mohameed J., Akhmedov A. T., Meng F. J., Jilling T., Lewis E. R., Johnson M. D., Xu A., Kass D., Martino J. M., Bellmeyer A., Albazi J. S., Emala C., Lee H. T., Dobbs L. G., Matalon S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4083–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chappe V., Mettey Y., Vierfond J. M., Hanrahan J. W., Gola M., Verrier B., Becq F. (1998) Br. J. Pharmacol. 123, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulteau L., Dérand R., Mettey Y., Métayé T., Morris M. R., McNeilly C. M., Folli C., Galietta L. J., Zegarra-Moran O., Pereira M. M., Jougla C., Dormer R. L., Vierfond J. M., Joffre M., Becq F. (2000) Am. J. Physiol. Cell Physiol. 279, C1925–C1937 [DOI] [PubMed] [Google Scholar]
- 39.Esther C. R., Jr., Alexis N. E., Clas M. L., Lazarowski E. R., Donaldson S. H., Ribeiro C. M., Moore C. G., Davis S. D., Boucher R. C. (2008) Eur. Respir. J. 31, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esther C. R., Jr., Boysen G., Olsen B. M., Collins L. B., Ghio A. J., Swenberg J. W., Boucher R. C. (2009) Am. J. Physiol. Lung Cell. Mol. Physiol. 296, L987–L993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X. M., Wang X. T., Yue H., Leung S. W., Thibodeau P. H., Thomas P. J., Guggino S. E. (2003) J. Biol. Chem. 278, 51232–51242 [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez C. F., Proudfoot M., Brown G., Korniyenko Y., Mori H., Savchenko A. V., Yakunin A. F. (2006) J. Biol. Chem. 281, 14514–14522 [DOI] [PubMed] [Google Scholar]
- 43.Servetnyk Z., Krjukova J., Gaston B., Zaman K., Hjelte L., Roomans G. M., Dragomir A. (2006) Respir. Res. 7, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hudson V. M. (2004) Treat. Respir. Med. 3, 353–363 [DOI] [PubMed] [Google Scholar]
- 45.Hudson V. M. (2001) Free Radic. Biol. Med. 30, 1440–1461 [DOI] [PubMed] [Google Scholar]
- 46.Gao L., Kim K. J., Yankaskas J. R., Forman H. J. (1999) Am. J. Physiol. Lung Cell. Mol. Physiol. 277, L113–L118 [DOI] [PubMed] [Google Scholar]
- 47.Roum J. H., Buhl R., McElvaney N. G., Borok Z., Crystal R. G. (1993) J. Appl. Physiol. 75, 2419–2424 [DOI] [PubMed] [Google Scholar]
- 48.Ballatori N., Krance S. M., Notenboom S., Shi S., Tieu K., Hammond C. L. (2009) Biol. Chem. 390, 191–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C., Krishnamurthy P. C., Penmatsa H., Marrs K. L., Wang X. Q., Zaccolo M., Jalink K., Li M., Nelson D. J., Schuetz J. D., Naren A. P. (2007) Cell 131, 940–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Day B. J., van Heeckeren A. M., Min E., Velsor L. W. (2004) Infect. Immun. 72, 2045–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tirouvanziam R., Conrad C. K., Bottiglieri T., Herzenberg L. A., Moss R. B., Herzenberg L. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4628–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ciofu O., Riis B., Pressler T., Poulsen H. E., Høiby N. (2005) Antimicrob. Agents Chemother. 49, 2276–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visca A., Bishop C. T., Hilton S. C., Hudson V. M. (2008) J. Cyst. Fibros. 7, 433–436 [DOI] [PubMed] [Google Scholar]
- 54.Bishop C., Hudson V. M., Hilton S. C., Wilde C. (2005) Chest 127, 308–317 [DOI] [PubMed] [Google Scholar]
- 55.Dauletbaev N., Fischer P., Aulbach B., Gross J., Kusche W., Thyroff-Friesinger U., Wagner T. O., Bargon J. (2009) Eur. J. Med. Res. 14, 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pawlak K., Kowalewska A., Mysliwiec M., Pawlak D. (2009) Am. J. Med. Sci. 338, 293–300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.