Abstract
Proteolytic processing of the amyloid precursor protein (APP) generates large soluble APP derivatives, β-amyloid (Aβ) peptides, and APP intracellular domain. Expression of the extracellular sequences of APP or its Caenorhabditis elegans counterpart has been shown to be sufficient in partially rescuing the CNS phenotypes of the APP-deficient mice and the lethality of the apl-1 null C. elegans, respectively, leaving open the question as what is the role of the highly conserved APP intracellular domain? To address this question, we created an APP knock-in allele in which the mouse Aβ sequence was replaced by the human Aβ. A frameshift mutation was introduced that replaced the last 39 residues of the APP sequence. We demonstrate that the C-terminal mutation does not overtly affect APP processing and amyloid pathology. In contrast, crossing the mutant allele with APP-like protein 2 (APLP2)-null mice results in similar neuromuscular synapse defects and early postnatal lethality as compared with mice doubly deficient in APP and APLP2, demonstrating an indispensable role of the APP C-terminal domain in these development activities. Our results establish an essential function of the conserved APP intracellular domain in developmental regulation, and this activity can be genetically uncoupled from APP processing and Aβ pathogenesis.
Keywords: Alzheimer's Disease, Amyloid, Gene Knockout, Mouse, Neurobiology, Amyloid Precursor Protein, Neuromuscular Junction
Introduction
Genetic and biochemical evidence establishes a central role of APP3 in Alzheimer disease pathogenesis. Genetic mutations or gene amplification of APP are linked to a subset of cases of early onset familial Alzheimer disease (FAD); APP processing generates β-amyloid (Aβ) peptides, which are the principal components of the amyloid plaque pathology (reviewed in Ref. 1). APP represents the founding member of a family of conserved type I membrane proteins including APL-1 in Caenorhabditis elegans, APPL in Drosophila, and APP, APP-like protein 1 (APLP1), and APLP2 in mammals (reviewed in Ref. 2). Full-length APP is processed by at least three proteinases known as α-, β-, and γ-secretases. Both α-secretase and β-secretase cleave APP in the extracellular domain with α-secretase cleavage occurring inside the Aβ domain and β-secretase at the amino terminus of Aβ. These proteolytic events generate large soluble APP derivatives and the membrane-anchored APP carboxyl-terminal fragments, which serve as substrates for subsequent γ-secretase processing, producing either p3 (product of α- and γ-secretases) or Aβ peptides (product of β- and γ-secretase) and the APP intracellular domain. The intracellular sequences are most highly conserved among the APP family members. Of particular interest, phosphorylation at the threonine 668 residue (Thr668) and adaptor protein interactions through the YENPTY motif have been shown to regulate APP localization, trafficking, amyloidogenic processing, and possibly cell signaling (reviewed in Refs. 2 and 3).
Despite the high degree of sequence conservation and the well characterized biochemical and cellular properties, in vivo loss-of-function studies in mice and in C. elegans both argue against an important role of the APP intracellular domain. Specifically, the apl-1-null C. elegans is lethal, and the lethality can be rescued by neuronal expression of the APL-1 extracellular domain (4). Mice deficient in APP are viable but exhibit subtle phenotypes including reduced body weight, locomotor activity, and forelimb grip strength and impaired synaptic plasticity, spatial learning, and memory (5, 6). Expressing only the APP extracellular domain was shown to be sufficient in rescuing the anatomical and behavioral abnormalities (7). Nevertheless, a recent publication documented that acute knockdown of APP by in utero electroporation of an APP RNAi construct leads to neuronal migration defect, and the phenotype can only be rescued by expressing the full-length APP, but not the APP extracellular or intracellular domains, either individually or combined (8).
Gene knock-out studies reveal genetic redundancies among the APP proteins as mice doubly deficient in APP/APLP2, APLP1/APLP2, or missing all three APP members are early postnatal lethal (9, 10). Our analysis of APP/APLP2 double knock-out mice identified an essential role for the APP family of proteins in the patterning of neuromuscular junction (NMJ) (11). Further investigation of neuromuscular synapse and central synaptogenesis support the notion that APP is a synaptic adhesion protein, and that the synaptogenic function requires full-length APP (12, 13). The early postnatal lethality and the diffused synaptic distribution of the NMJ present in the APP/APLP2 double knock-out animals provide sensitive and specific readouts for us to definitely determine the role of the APP C-terminal domain in vivo.
By creating a strain of APP knock-in mice in which the APP intracellular domain was mutated by introducing a frameshift mutation, we report here that the neuromuscular synapse structure and animal viability require the highly conserved APP intracellular domain. In contrast, the C-terminal domain is dispensable for APP processing, secretion, and amyloidogenesis.
EXPERIMENTAL PROCEDURES
Animals
APLP2 knock-out mice (9), PS1M146V knock-in mice (14, 15), and APP/hAβ mice, which carry the Swedish and London mutations and humanized Aβ sequence (16), were described as cited. To generate APP/hAβ/mutC knock-in mice, a gene-targeting vector including the Swedish/Arctic/London FAD mutations, the humanized Aβ sequence, and a frameshift mutation at the sequence encoding Ile656 (APP695 numbering) was electroporated to R1 ES cells (detailed description can be found in the supplemental methods and supplemental Fig. S1). ES clones were screened by Southern blotting, and three clones were used to inject blastocysts to create chimeric mice. Chimeric mice were bred to C57BL/6 to establish germline transmission of the knock-in allele. These knock-in mice were then mated with transgenic mice expressing the Cre recombinase under the protamine promoter (17) to remove the neomycin resistance cassette and to produce the APP/hAβ/mutC allele. Genotyping was done by PCR using the following primer pairs (5′ to 3′): GTAATGCCTGTGTGGCCAAACACATG and AAGTAATGGATTTGTTCTCCCAGGTCG, which amplify the loxP insertion site. The expected PCR product from the wild-type allele is 230 bp, and the expected PCR product from the knock-in allele is 270 bp.
Antibodies and Reagents
22C11 and 6E10 monoclonal antibodies are available from Covance. The polyclonal anti-APP C-terminal antibody APPc was described previously (12). Anti-FLAG (rabbit polyclonal), anti-synaptophysin, and anti-choline transporter (CHT) antibodies were purchased from Sigma, DAKO, and Chemicon, respectively. α-Bungarotoxin was from Molecular Probes.
Quantitative Real-time PCR
Total RNA was isolated from mice brains using the RNeasy lipid tissue mini kit (Invitrogen) and subjected to DNase I digestion to remove contaminating genomic DNA. Reverse transcription was performed using a SuperScript III RNase H-reverse transcriptase (Invitrogen), and the reaction mix was subjected to quantitative real-time PCR using an ABI PRISM sequence detection system 7000 (Applied Biosystems, Inc.). Primers were designed with Primer Express Version 2.0 software (Applied Biosystems) using sequence data from the National Center for Biotechnology Information (NCBI). The sets of GAPDH and hypoxanthine-guanine phosphoribosyltransferase primers were used as an internal control for each specific gene amplification. The relative levels of expression were quantified and analyzed by using the ABI PRISM sequence detection system 7000 software. The real-time value for each sample was averaged and compared using the comparative threshold cycle method. The relative amount of target RNA was calculated relative to the expression of endogenous reference and relative to a calibrator, which was the mean threshold cycle of control samples.
Neuronal Culture
Postnatal day 0 (P0) pups from APP/hAβ/mutC heterozygous breeding were genotyped using the REDExtract-N-AmpTM tissue PCR kit (Sigma). Homozygous knock-in pups and their wild-type littermates were selected, and their hippocampi were dissected. The tissue was trypsinized, mechanically dissociated, washed, resuspended in Neurobasal medium with B27 supplement (Invitrogen), and plated on poly-d-lysine-coated 60-mm dishes. Conditioned medium and total cell lysates (in PBS with Complete protease inhibitor cocktail) were collected at 14 days in vitro.
Biotinylation Assay
HEK293 cells were transiently transfected with APP constructs. Cells were washed with ice-cold PBS containing 1.0 mm MgCl2 and 0.1 mm CaCl2 (PBS/Ca-Mg) and treated with sulfo-NHS-SS-biotin (1.5 mg/ml; Pierce) for 1 h on ice in PBS/Ca-Mg. Biotinylating reagents were removed by incubating with cold 100 mm glycine in PBS/Ca-Mg for 30 min followed by three washes with cold PBS/Ca-Mg. Cells were incubated at 37 °C to allow internalization to occur, which was subsequently terminated by transferring culture plates to ice. Residual cell surface biotin was stripped with freshly prepared 50 mm mercaptoethanesulfonic acid (Sigma) in TE buffer (150 mm NaCl, 1 mm EDTA, 0.2% bovine serum albumin, 20 mm Tris, pH 8.6) for 30 min and quenched with iodoacetamide (5 mg/ml) in PBS/Ca-Mg. Cells were then lysed in 1% CHAPS lysis buffer containing 50 mm Tris, pH 7.4, 150 mm NaCl, and protease inhibitors (Roche Applied Science). Biotinylated proteins and non-biotinylated proteins were separated by incubation with an UltraLink-NeutrAvidin bead (Pierce). Samples were subject to Western blot analysis.
Western Blotting
To prepare total tissue/cell lysate, mouse brain, spinal cord, or cultured neurons were homogenized using radioimmune precipitation buffer (1% Nonidet P-40, 50 mm Tris, pH 8.0, 150 mm NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 2 mm EDTA) containing Complete protease inhibitor cocktail (Roche Applied Science). After three sets of 10 pulses of sonication, the homogenates were spun at 20,000 × g for 15 min. To prepare the PBS-soluble fraction for soluble APP quantification, brain/spinal cord was homogenized in PBS with Complete protease inhibitor cocktail, and then tissue lysates were centrifuged at 100,000 × g at 4 °C for 1 h to collect supernatants. Protein concentrations were determined using the Bio-Rad protein dye assay. 10 μg of protein were loaded on a 10% SDS-PAGE gel run at 100 V for 2 h at room temperature and transferred onto a nitrocellulose membrane (Bio-Rad) at 100 V for 1 h. Membranes were blocked 1 h using 5% nonfat dry milk in TBS containing 0.1% Tween 20 (TBST, Sigma). After three washes with TBST, secondary antibody application was performed at room temperature for 1 h using 5% milk in TBST followed by three additional washes with TBST. Signals of Western blots were detected by enhanced chemiluminescence (GE Healthcare), scanned, and analyzed using ImageJ software from the National Institutes of Health. Data were represented as mean ± S.E. of three samples/group.
Sandwich ELISA
Brain halves were weighted and homogenized in 4× (w/v) homogenization buffer composed of PBS, 1% Triton X-100, and Complete protease inhibitor cocktail (Roche Applied Science). Total brain lysates were centrifuged at 20,000 × g, and supernatants were diluted two times and then applied to Aβ40 ELISA kit (Invitrogen) following the company's protocol. Each sample was analyzed in duplicates. 4–5 animals were used for each genotype.
Mass Spectrometry
The measurement of Aβ peptides using immunoprecipitation/mass spectrometry (IP/MS) was carried out essentially as described except that the 6E10 antibody was used to precipitate Aβ from PBS extractions due to the disruption of the 4G8 site upon introducing the Arctic mutation (18, 19). Mass spectra were collected using a TOF/TOF 5800 mass spectrometer (ABSciex). Each mass spectrum was averaged from 4000 laser shots and calibrated using bovine insulin as an internal mass celebrant. Peak areas were used for relative quantification.
Immunohistochemistry and Amyloid Load Quantification
Antibody staining on paraffin-embedded brain sections was performed as described (20). In particular, paraformaldehyde-perfused brains were cut into 10-μm paraffin sections. The sections were deparaffinized in xylene, rinsed with ethanol, and rehydrated through ethanol gradient. Antigen-retrieval was done by incubating sections with 70% formic acid for 6 min. Endogenous peroxidase activity was quenched by incubating the slides in 0.3% H2O2 for 30 min. The slides were rinsed with Tris-buffered saline (TBS; 50 mm Tris-Cl, pH 7.5 and 250 mm NaCl). Nonspecific epitopes were blocked for 30 min with 3% normal goat serum, 0.4% Triton X-100 in TBS. Primary antibody 6E10 was diluted 1:1000 in blocking buffer and incubated overnight at 4 °C in a humid chamber. Sections were then washed three times for 5 min each in TBS and incubated with anti-mouse secondary antibody/peroxidase-conjugated streptavidin (VECTASTAIN ABC kit; Vector Laboratories) following the company's protocol. Pictures were taken with a Zeiss Axioskop 2 Plus microscope equipped with the Axiocam MRC digital camera, and the images were processed with the Axiovision 3.1 software. Images covering the entire cortex were taken. The number of Aβ plaques (>5 μm) was counted manually under an Olympus microscope (CX 31). Data were reported as the total number of plaques divided by the total cortical area for each animal.
Immunofluorescence Staining of Neuromuscular Junction
Whole-mount immunostaining of the diaphragm muscle and quantification of neuromuscular phenotypes were carried out as described (11, 12). Confocal images were obtained with a Zeiss 510 laser scanning microscope, and quantification was done using the ImageJ program from the National Institutes of Health.
Statistical Analysis
Genotyping analysis of the offspring from APPki/−APLP2+/− male and female intercrosses was performed using chi-square analysis. Aβ levels and plaque load were analyzed by analysis of variance. Student's t test was used for all other analysis. *, p < 0.05, **, p < 0.01, ***, p < 0.001. Data were presented as average ± S.E. (standard error of the mean).
RESULTS
Generation and Expression Analysis of APP/hAβ/mutC Knock-in Animals
To investigate the role of the highly conserved APP C-terminal domain in survival, neuromuscular synapse development, and amyloid pathology in vivo, we created a knock-in allele in which the mouse Aβ was replaced by the human Aβ sequence with simultaneous introduction of the Swedish/London/Arctic FAD mutations. In addition, the two cytosines in ATC CAT encoding residues Ile656–His657 of APP (695 isoform numbering) were deleted, resulting in a frameshift starting at His657 and deletion of the last 39 amino acids of the APP C-terminal sequences including the highly conserved Thr668 residue and the YENPTY sequence (Fig. 1A and supplemental Fig. S1). A 10-amino acid out-of-frame spacer sequence was simultaneously introduced downstream of His657 to ensure proper membrane anchoring and γ-secretase processing of the C-terminal deleted APP. This knock-in allele is herein termed as APP/hAβ/mutC or ki.
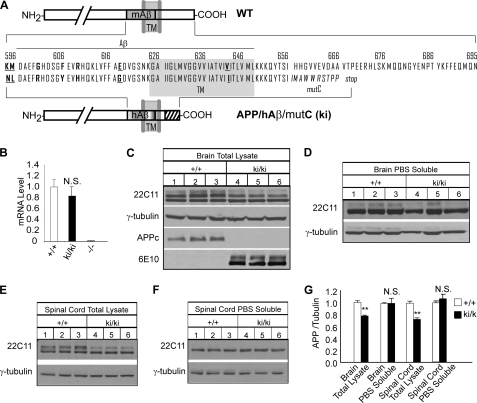
FIGURE 1.
Generation and biochemical characterization of APP/hAβ/mutC ki mice. A, schematic representation of wild-type (WT) and APP/hAβ/mutC (ki) alleles. TM stands for transmembrane region (also marked by gray shading), and mAβ and hAβ represent mouse and human Aβ sequences, respectively. Amino acid sequences from the Aβ region to the end of the C terminus of both the wild-type allele and the ki allele are listed. Residues corresponding to Swedish (K595N and M596L), Arctic (E618G), and London (V642I) mutation sites are shown in bold and underlined letters. Residues different between mouse and human Aβ are shown in bold letters. Frameshift mutations of the ki allele, which starts at the coding sequence for residue Ile656 and results in stop codon after 10 amino acids, are indicated by italic letters. B, quantitative real-time PCR of APP mRNA from 2-month-old wild-type (+/+), homozygous APP/hAβ/mutC knock-in (ki/ki), and APP knock-out (−/−) mouse brains. APP−/− was used as a negative control. C–F, representative Western blot analysis of 2-month-old ki/ki mice and their wild-type (+/+) littermates of APP expression in total brain lysate, PBS-soluble fraction of the brain lysate, total spinal cord lysate, and PBS-soluble fraction of spinal cord lysate, respectively, using the 22C11, 6E10, and APPc antibodies. γ-Tubulin blot was used as protein loading control. G, quantification of the relative ratio of 22C11/γ-tubulin blots. Both the upper and the lower bands were included in the brain and spinal cord total lysate quantification. **, p < 0.01; N.S., non-significant (p > 0.05) (Student's t test).
Homozygous APP/hAβ/mutC (ki/ki) mice are viable, fertile, and exhibit a normal body weight as compared with their wild-type littermates (not shown). To evaluate the effect of the C-terminal mutation on APP expression, processing, and secretion, we first compared the mRNA levels of APP in brains of 2-month-old ki/ki mice and their wild-type littermates by quantitative real-time PCR using the APP knock-out mouse brains as a negative control and found no statistically significant differences between the two genotypes (Fig. 1B). Western blot analysis of total brain lysates using the 22C11 antibody (which recognizes an extracellular epitope) revealed two major bands with the upper band presumably full-length APP and the lower band soluble extracellular processed APP derivatives. Quantification of the total APP showed a slight reduction (∼20%) in the ki/ki samples (Fig. 1C, 22C11, quantified in Fig. 1G). Because both the lower band of total APP (Fig. 1C) and the production of PBS-extractable soluble APP ectodomains (Fig. 1D) were similar between the ki/ki and wild-type brains, the lower levels of total APP in the ki/ki samples must be due to the preferential reduction of full-length APP/hAβ/mutC protein. Similar results were obtained when spinal cord tissue was analyzed (Fig. 1, E and F). The absence of the APP intracellular domain and the introduction of human Aβ sequence in the APP/hAβ/mutC protein were confirmed by Western blotting using the C-terminal specific APPc antibody and human Aβ-specific 6E10 antibody, respectively (Fig. 1C, APPc and 6E10). This corresponds to the absence of APP/Fe65 interaction using the bimolecular fluorescence complementation assay (supplemental Fig. S2) (21) The reduced full-length APP/hAβ/mutC but normal soluble secreted APP is consistent with the observation that deletion of the APP intracellular domain leads to its reduced internalization and elevated secretion (Fig. 2D) (22).
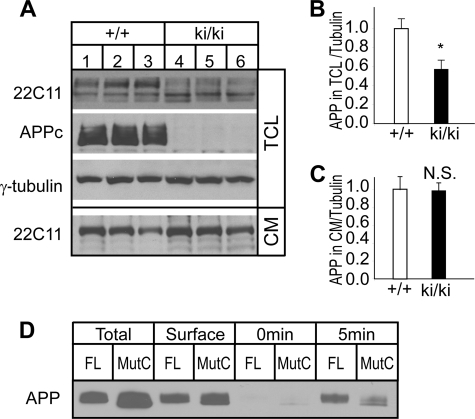
FIGURE 2.
Measurement of APP secretion from APP/hAβ/mutC knock-in neurons. A, Western blot analysis of APP expressed in total cell lysate (TCL) and conditioned medium (CM) of hippocampal neuronal cultures of wild-type (+/+) and homozygous ki/ki pups. Antibodies 22C11 and APPc recognize the APP N-terminal region and APP C-terminal region, respectively. γ-Tubulin blot was used as protein loading control. B and C are quantifications of 22C11 blots of APP to tubulin ratio in total cell lysate and conditioned medium, respectively. *, p < 0.05; N.S., non-significant (p > 0.05) (t test.). D, representative biotinylation assay showing a slower rate of APP internalization in the absence of the intracellular domain. HEK293 cells expressing full-length APP (FL) or APP/hAβ/mutC (MutC) constructs were cell surface biotinylated and incubated at 37 °C for 5 min to allow endocytosis of biotinylated APP. After stripping the remaining biotin from the cell surface (0 min), internalized biotinylated APP was isolated and detected by immunoblot using the 22C11 antibody (5 min). 10% of the lysate was reserved as a representative of the total protein expressed (Total) prior to the isolation of biotinylated surface proteins (Surface).
We next investigated APP processing and secretion using primary neuronal cultures prepared from newborn littermate ki/ki pups and their wild-type controls. At 14 days in vitro, conditioned medium and total cell lysates were collected and analyzed by Western blotting (Fig. 2A). Similar to that of the adult mouse brain and spinal cord, there was a significant reduction of APP in total cell lysates between the ki/ki and wild-type cultures as blotted by the N-terminal antibody 22C11, but the secretion of APPs into the conditioned medium was comparable (quantification in Fig. 2, B and C). Transfection of full-length or APP/hAβ/mutC constructs followed by examination of internalization of surface APP by biotinylation assay showed that, consistent with the published report (22), APP/hAβ/mutC exhibited reduced internalization, thus leading to elevated secretion (Fig. 2D). Overall, the results suggest that the highly conserved APP C-terminal domain is dispensable in APP extracellular processing and soluble APP production and secretion.
Analysis of Aβ Production and Amyloid Pathology in APP/hAβ/mutC Knock-in Animals
Having established that the APP C-terminal domain is dispensable for APP secretion, we next asked whether it is required for Aβ production and development of β-amyloid pathology. We used a sandwich ELISA to measure Aβ40 levels in 3-month-old APP/hAβ/mutC animals in comparison with another strain of full-length APP knock-in mice with humanized Aβ and Swedish and London FAD mutations (APP/hAβ) at the same age (16). This young age was chosen to avoid the confounding effects due to amyloid deposition. The results showed similar Aβ40 levels and dose-dependent increases in both strains, suggesting that the C-terminal replacement does not have significant impact on γ-secretase processing and Aβ production (Fig. 3A). Because studies of the APP/hAβ knock-in animals have shown that these mice develop minimal amyloid plaque pathology in their lifetime (16), to facilitate the development of amyloid pathology, we crossed the APP/hAβ/mutC and APP/hAβ animals with the PS1 knock-in mice carrying the M146V FAD mutation and created animals that are doubly homozygous for APP/hAβ/mutC or APP/hAβ and PS1M146V. The addition of the PS1M146V mutation resulted in a similar increase of Aβ levels in both APP knock-in alleles (Fig. 3A), suggesting that the C-terminal sequence does not affect the modification of γ-secretase processing by the PS1 FAD mutation.
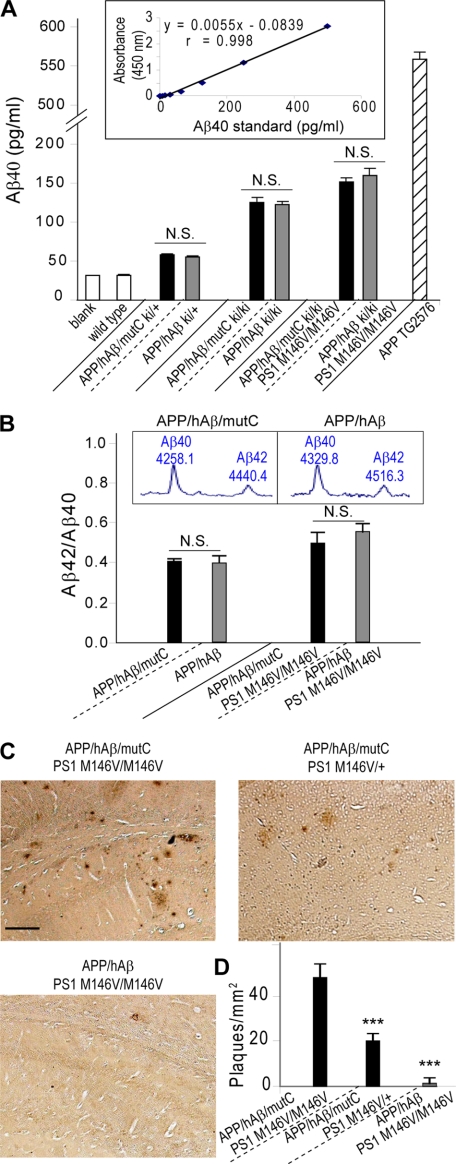
FIGURE 3.
Analysis of Aβ levels and plaque pathology in APP/hAβ/mutC mice. A, sandwich ELISA measurement of Aβ40 levels in the brains of APP/hAβ/mutC and APP/hAβ knock-in mice at 3 months of age. APP/hAβ/mutC and APP/hAβ mice are represented by black and gray bars, respectively. PS1 is wild type unless otherwise indicated, and blank represents buffer blank control of ELISA plate. The ELISA kit does not recognize mouse Aβ sequence in wild-type animals and was used as an additional negative control. The APP transgenic mice Tg2576 were used as positive control. n = 5/genotype. The Aβ40 peptide standard curve is shown in the inset. B, Aβ42/Aβ40 ratio determined by IP/MS in brains of 3-month-old APP/hAβ/mutC (black bars) or APP/hAβ (gray bars) knock-in mice with or without the PS1M146V mutation. n = 3 for each genotype. Typical mass spectrum traces for Aβ40 and Aβ42 are shown in the inset. C, representative plaque images in the hippocampus area of 13-month-old APP/hAβ/mutC ki/ki; PS1M146V/+, APP/hAβ/mutC ki/ki; PS1M146V/M146V, and APP/hAβ ki/ki; and PS1M146V/M146V mice. Scale bar, 100 μm. D, quantification of plaque load in the cortex and hippocampus of the above animals. ***, p < 0.001; N.S., non-significant (analysis of variance).
Due to the physiological expression of the APP alleles in the knock-in animals, the Aβ42 level is below the detection limit of the ELISA kit. We therefore resorted to a more sensitive immunoprecipitation/mass spectrometry method to measure the levels of Aβ42 and normalized the values to Aβ40 (Fig. 3B). Comparing the Aβ42/Aβ40 ratios from 3 month-old APP/hAβ/mutC and APP/hAβ knock-in mice, with or without the PS1M146V FAD mutation, we did not find any significant differences between the two knock-in lines, again suggesting that C-terminal region does not play a critical role in regulating the pathological processing of APP.
Immunostaining of 13-month-old APP/hAβ/mutC and PS1M146V double knock-in mice with the 6E10 antibody revealed abundant Aβ plaque deposits in cortex and hippocampus. The degree of Aβ pathology was dependent on the PS1M146V dosage (Fig. 3C and quantified in Fig. 3D). Consistent with the fact that the Arctic variants of Aβ are more robust in developing parenchymal amyloidosis (23, 24), the amyloid loads of APP/hAβ/mutC and PS1M146V double knock-in animals are much higher than the corresponding APP/hAβ and PS1M146V double knock-in animals without the Arctic mutation (Fig. 3, C and D).
Analysis of Survival and Neuromuscular Synapse Development in APP/hAβ/mutC Knock-in Animals
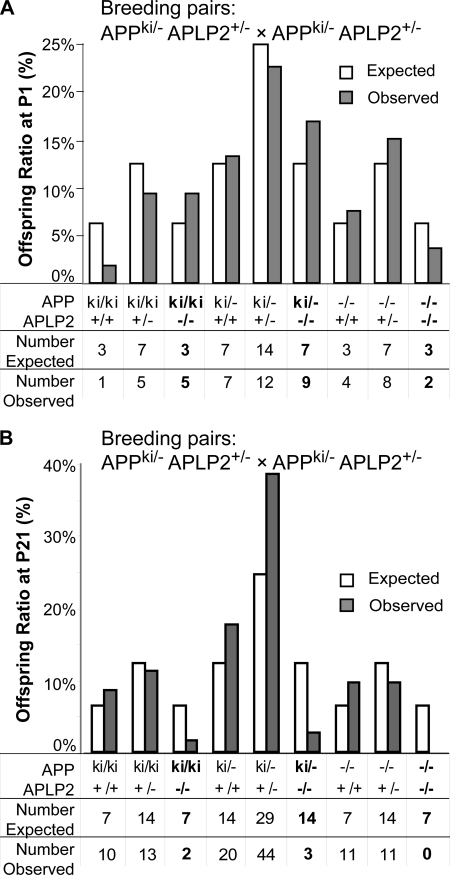
Our previous studies established that APP and APLP2 play essential yet redundant roles in animal viability and neuromuscular synapse assembly (11). To determine whether these developmental activities require the APP C-terminal domain, we performed intercrosses of mice with one copy of each of the APP/hAβ/mutC and APLP2-null mutation (APPki/−APLP2+/−). We determined the genotypes of the surviving offspring at postnatal day 1 (P1) and at weaning age (P21) and compared the number observed against the number expected (Fig. 4). The 54 newborn mice from the intercrossing analyzed showed a close to Mendelian ratio for all genotypes (Fig. 4A). However, few of the APPki/kiAPLP2−/− and APPki/−APLP2−/− mice survived to adulthood (Fig. 4B). Of the 113 offspring genotyped at weaning age, only ∼25% of the expected APPki/kiAPLP2−/− and APPki/−APLP2−/− mice were recovered (Fig. 4B), which was significantly different from the predicted Mendelian ratio (chi-square = 33.8; degrees of freedom (df), 8; p < 0.001). These results demonstrate that, in contrast to the reported dispensable role of the APP intracellular domain in APP-mediated growth, anatomical, and synaptic properties (7), the highly conserved APP C-terminal sequences are essential in postnatal viability.
FIGURE 4.
Postnatal lethality of APP/hAβ/mutC knock-in mice on APLP2-null background. A, analysis of genotypes of 53 offspring collected at P1 derived from mating of APPki/−/APLP2+/− males and females. Gray bars represent observed frequency of various genotypes as the percentage of total, and open bars are expected frequency based on Mendelian inheritance. Chi-square = 4.9; df, 8; p > 0.1. B, analysis of genotypes of 114 offspring collected at P21 derived from the same breeding as in A. Genotypes with impaired survival are highlighted in bold. Chi-square = 33.8; df, 8; p < 0.001.
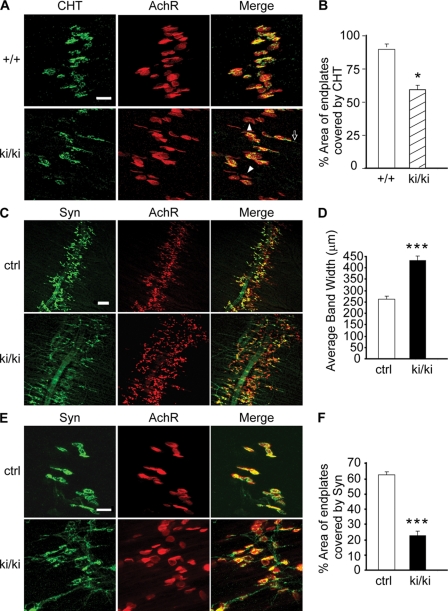
Our studies of APP-mediated NMJ development suggest that this activity correlates with a change in the presynaptic localization of the high affinity CHT and that these two proteins may physically interact via their intracellular sequences (12). The creation of the APP/hAβ/mutC mice allows testing of the role of the APP cytoplasmic tail in NMJ development. Indeed, similar to the APP-null mutant, immunostaining of CHT in the NMJ of APP/hAβ/mutC mice revealed clear mislocalization of CHT (Fig. 5, A and B). Moreover, whole-mount staining of diaphragm of newborn APPki/−APLP2−/− or APPki/kiAPLP2−/− pups with anti-synaptophysin antibody and α-bungarotoxin showed diffused presynaptic and postsynaptic distribution (Fig. 5, C and D) and reduced pre- and postsynaptic apposition (Fig. 5, E and F) indistinguishable from the APP/APLP2 double deficient mice. The combined results demonstrate an indispensable role of the highly conserved APP intracellular sequences in survival and proper CHT targeting and neuromuscular synapse development.
FIGURE 5.
Neuromuscular synapse defects in APP/hAβ/mutC mice. A, double labeling of P0 diaphragm muscles of wild-type (+/+) and APP/hAβ/mutC ki/ki littermates with the anti-CHT antibody and α-bungarotoxin that recognizes the postsynaptic acetylcholine receptors (AchR). Merge, overlay of CHT and AchR images. The open arrow marks the CHT staining beyond the end plates, and the arrowheads label the synaptic sites with sparse CHT staining. B, quantification of the percentage of AchR-positive endplates covered by CHT immunoreactivity (average ± S.E. of 20 endplates per genotype). C, whole-mount staining of P0 diaphragm muscles of APPki/kiAPLP2−/− mutants (ki/ki) and littermate APP+/+APLP2−/− controls (ctrl) with an anti-synaptophysin (Syn) antibody and α-bungarotoxin (AchR), showing diffused pre- and postsynaptic distribution in the ki/ki mutant. Merge, overlay of Syn and AchR images. D, quantification of the average bandwidth of AchR-positive endplates. E, higher magnification images of synapse structures showing axonal staining of Syn and poorly covered endplates by Syn and extrasynaptic Syn staining in the ki/ki mutant. Merge, overlay of Syn and AchR images. F, quantification of the percentage of AchR-positive endplates covered by Syn (average ± S.E. of 20 endplates/genotype). ***, p < 0.001; *, p < 0.05 (Student's t test). Scale bars in A and E, 20 μm; scale bar in C, 100 μm.
DISCUSSION
Genetic studies in C. elegans and mammals have established essential functions of APP proteins in development (4, 9, 26). Interestingly, expression of the APL-1 extracellular domain has been shown to be sufficient in rescuing the apl-1-null lethality (4). Likewise, expressing the soluble, α-secretase-cleaved APP ectodomain complements the anatomical and behavioral abnormalities of the APP-deficient mice (7). Both results argue for a dispensable role of the APP intracellular domain. However, our previous in vivo and HEK293/hippocampal mixed culture studies support an important activity of the APP C-terminal domain in modulating neuromuscular synapse and central synaptogenesis (12, 13). By creating mice with mutated APP intracellular sequences, we demonstrate here for the first time that the highly conserved APP intracellular sequences are required for APP-mediated survival and neuromuscular synapse assembly in vivo. It needs to be pointed out that to simultaneously determine the role of the APP intracellular domain in developmental function and Aβ pathogenesis, we introduced both the human Aβ sequence with FAD mutations and the C-terminal mutation in APP/hAβ/mutC knock-in animals. Therefore, it is conceivable that changes in the Aβ region contribute to the lethality and NMJ defects. We believe that it is highly unlikely because analysis of another APP knock-in strain in which the first Tyr residue of the YENPTY sequence was mutated, whereas the Aβ region was not altered, revealed similar developmental defects.4
Although the reason for the distinct domain requirement for C. elegans and mouse viability is not known, it is worth noting that the lethality of the apl-1-null worm is likely caused by a molting defect not relevant to mammals. Indeed, expression of the corresponding APP extracellular domain is not able to rescue the apl-1 deficiency (4). Phenotypes present in APP-null mice are rather diverse, and the underlying mechanisms are not established. As such, it is difficult to explain the apparent differential pathways mediating the APP activity in synaptic plasticity and synaptogenesis. The fact that APP exists both as a full-length protein and in multiple processed forms and that the cleavage products can be differentially sorted and independently transported makes it plausible that these APP isoforms confer distinct APP activities (27). Nevertheless, our previous mixed culture studies (13), combined with the current NMJ investigation, make a strong argument that full-length APP may play an important role not only in neuromuscular synapse development but also a central synaptogenesis and synaptic function as well.
We reported that APP is targeted to the synaptic sites of the NMJ, where it may modulate CHT activity through physical interaction mediated by the APP intracellular domain (12, 13). Our finding that expression of APP with mutated C-terminal sequences leads to aberrant CHT localization and impaired NMJ patterning strengthens this notion. However, APP is known to undergo kinesin-dependent trafficking via the C-terminal sequences (28). Although a recent report documented that the fast anterograde transport of APP does not require the intracellular domain or any sorting signal (29), we cannot exclude the possibility that the neuromuscular synapse defect seen in APP/hAβ/mutC mice is primarily caused by defective APP trafficking. Furthermore, APP intracellular sequences mediate additional activities, including interactions with multiple proteins (reviewed in Ref. 3) and transcriptional regulation via binding to Fe65 (30, 31), so it is therefore conceivable that defective adaptor protein interactions and intracellular signaling pathway may contribute to the developmental phenotypes seen in the APP/hAβ/mutC mice.
In contrast to its critical role in survival and neuromuscular synapse organization during development, we show here that the highly conserved APP intracellular domain does not overtly affect APP expression, processing, or secretion in adult brain or in primary neuronal cultures. The Aβ40 levels, Aβ42/Aβ40 ratio, and modulation by the PS1M146V FAD mutation are all comparable with a similar APP knock-in strain expressing the full-length protein. Although subtle effects of the Arctic mutation on APP localization or β-secretase cleavage as reported by Sahlin et al. (25) cannot be formally excluded, our results are consistent with the published reports that introduction of the Arctic mutation leads to a more aggressive amyloid pathology, likely due to enhanced Aβ fibrilization (23, 24). In light of the extensively published reports addressing the various effects of the Thr668 residue and the YENPTY sequence on APP localization, trafficking, and processing (reviewed in Refs. 2 and 3), the relatively normal APP and Aβ metabolism in the APP/hAβ/mutC mice is therefore unexpected. Differences in the model systems (in vivo versus in vitro), expression levels (physiological versus overexpression), cell types (neurons versus non-neuronal cells), and the nature of the systems (chronic versus acute) could all contribute to the contrasting findings between our work and the published studies.
Because of the central role of APP in Alzheimer disease, it is essential to understand the mechanisms mediating its physiological function and pathogenesis. By creating a novel APP knock-in allele that allows us to examine the in vivo function of the highly conserved APP intracellular domain in developmental regulation and Aβ pathology, we report here that the two pathways can be genetically uncoupled. Because the APP intracellular domain is critical for its physiological function but dispensable for Aβ production, targeting this region may thus lead to undesirable physiological impairment rather than anticipated Aβ modulation.
Acknowledgments
We thank N. Aithmitti and X. Chen for expert technical assistance and members of the Zheng laboratory for stimulating discussions. We are grateful to the Baylor College of Medicine Eunice Kennedy Shriver Intellectual and Developmental Disabilities Research Center (HD024064) for support in confocal imaging.
This work was supported, in whole or in part, by National Institutes of Health Grants AG020670 and AG032051 (to H. Z.), NS061777 (to R. W.), and MH52804 (to T. C. S.). This work was also supported by Alzheimer's Association Grant IIRG-05-14824 (to R. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental methods and references and Figs. S1 and S2.
This article was selected as a Paper of the Week.
Z. Wang, H. Zheng, A. Barbagallo, and L. D'Adamio, unpublished data.
- APP
- amyloid precursor protein
- Aβ
- β-amyloid
- hAβ
- humanized Aβ
- FAD
- familial Alzheimer disease
- NMJ
- neuromuscular junction
- CHT
- choline transporter
- df
- degrees of freedom
- ki
- knock-in
- AchR
- acetylcholine receptors
- Syn
- synaptophysin
- P
- postnatal day.
REFERENCES
- 1.Hardy J. (2006) Neuron 52, 3–13 [DOI] [PubMed] [Google Scholar]
- 2.Zheng H., Koo E. H. (2006) Mol. Neurodegener. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King G. D., Scott Turner R. (2004) Exp. Neurol. 185, 208–219 [DOI] [PubMed] [Google Scholar]
- 4.Hornsten A., Lieberthal J., Fadia S., Malins R., Ha L., Xu X., Daigle I., Markowitz M., O'Connor G., Plasterk R., Li C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1971–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng H., Jiang M., Trumbauer M. E., Sirinathsinghji D. J., Hopkins R., Smith D. W., Heavens R. P., Dawson G. R., Boyce S., Conner M. W., Stevens K. A., Slunt H. H., Sisoda S. S., Chen H. Y., Van der Ploeg L. H. (1995) Cell 81, 525–531 [DOI] [PubMed] [Google Scholar]
- 6.Dawson G. R., Seabrook G. R., Zheng H., Smith D. W., Graham S., O'Dowd G., Bowery B. J., Boyce S., Trumbauer M. E., Chen H. Y., Van der Ploeg L. H., Sirinathsinghji D. J. (1999) Neuroscience 90, 1–13 [DOI] [PubMed] [Google Scholar]
- 7.Ring S., Weyer S. W., Kilian S. B., Waldron E., Pietrzik C. U., Filippov M. A., Herms J., Buchholz C., Eckman C. B., Korte M., Wolfer D. P., Müller U. C. (2007) J. Neurosci. 27, 7817–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young-Pearse T. L., Bai J., Chang R., Zheng J. B., LoTurco J. J., Selkoe D. J. (2007) J. Neurosci. 27, 14459–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Koch C. S., Zheng H., Chen H., Trumbauer M., Thinakaran G., van der Ploeg L. H., Price D. L., Sisodia S. S. (1997) Neurobiol. Aging 18, 661–669 [DOI] [PubMed] [Google Scholar]
- 10.Heber S., Herms J., Gajic V., Hainfellner J., Aguzzi A., Rülicke T., von Kretzschmar H., von Koch C., Sisodia S., Tremml P., Lipp H. P., Wolfer D. P., Müller U. (2000) J. Neurosci. 20, 7951–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P., Yang G., Mosier D. R., Chang P., Zaidi T., Gong Y. D., Zhao N. M., Dominguez B., Lee K. F., Gan W. B., Zheng H. (2005) J. Neurosci. 25, 1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B., Yang L., Wang Z., Zheng H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14140–14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Wang B., Yang L., Guo Q., Aithmitti N., Songyang Z., Zheng H. (2009) J. Neurosci. 29, 10788–10801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Q., Fu W., Sopher B. L., Miller M. W., Ware C. B., Martin G. M., Mattson M. P. (1999) Nat. Med. 5, 101–106 [DOI] [PubMed] [Google Scholar]
- 15.Wang R., Wang B., He W., Zheng H. (2006) J. Biol. Chem. 281, 15330–15336 [DOI] [PubMed] [Google Scholar]
- 16.Köhler C., Ebert U., Baumann K., Schröder H. (2005) Neurobiol Dis. 20, 528–540 [DOI] [PubMed] [Google Scholar]
- 17.O'Gorman S., Dagenais N. A., Qian M., Marchuk Y. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14602–14607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uljon S. N., Mazzarelli L., Chait B. T., Wang R. (2000) Methods Mol. Biol. 146, 439–452 [DOI] [PubMed] [Google Scholar]
- 19.Lehman E. J., Kulnane L. S., Gao Y., Petriello M. C., Pimpis K. M., Younkin L., Dolios G., Wang R., Younkin S. G., Lamb B. T. (2003) Hum. Mol. Genet. 12, 2949–2956 [DOI] [PubMed] [Google Scholar]
- 20.Dineley K. T., Xia X., Bui D., Sweatt J. D., Zheng H. (2002) J. Biol. Chem. 277, 22768–22780 [DOI] [PubMed] [Google Scholar]
- 21.Kerppola T. K. (2006) Nat. Rev. Mol. Cell Biol. 7, 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koo E. H., Squazzo S. L., Selkoe D. J., Koo C. H. (1996) J. Cell Sci. 109, 991–998 [DOI] [PubMed] [Google Scholar]
- 23.Nilsberth C., Westlind-Danielsson A., Eckman C. B., Condron M. M., Axelman K., Forsell C., Stenh C., Luthman J., Teplow D. B., Younkin S. G., Näslund J., Lannfelt L. (2001) Nat. Neurosci. 4, 887–893 [DOI] [PubMed] [Google Scholar]
- 24.Cheng I. H., Palop J. J., Esposito L. A., Bien-Ly N., Yan F., Mucke L. (2004) Nat. Med. 10, 1190–1192 [DOI] [PubMed] [Google Scholar]
- 25.Sahlin C., Lord A., Magnusson K., Englund H., Almeida C. G., Greengard P., Nyberg F., Gouras G. K., Lannfelt L., Nilsson L. N. (2007) J. Neurochem. 101, 854–862 [DOI] [PubMed] [Google Scholar]
- 26.Herms J., Anliker B., Heber S., Ring S., Fuhrmann M., Kretzschmar H., Sisodia S., Müller U. (2004) EMBO J. 23, 4106–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muresan V., Varvel N. H., Lamb B. T., Muresan Z. (2009) J. Neurosci. 29, 3565–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koo E. H., Sisodia S. S., Archer D. R., Martin L. J., Weidemann A., Beyreuther K., Fischer P., Masters C. L., Price D. L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Back S., Haas P., Tschäpe J. A., Gruebl T., Kirsch J., Müller U., Beyreuther K., Kins S. (2007) J. Neurosci. Res. 85, 2580–2590 [DOI] [PubMed] [Google Scholar]
- 30.Cao X., Südhof T. C. (2001) Science 293, 115–120 [DOI] [PubMed] [Google Scholar]
- 31.Cao X., Südhof T. C. (2004) J. Biol. Chem. 279, 24601–24611 [DOI] [PubMed] [Google Scholar]