Abstract
Resident tissue macrophages are activated by the fungal pathogen Candida albicans to release eicosanoids, which are important modulators of inflammation and immune responses. Our objective was to identify the macrophage receptors engaged by C. albicans that mediate activation of group IVA cytosolic phospholipase A2 (cPLA2α), a regulatory enzyme that releases arachidonic acid (AA) for production of prostaglandins and leukotrienes. A comparison of peritoneal macrophages from wild type and knock-out mice demonstrates that the β-glucan receptor Dectin-1 and MyD88 regulate early release of AA and eicosanoids in response to C. albicans. However, cyclooxygenase 2 (COX2) expression and later phase eicosanoid production are defective in MyD88−/− but not Dectin-1−/− macrophages. Furthermore, C. albicans-stimulated activation of MAPK and phosphorylation of cPLA2α on Ser-505 are regulated by MyD88 and not Dectin-1. In contrast, Dectin-1 mediates MAPK activation, cPLA2α phosphorylation, and COX2 expression in response to particulate β-glucan suggesting that other receptors engaged by C. albicans preferentially mediate these responses. Results also implicate the mannan-binding receptor Dectin-2 in regulating cPLA2α. C. albicans-stimulated MAPK activation and AA release are blocked by d-mannose and Dectin-2-specific antibody, and overexpression of Dectin-2 in RAW264.7 macrophages enhances C. albicans-stimulated MAPK activation, AA release, and COX2 expression. In addition, calcium mobilization is enhanced in RAW264.7 macrophages overexpressing Dectin-1 or -2. The results demonstrate that C. albicans engages both β-glucan and mannan-binding receptors on macrophages that act with MyD88 to regulate the activation of cPLA2α and eicosanoid production.
Keywords: Arachidonic Acid, Eicosanoid, Innate Immunity, Macrophage, MAPKs, Candida albicans, Dectin-1, MyD88, Cytosolic Phospholipase A2
Introduction
The innate immune system is an important first line of defense against invasive pathogens (1). Myeloid cells, including mononuclear phagocytes and granulocytes, that first encounter microorganisms initiate innate immune responses to contain infections. Professional phagocytes possess a number of receptors, such as Toll-like receptors and C-type lectin receptors, which recognize specific molecular components on microorganisms (2–4). Engagement of these receptors initiates signaling pathways that trigger the production of chemokines and cytokines that are important for recruiting other myeloid cells to the site of infection and for initiating adaptive immunity (5, 6).
Candida albicans is a human commensal that colonizes the gastrointestinal tract, skin, and mucosal surfaces. It is an opportunistic fungal pathogen in immunocompromised hosts and the critically ill, and is the principal cause of mycoses worldwide (7). C. albicans is responsible for a large proportion of nosocomial bloodstream infections with a crude mortality rate of over 40% (7). It invades through injuries in the skin or mucosa and can colonize most tissues particularly the gastrointestinal tract, lung, kidney, and brain. Toll-like receptors and C-type lectin receptors have been identified on macrophages that recognize cell wall components of C. albicans (8–10). The cell wall of C. albicans is composed of polysaccharides of glucose (β-1,3- and -1,6-glucans), N-acetyl-d-glucosamine (chitin), and mannose (mannans) (11–14). TLR4, the C-type lectin receptors Dectin-2 and Mincle, and the macrophage mannose receptor have been implicated in the recognition of C. albicans mannans (8, 10, 15–17). A number of receptors have been reported to bind to β-glucan, including Dectin-1, lactosylceramide, complement receptor 3, and scavenger receptors (18–21). However, there is considerable evidence implicating the phagocytic receptor Dectin-1 in mediating macrophage responses to fungal agents and in regulating immune defense to fungal infection in mice and humans (22–30). Dectin-1 contains a C-type lectin-like extracellular domain and an immunoreceptor tyrosine-based activation-like motif in the cytoplasmic tail that signals through spleen tyrosine kinase (Syk) and CARD-9 (26, 31).
We previously reported that C. albicans activates group IVA cytosolic phospholipase A2 (cPLA2α)3 in resident mouse peritoneal and alveolar macrophages (32, 33). cPLA2α releases arachidonic acid (AA) that is metabolized to a number of bioactive lipid mediators such as prostaglandins and leukotrienes. Eicosanoids are secreted by cells and regulate acute inflammation and innate immune responses (34). They act locally in an autocrine or paracrine manner by binding to specific G-protein-coupled receptors.
Considerable progress has been made in identifying the receptors engaged by C. albicans and the signaling pathways that promote cytokine production, but the regulation of cPLA2α activation and lipid mediator production is poorly understood. cPLA2α is regulated post-translationally by increases in intracellular calcium and phosphorylation (35). Calcium binds to the C2 domain of cPLA2α and promotes translocation from the cytosol to intracellular membranes for accessing phospholipid substrate (36–38). Phosphorylation of Ser-505 by MAPK enhances the hydrolytic activity of cPLA2α (39, 40). Our previous results implicated a β-glucan receptor in mediating the activation of cPLA2α by live C. albicans in resident peritoneal macrophages (32). Results of this study suggest a role for Dectin-1, -2, and MyD88-dependent pathways in regulating cPLA2α activation and the production of eicosanoids in macrophages.
EXPERIMENTAL PROCEDURES
Materials
Zymosan was purchased from Sigma and boiled in PBS three times before use. Particulate β-glucan was purified from Saccharomyces cerevisiae and structurally characterized by NMR (41). Endotoxin-free water-soluble glucan phosphate (soluble glucan-P) was prepared from particulate β-glucan as described previously (42). [5,6,8,9,11,12,14,15-3H]AA (specific activity 100 Ci/mmol) was from PerkinElmer Life Sciences. Fetal bovine serum (FBS) (Gemini Bio-Products) was heat-inactivated at 56 °C for 30 min before use. Dulbecco's modified Eagle's medium (DMEM) was from Cambrex BioScience. Human serum albumin was obtained from Intergen. MAPK inhibitors U0126 and SB202190 were obtained from Calbiochem. Polyclonal antibodies to murine COX2 and β-tubulin were from Cayman Chemical Co. Polyclonal antibody to cPLA2α was raised as described previously (43). Antibodies to phosphorylated ERKs, p38, and cPLA2α (Ser-505) were obtained from Cell Signaling Technology, Inc. Anti-Dectin-2 monoclonal antibody D2.11E4 was generated as described previously (15), and isotype control rat-IgG2a was obtained from BioLegend. Fluo-4-AM was from Invitrogen. Zeocin was purchased from InvivoGen and G418 from Mediatech, Inc.
Mouse Strains
Pathogen-free BALB/c mice were obtained from Harlan Sprague-Dawley. cPLA2α−/− mice were generated using 129 embryonic stem cells in a C57BL/6 strain as described previously (44). The mixed strain was backcrossed onto a BALB/c background and used after 10 generations. The TLR4 mutant mouse strain C3H/HeJ and control strain C3H/HeOuJ were obtained from The Jackson Laboratory. TLR2−/− (C57BL/6) and MyD88−/− mice (C57BL/6/129) were generated as described previously (45). MyD88+/− C57BL/6/129 mice were crossed to generate MyD88−/− mice and MyD88+/+ littermate controls. C57BL/6 control mice were obtained from The Jackson Laboratory. TLR9-deficient mice (BALB/c) were provided by Dr. Ted Standiford (University of Michigan). Dectin-1−/− mice (129sv/ev) were produced as described previously (28), and age- and strain-matched controls were obtained from Taconic Farms, Inc. Mice were used for macrophage isolation at 7–12 weeks of age.
C. albicans Strains and Culture
C. albicans (ATCC 10261) was grown on Sabouraud dextrose agar plates and maintained at 4 °C. The day before the experiment, it was streaked onto a fresh plate and incubated overnight at 37 °C. C. albicans was scraped from the plate and washed twice in endotoxin-free PBS.
Peritoneal Macrophage Isolation and AA Release Assay
Resident mouse peritoneal macrophages were obtained by peritoneal lavage as described previously (32). Cells were plated at a density of 5 × 105/cm2 (48-well plate) and incubated for 2 h at 37 °C in a humidified atmosphere of 5% CO2 in air. After washing the cultures to remove nonadherent cells, the adherent macrophages were incubated in DMEM containing 10% heat-inactivated FBS, 100 μg/ml streptomycin sulfate, 100 units/ml penicillin G, 0.29 mg/ml glutamine, and [3H]AA (0.1 μCi/250 μl/well) for 16–18 h at 37 °C. The cells were washed twice with serum-free DMEM containing 0.1% human serum albumin (stimulation medium) to remove unincorporated [3H]AA and then incubated in stimulation medium. After stimulation of the macrophages, the culture medium was removed and centrifuged, and the amount of radioactivity released into the medium was measured by scintillation counting. The cell-associated radioactivity was measured following solubilization of the monolayer with 0.1% Triton X-100. The amount of radioactivity released is expressed as percent of the total radioactivity incorporated (cell-associated plus medium).
RAW264.7 Macrophages Stably Expressing Dectin-1 or -2
Dectin-1 and -2 were cloned into pFBNeo (Stratagene) or pMXs-IZ vectors, respectively (46). They were transfected into the HEK293T-based Phoenix ecotropic packaging cells using FuGENE 6, and retroviral supernatants were collected after 48 h to transduce RAW264.7 cells expressing previously either pMXs-IZ or pFBNeo, respectively. RAW264.7 cells expressing both empty vectors were also generated. Cells were selected and maintained in 0.6 mg/ml geneticin (Sigma) and 0.4 mg/ml Zeocin (Invitrogen). For FACS analysis, RAW264.7 were cultured in a 24-well plate at 5 × 104 cells per well. The next day, the medium was removed, and cells were incubated in blocking buffer (PBS containing 5% heat-inactivated rabbit serum, 0.5% BSA, 5 mm EDTA, 2 mm NaN3) for 1 h at 4 °C. Cells were incubated with a 1:50 dilution of Alexa647-labeled anti-Dectin-1 or isotype control (AbD Serotec) for 1 h at 4 °C, washed three times with washing buffer (PBS containing 0.5% BSA, 5 mm EDTA, and 2 mm NaN3), and resuspended in 1% formaldehyde (in PBS). The anti-Dectin-2 antibody (clone D2.11E4) or its IgG2a isotype control (OX11) was added at 10 μg/ml and incubated for 1 h at 4 °C. After two washes, a phycoerythrin-conjugated anti-rat antibody (Jackson ImmunoResearch) was added for a further 1 h at 4 °C. After this time, cells were treated as those incubated with directly labeled antibodies. Data were acquired on a CyAn ADP analyzer (Beckman-Coulter) and analysis was performed using Summit software (Beckman-Coulter).
For measuring AA release, RAW264.7 cells were plated at 7.5 × 104/cm2 well (48-well plates) in RPMI 1640 medium containing 10% heat-inactivated FBS, 0.6 mg/ml G418, 0.4 mg/ml Zeocin, 100 μg/ml streptomycin sulfate, 100 units/ml penicillin G, and 0.29 mg/ml glutamine. After ∼6 h, they were washed once and incubated in RPMI containing [3H]AA, and release of AA was measured as described above for mouse peritoneal macrophages.
C. albicans Recognition Assay
Peritoneal macrophages cultured overnight as described above were incubated with C. albicans (m.o.i. 5) in stimulation medium for 1 h. Macrophages were washed three times with PBS and then lysed with 0.1% Triton X-100 in PBS. C. albicans in the lysate was plated on Sabouraud dextrose agar plates. The number of colony-forming units was determined after incubation of plates for 24 h at 37 °C. The phagocytic index was determined by using microscopy to quantify C. albicans phagocytosis by macrophages as described previously (47).
Mass Spectrometry Eicosanoid Analysis
The culture medium was collected 1 and 6 h after macrophage stimulation, centrifuged, and then stored at −80 °C. The media were thawed and mixed with an equal volume of cold methanol prior to analysis. The samples were diluted in water to a final methanol concentration of <15% and then extracted using a solid phase extraction cartridge (Strata Polymeric Reversed Phase 60 mg/ml, Phenomenex, Torrance, CA). The eluate (1 ml of methanol) was dried and reconstituted in 75 μl of HPLC solvent A (8.3 mm acetic acid and buffered to pH 5.7 with NH4OH) and 25 μl of solvent B (acetonitrile/methanol, 65:35, v/v). An aliquot of each sample (50 μl) was injected into an HPLC, and metabolites were separated on a C18 column (Ascentis 15 cm × 2.1 mm, 5 μm, Supelco), eluted at a flow rate of 200 μl/min with a linear gradient from 25 to 75% solvent B in 13 min, and then increased to 98% in 2 min and held for 11 min. The HPLC system was directly interfaced into the electrospray ionization source of a triple quadrupole mass spectrometer (Sciex API 3000, PE-Sciex, Thornhill Ontario, Canada). Mass spectrometric analyses were performed in the negative ion mode using multiple reaction monitoring for specific analytes. Deuterated internal standards were detected using the following transitions: m/z 355→275 for d4-prostaglandin (PG) E2; m/z 373→173 for d4-thromboxane B2; m/z 373→167 for d4-6-keto-PGF1α; m/z 311→213 for d8-AA; m/z 629→272 for d4-LTC4; and m/z 327→116 for d8-5-HETE. Eicosanoids were detected centered in specific retention time (RT) windows using the following transitions and limits of quantitation: PGE2, RT 9.3 min, m/z 351→271, 8 pg/ml; thromboxane B2, RT 8.3 min, m/z 369→169, 40 pg/ml; AA, RT 20.5 min, m/z 303→259, 24 pg/ml; 6-keto-PGF1α, RT 6.4 min, m/z 369→163, 40 pg/ml; LTC4, RT 10.1 min, m/z 624→272, 40 pg/ml; 5-HETE, RT 17.2 min, m/z 319→115, 8 pg/ml; 15-HETE, RT 16.5 min, m/z 319→219, 8 pg/ml. Multiple reaction monitoring chromatograms using a similar analytic scheme have been described previously (48). Quantitative results were calculated by determining the ratio of the signal of an analyte to that for an internal standard and comparing with a standard isotope dilution curve (49).
Western Blots
To prepare lysates for Western blots, cell monolayers were washed twice in ice-cold PBS and then scraped in lysis buffer: 50 mm Hepes, pH 7.4, 150 mm sodium chloride, 10% glycerol, 1% Triton X-100, 1 mm EGTA, 1 mm EDTA, 200 μm sodium vanadate, 10 mm tetrasodium pyrophosphate, 100 mm sodium fluoride, 300 nm p-nitrophenyl phosphate, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. After incubation on ice for 30 min, lysates were centrifuged at 15,000 rpm for 15 min, and protein concentration in the supernatant was determined by the bicinchoninic acid method. Lysates were boiled for 5 min after addition of Laemmli electrophoresis sample buffer, and proteins were then separated on 10% SDS-polyacrylamide gels. After transfer to nitrocellulose membrane, samples were incubated in blocking buffer (20 mm Tris-HCl, pH 7.6, 137 mm NaCl, 0.05% Tween (TTBS)) containing 5% nonfat milk for 1 h, and then incubated overnight at 4 °C with primary antibodies in TTBS. The membranes were incubated with anti-rabbit IgG horseradish peroxidase antibody (1:5000) in TTBS for 30 min at room temperature. The immunoreactive proteins were detected using the ECL system from Amersham Biosciences.
Calcium Imaging
RAW264.7 cells expressing Dectin-1 or -2 and vector control cells were cultured overnight in glass-bottomed MatTek plates in supplemented RPMI 1640 medium. Cells were washed with Hanks' balanced salt solution containing 25 mm Hepes, pH 7.4, and incubated with 5 μm Fluo-4-AM in the presence of 0.02% pluronic acid for 45 min at 25 °C. After loading, the cells were washed with and incubated in phenol red-free DMEM containing 25 mm Hepes, pH 7.4, for 30 min. Microscopy was conducted on an inverted Zeiss 200 M microscope driven by Intelligent Imaging Innovations Inc. software (Slidebook 4.2). Images were collected every 5 s after adding zymosan. Fluorescent traces of individual cells represent the fold fluorescence over base line at time 0 (FT/F0) after background subtraction.
Statistics
Statistics were calculated in GraphPad using unpaired t test to obtain two-tailed p values.
RESULTS
Role of Dectin-1 in Regulating AA Release and Eicosanoid Production
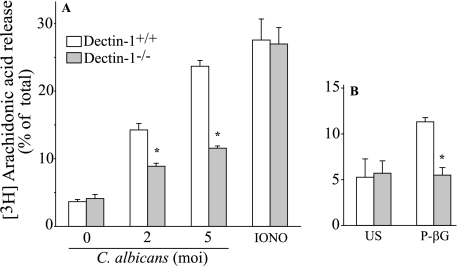
We reported that C. albicans-stimulated AA release is blocked by soluble glucan-P implicating a β-glucan receptor (32). To determine whether Dectin-1 is the β-glucan receptor that mediates cPLA2α activation and eicosanoid production in resident peritoneal macrophages in response to live C. albicans, we compared Dectin-1+/+ and Dectin-1−/− macrophages (Fig. 1). Dectin-1−/− macrophages release ∼55–62% less AA in response to C. albicans (m.o.i. 2–5) than wild type macrophages. Similar results are obtained with zymosan (data not shown). In contrast, AA release is identical in Dectin-1+/+ and Dectin-1−/− macrophages stimulated with ionomycin, which activates cPLA2α in a receptor-independent manner by increasing intracellular calcium. Purified particulate β-glucan stimulates AA release from Dectin-1+/+ macrophages but not from Dectin-1−/− macrophages indicating that it acts specifically through Dectin-1 (Fig. 1B).
FIGURE 1.
Role of Dectin-1 in regulating AA release in peritoneal macrophages treated with C. albicans. [3H]AA-labeled wild type and Dectin-1−/− macrophages were stimulated with C. albicans (ATCC 10261, serotype A), 1 μm ionomycin (IONO) (A) or 100 μg/ml particulate β-glucan (P-βG) (B). The amount of [3H]AA released into the medium from stimulated and unstimulated (US) cells is expressed as a % of the total incorporated radioactivity (cells plus medium). The data are the average of three experiments ±S.E. The asterisk indicates a significant decrease (p < 0.05) for Dectin-1−/− macrophages compared with wild type macrophages.
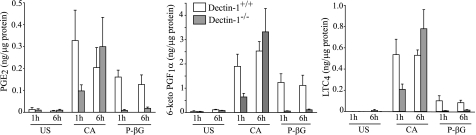
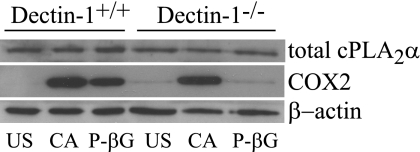
Resident peritoneal macrophages stimulated with C. albicans for 3 and 6 h produce PGE2, PGI2 (measured as the stable metabolite 6-keto-PGF1α), and LTC4 as the major oxygenated metabolites of AA from the cyclooxygenase and lipoxygenase pathways, and lower levels of thromboxane B2, 5-HETE, and 15-HETE (Table 1). Their levels are reduced to base line in cPLA2α−/− macrophages (or were below the level of detection) confirming an essential role for cPLA2α. The role of Dectin-1 in mediating eicosanoid production at early (1 h) and later (6 h) times after adding C. albicans was determined. Levels of PGE2, 6-keto-PGF1α, and LTC4 1 h after C. albicans (m.o.i. 5) addition are ∼65% lower in Dectin-1−/− macrophages (Fig. 2) but by 6 h levels are similar in Dectin-1−/− and Dectin-1+/+ macrophages (Fig. 2). For determining the levels of eicosanoids produced 6 h after stimulation, lower amounts of C. albicans (m.o.i. 2) are initially added to avoid overgrowth of the yeast in the culture medium. In contrast to C. albicans, particulate β-glucan-stimulated eicosanoid production is dependent on Dectin-1 at both 1 and 6 h (Fig. 2). When normalized to the amount of eicosanoids produced by wild type macrophages, Dectin-1−/− macrophages produce significantly less eicosanoids when stimulated with C. albicans for 1 h, but not at 6 h, and significantly less with particulate β-glucan at both 1 and 6 h (supplemental Table 1). C. albicans and particulate β-glucan stimulate expression of COX2 in peritoneal macrophages that occurs maximally by 6 h (32). COX2 expression is completely attenuated in Dectin-1−/− macrophages stimulated with particulate β-glucan but is not significantly decreased in Dectin-1−/− macrophages treated with C. albicans (Fig. 3). Levels of total cPLA2α are similar in unstimulated Dectin-1+/+ and Dectin-1−/− macrophages and are not affected by infection with C. albicans for 6 h.
TABLE 1.
Levels of eicosanoids produced by wild type and cPLA2α−/− peritoneal macrophages stimulated with C. albicans for 3 and 6 h
The lower limit of quantitation for the eicosanoids is specified under “Experimental Procedures.” The following abbreviations are used: US, unstimulated; TXB2, thromboxane B2; ND, not detected.
| Time | Eicosanoid | cPLA2α+/+ |
cPLA2α−/− |
||
|---|---|---|---|---|---|
| US | C. albicans | US | C. albicans | ||
| ng/ml | ng/ml | ||||
| 3 h | PGE2 | ND | 2.44 ± 0.834 | ND | ND |
| 6-Keto-PGF1α | ND | 1.90 ± 0.618 | ND | ND | |
| TXB2 | 0.01 ± 0.000 | 0.11 ± 0.022 | ND | ND | |
| LTC4 | ND | 3.33 ± 0.237 | 0.07 ± 0.091 | 0.01 ± 0.004 | |
| 5-HETE | 0.02 ± 0.000 | 0.26 ± 0.022 | 0.02 ± 0.005 | 0.02 ± 0.000 | |
| 15-HETE | 0.03 ± 0.015 | 0.19 ± 0.042 | 0.04 ± 0.004 | 0.04 ± 0.009 | |
| 6 h | PGE2 | ND | 3.07 ± 0.208 | ND | ND |
| 6-Keto-PGF1α | ND | 2.90 ± 0.337 | ND | ND | |
| TXB2 | ND | 0.16 ± 0.002 | ND | ND | |
| LTC4 | 0.01 ± 0.004 | 5.74 ± 0.497 | 0.03 ± 0.005 | 0.02 ± 0.007 | |
| 5-HETE | 0.02 ± 0.004 | 0.30 ± 0.043 | 0.02 ± 0.001 | 0.02 ± 0.010 | |
| 15-HETE | 0.06 ± 0.003 | 0.26 ± 0.004 | 0.04 ± 0.010 | 0.05 ± 0.002 | |
FIGURE 2.
Role of Dectin-1 in mediating eicosanoid production in peritoneal macrophages. Wild type (open bars) and Dectin-1−/− (shaded bars) macrophages were incubated with live C. albicans for 1 (m.o.i. 5) or 6 h (m.o.i. 2) or with 100 μg/ml P-βG. A lower m.o.i. of C. albicans is used for the longer time point to avoid overgrowth in the culture medium. The culture medium was analyzed for eicosanoids by mass spectrometry. The data are the average of three experiments ± S.E. US, unstimulated; CA, C. albicans.
FIGURE 3.
Role of Dectin-1 in COX2 induction. Expression of COX2 and total cPLA2α in wild type and Dectin-1−/− peritoneal macrophages was determined by Western blotting. Cells were unstimulated (US) or stimulated with C. albicans (CA, m.o.i. 2) or 100 μg/ml P-βG for 6 h. Sample loading was determined using antibodies to β-actin. Results are representative of three independent experiments.
MyD88 Contributes to cPLA2α Activation and Eicosanoid Production
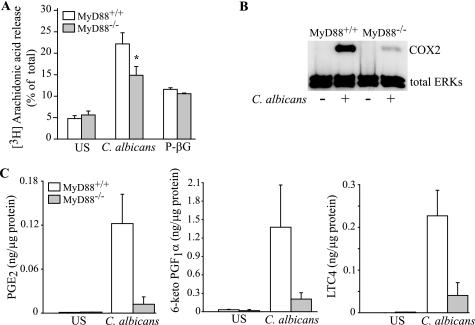
The results above demonstrate that particulate β-glucan acts through Dectin-1 to activate cPLA2α, COX2 expression, and eicosanoid production. However, in C. albicans-stimulated macrophages, Dectin-1 contributes to the early activation of cPLA2α, but other receptors promote COX2 expression and delayed phase eicosanoid production. We previously reported that TLR2 and TLR4, which are implicated in mediating fungal responses, do not play a role in regulating short term AA release in resident peritoneal macrophages stimulated with zymosan and C. albicans, but TLR2 partially mediates up-regulation of COX2 and prostanoid production (32). We investigated the role of MyD88, the adaptor protein necessary for transducing signals of several other Toll-like receptors and the interleukin-1 receptor (3). MyD88−/− macrophages stimulated for 1 h with C. albicans release ∼50% less AA than wild type cells (Fig. 4A). In contrast, wild type and MyD88−/− macrophages release similar amounts of AA in response to particulate β-glucan. Eicosanoid production at 1 h after adding C. albicans is also partially decreased in MyD88−/− macrophages (data not shown). The up-regulation of COX2 6 h after adding C. albicans is almost completely ablated in MyD88−/− peritoneal macrophages (Fig. 4B), and the production of PGE2, PGI2, and LTC4 at 6 h is reduced by 80–90% (Fig. 4C). In contrast to Dectin-1−/− macrophages, eicosanoid production in MyD88−/− macrophage stimulated with C. albicans for 6 h is significantly lower than in wild type macrophages (supplemental Table 2). The results suggest a role for a receptor that uses the MyD88 adaptor for signaling particularly for regulating late phase eicosanoid production and COX2 expression.
FIGURE 4.
MyD88 regulates AA release, eicosanoid production, and COX2 expression. Wild type and MyD88−/− peritoneal macrophages were stimulated with C. albicans (m.o.i. 5) and 100 μg/ml P-βG for 60 min to measure [3H]AA release (A), and with C. albicans (m.o.i. 2) for 6 h to evaluate COX2 expression on Western blots using antibodies to total ERKs to evaluate sample loading (B), and to measure eicosanoids in the culture medium by mass spectrometry (C). The data are the average of nine experiments for C. albicans and five experiments for P-βG ± S.E. (A), the average of three experiments ± S.E. (C), and a representative of three independent experiments (B). The asterisk in A indicates a significant decrease (p < 0.05) for MyD88−/− macrophages compared with wild type macrophages. US, unstimulated.
Role of a Mannan-binding Receptor
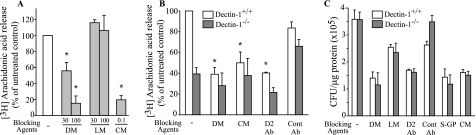
Blocking agents were used to investigate if a C. albicans mannan-binding receptor regulates cPLA2α activation in peritoneal macrophages. Mannans from C. albicans and d-mannose, but not l-mannose, block C. albicans-stimulated AA release from peritoneal macrophages (BALB/c) (Fig. 5A). The release of AA stimulated by ionomycin is not blocked by d-mannose or C. albicans mannan (data not shown) suggesting that blocking is not due to a nonspecific effect on activation of cPLA2α. Resident peritoneal macrophages poorly express the mannose receptor suggesting a role for other mannan-binding receptors (50). The receptor galectin-3, which binds to C. albicans β-1,2-oligomannosides, was ruled out because C. albicans-induced AA release is not blocked by lactose, which prevents binding of C. albicans to galectin-3 (51, 52).
FIGURE 5.
Role of a C. albicans mannan-binding receptor. A, [3H]AA-labeled peritoneal macrophages (BALB/c) were preincubated with d-mannose (DM), l-mannose (LM) (mm), or mannans from C. albicans (CM) (mg/ml). B, [3H]AA-labeled peritoneal macrophages from wild type or Dectin-1−/− mice were preincubated with d-mannose (100 mm), mannans from C. albicans (0.1 mg/ml), 10 μg/ml Dectin-2-blocking antibody (D2 Ab) or control antibody (Cont Ab). Macrophages were then stimulated with C. albicans (m.o.i. 5) for 60 min. The amount of [3H]AA release is expressed as a % of the total incorporated radioactivity (cells plus medium) after subtracting background release. The blockers did not significantly affect background release. The asterisk indicates a significant decrease (p < 0.05) compared with untreated macrophages. C, macrophages were incubated for 30 min with 100 mm d-mannose or l-mannose, 10 μg/ml Dectin-2 antibody or control antibody, 100 μg/ml soluble glucan-P (S-GP), and 100 μg/ml mannans from C. albicans and then stimulated for 60 min with C. albicans (m.o.i. 5). Macrophages were washed three times, and lysates were plated on Sabouraud agar plates to determine colony-forming units (CFU). A representative of three independent experiments is shown.
Dectin-2 is a mannan-binding C-type lectin expressed in myeloid cells that, together with Dectin-1, triggers cell activation through Syk and CARD 9 (16, 53). To investigate the role of this mannan-binding receptor, the effect of Dectin-2-blocking antibody on C. albicans-stimulated AA release from wild type and Dectin-1−/− macrophages (129sv/ev) was determined. In wild type macrophages, Dectin-2 antibody blocked AA release as effectively as d-mannose and Candida mannans, but control antibodies had little effect (Fig. 5B). Dectin-2 antibodies further reduced AA release from Dectin-1−/− macrophages, although this did not reach statistical significance. Although control antibodies had little effect on blocking AA release from C. albicans-stimulated Dectin-1+/+ macrophages, we consistently observed that they enhanced AA release from C. albicans-stimulated Dectin-1−/− macrophages (Fig. 5B).
We also tested the effect of blocking agents on recognition (binding and internalization) of C. albicans by wild type and Dectin-1−/− macrophages (129sv/ev) using an assay that measures colony-forming units in the macrophage cultures. The recognition of C. albicans is similar in wild type and Dectin-1−/− macrophages implicating multiple receptors in mediating C. albicans binding (Fig. 5C). The recognition of C. albicans is partially blocked by d-mannose (but not l-mannose), by a Dectin-2-blocking antibody (but not by control IgG), by mannans purified from C. albicans, and by soluble glucan-P (Fig. 5C). Surprisingly, there was a similar amount of blocking in wild type and Dectin-1−/− macrophages. The ability of soluble glucan-P to block recognition of C. albicans by macrophages lacking Dectin-1 suggests the presence of other receptors that contribute to β-glucan binding. These results were corroborated using microscopy to determine the effect of the blocking agents on phagocytosis of C. albicans. There was no significant difference in the phagocytic index of wild type and Dectin-1−/− macrophages (data not shown). Treating the macrophages with d-mannose, Candida mannans, the dectin-2-blocking antibody, and soluble-glucan-P reduced the phagocytic index to a similar extent (45–55%) in wild type and Dectin-1−/− macrophages (data not shown). The effect of soluble glucan-P and Candida mannans on C. albicans recognition by macrophages from BALB/c mice was also tested. Soluble glucan-P and mannans reduce recognition (colony-forming units) to 58.0 ± 5.6% and 28.7 ± 0.9% of levels observed in untreated macrophages (100%), respectively. When the peritoneal macrophages (BALB/c) are treated with both soluble glucan-P and Candida mannans, recognition is further reduced to 20.2 ± 0.8% of controls. Therefore, β-glucan and mannan-binding receptors contribute to C. albicans recognition in 129sv/ev and BALB/c macrophages.
Regulation of MAPK Activation and Phosphorylation of cPLA2α on Ser-505
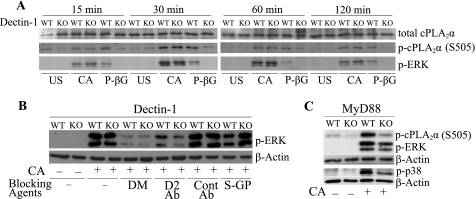
We investigated the role of Dectin-1 and MyD88 in regulating activation of MAPKs, which are important in regulating cPLA2α. We first confirmed that C. albicans-stimulated AA release is blocked by 86% with the MEK1 inhibitor U0126 and 63% by the p38 inhibitor SB203560, similar to previous results using zymosan (54). We compared the time course of ERK activation and phosphorylation of cPLA2α on Ser-505 in Dectin-1−/− and wild type macrophages stimulated with C. albicans and particulate β-glucan. Activation of ERKs in response to C. albicans is evident by 15 min, peaks at 30 min, and then remains activated at a lower level for 120 min (Fig. 6A). Surprisingly, the extent of ERK activation and phosphorylation of cPLA2α on Ser-505 is similar in Dectin-1−/− and wild type macrophages in response to C. albicans. In contrast, the activation of ERKs and Ser-505 phosphorylation in macrophages stimulated with particulate β-glucan is dependent on Dectin-1 (Fig. 6A). In addition, the p38 MAPK is activated to a similar extent by C. albicans in wild type and Dectin-1−/− macrophages (data not shown). We also compared the effect of β-glucan and mannan-blocking agents on C. albicans-stimulated ERK activation. C. albicans-stimulated ERK activation is blocked almost completely by d-mannose in both wild type and Dectin-1−/− macrophages (Fig. 6B), but l-mannose has no effect (data not shown). The Dectin-2-blocking antibody, but not control antibody, inhibits ERK activation in Dectin-1+/+ macrophages and to a slightly greater extent in Dectin-1−/− macrophages. In contrast, soluble glucan-P weakly blocks ERK activation in Dectin-1+/+ macrophages and has no effect on ERK activation in Dectin-1−/− macrophages (Fig. 6B). Activation of ERKs and p38 in response to C. albicans is also partially dependent on MyD88 (Fig. 6C). The reduced activation of these kinases in MyD88−/− macrophages compared with wild type cells correlates with a lower level of phosphorylation of cPLA2α on Ser-505. The results suggest that Dectin-2 and MyD88 pathways regulate MAPK activation by C. albicans in peritoneal macrophages.
FIGURE 6.
Regulation of ERK activation and cPLA2α Ser-505 phosphorylation. Wild type (WT) and Dectin-1−/− (KO) macrophages were unstimulated (US) or stimulated with C. albicans (CA, m.o.i. 5) or 100 μg/ml P-βG for the indicated times (A) or treated with d-mannose (DM, 100 mm), 10 μg/ml Dectin-2-blocking antibody (D2 Ab), control antibody (Cont Ab), or 100 μg/ml soluble glucan-P (S-GP) and then stimulated with C. albicans (m.o.i. 5) for 30 min (B). C, wild type or MyD88 knock-out macrophages were treated with C. albicans (m.o.i. 5) for 30 min. Activation of ERKs (B, C) and cPLA2α Ser-505 (S505) (A, C) phosphorylation was determined on Western blots using phospho-specific antibodies. Sample loading was evaluated using antibodies to total cPLA2α (A) or β-actin (B and C). A representative of three (A and B) or two (C) independent experiments is shown.
Comparison of RAW264.7 Macrophages Expressing Dectin-1 and -2
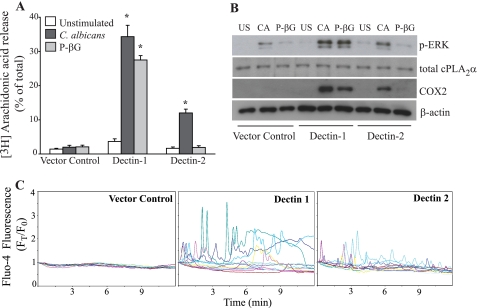
Our results suggest a role for Dectin-1 and -2 in regulating C. albicans-stimulated AA release in peritoneal macrophages. The partial decrease in AA release observed in Dectin-1−/− macrophages is not due to a defect in activation of MAPKs and cPLA2α Ser-505 phosphorylation. Dectin-1 may regulate cPLA2α by mediating calcium mobilization. The ability of the Dectin-2 antibody to block ERK activation and AA release suggests a role for Dectin-2 in regulating cPLA2α activation in peritoneal macrophages. Another approach to determine whether Dectin-2 can promote AA release was to use RAW264.7 macrophages overexpressing Dectin-2. FACS analysis confirmed the increased surface expression of Dectin-1 and -2 in RAW264.7 macrophages stably expressing these receptors (supplemental Fig. 1). Vector controls express a low level of Dectin-1 but not Dectin-2. As shown in Fig. 7A, C. albicans and particulate β-glucan do not stimulate AA release from RAW264.7 vector controls. Overexpressing Dectin-1 greatly increases AA release in response to C. albicans, as reported previously (32), and in response to particulate β-glucan. RAW264.7 cells overexpressing Dectin-2 also release significantly higher levels of AA than vector controls in response to C. albicans but not to particulate β-glucan (Fig. 7A). Similar results are observed using zymosan (data not shown). In RAW264.7 cells overexpressing Dectin-1 or -2, there is an increase in ERK activation and COX2 expression in response to C. albicans compared with vector controls (Fig. 7B). Particulate β-glucan increases these responses only in the Dectin-1-overexpressing cells. cPLA2α is expressed at similar levels in vector controls and RAW264.7 cells overexpressing either Dectin-1 or -2 (Fig. 7B).
FIGURE 7.
Responses of RAW264.7 cells expressing Dectin-1 and Dectin-2. Vector control cells and RAW264.7 expressing Dectin-1 or -2 were stimulated with C. albicans (m.o.i. 5) or particulate β-glucan for 60 min to measure [3H]AA release (A) or for 30 min to measure ERK activation or 6 h to measure COX2 expression on Western blots (B). Sample loading was evaluated using antibodies to total cPLA2α or β-actin. A, results are the average of three experiments ± S.E. The asterisk indicates a significant increase (p < 0.05) compared with vector controls. B, representation of three independent experiments is shown. C, live cell calcium imaging of RAW264.7 cells loaded with Fluo-4 was determined over time after addition of zymosan (10 particles/cell). Data are presented (FT/F0) relative to time 0, and starting F0 for each cell is set at 1. Each colored tracing represents data from an individual cell, and analysis of 15 cells for each condition from a representative of three experiments is shown.
Live cell imaging of RAW264.7 cells loaded with Fluo-4 was used to determine whether Dectin-1 and -2 mediate increases in [Ca2+]i in response to zymosan. We used zymosan for these experiments because it was previously used to define the mode of calcium mobilization in resident mouse peritoneal macrophages (47). Zymosan does not induce increases in [Ca2+]i in vector control cells (Fig. 7C). In RAW264.7 cells expressing Dectin-1, many cells exhibit increases in [Ca2+]i in response to zymosan. The timing and pattern of calcium responses of single cells (each depicted as a different colored line) in the population is very heterogeneous and may reflect differences in the timing of zymosan particle interaction with the cell surface. RAW264.7 cells expressing Dectin-2 also exhibit increases in [Ca2+]i, although responses are generally lower than in cells expressing Dectin-1. In contrast to zymosan, ionomycin stimulates increases in [Ca2+]i in vector controls as well as in RAW264.7 cells expressing Dectin-1 and -2 (supplemental Fig. 2).
DISCUSSION
This study demonstrates that C. albicans stimulates cPLA2α-dependent production of AA metabolites from cyclooxygenase and lipoxygenase pathways in resident peritoneal macrophages. The results show that recognition of C. albicans cell wall components by peritoneal macrophages involves multiple receptors that differentially regulate cPLA2α-mediated AA release, COX2 expression, and eicosanoid production as summarized in Fig. 8. Comparisons of wild type and knock-out macrophages demonstrate that Dectin-1 and MyD88 contribute to cPLA2α activation and eicosanoid production, and blocking experiments suggest the involvement of the mannan-binding receptor Dectin-2. In addition to the recognition of fungal β-glucans by Dectin-1, a number of mannose-binding C-type lectins such as DC-SIGN (SIGNRs), Mincle, and Dectin-2 contribute to fungal recognition and in some cases the induction of responses in innate immune cells (55–58).
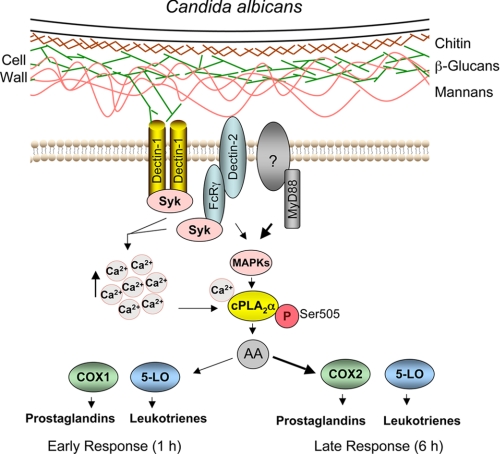
FIGURE 8.
Summary of signaling pathways for cPLA2α activation and eicosanoid production stimulated by engagement of C. albicans cell wall components with receptors in resident peritoneal macrophages. Our results suggest that β-glucans (green) and mannans (orange) of the C. albicans cell wall engage Dectin-1 and -2 on mouse peritoneal macrophages to promote increases in [Ca2+]i and activation of MAPKs, respectively. MyD88, through an as yet unidentified receptor, also contributes to the early activation of MAPKs and phosphorylation of cPLA2α on Ser-505. Calcium binds to the N-terminal C2 domain on cPLA2α and promotes translocation of cPLA2α from the cytosol to intracellular membrane to access phospholipid substrate. Phosphorylation on Ser-505 enhances the release of AA by increasing the catalytic activity of cPLA2α. Dectin-1, -2, and MyD88 contribute to the early release of eicosanoids, but the MyD88 pathway is preferentially used to mediate COX2 expression and the late response of eicosanoid production in resident peritoneal macrophages.
The results demonstrate that AA release and production of eicosanoids that occur early after infection with C. albicans are mediated in part by binding of β-glucan to Dectin-1 (Fig. 8). Resident mouse peritoneal macrophages are unique in their ability to rapidly produce relatively large quantities of prostanoids within minutes of stimulation through the COX1 pathway (59, 60). However, the more delayed production of eicosanoids and COX2 expression is similar in wild type and Dectin-1−/− macrophages. It has been reported that Dectin-1 induces signals for cytokine production primarily during the initial interaction with β-glucan at the cell surface, and ligand binding promotes internalization of Dectin-1 and degradation in lysosomes (27, 46, 61). However, results using the Dectin-1 agonist particulate β-glucan revealed that Dectin-1 can promote COX2 expression in peritoneal macrophages, but in response to C. albicans, other receptors that act in part through MyD88 preferentially mediate COX2 expression and delayed eicosanoid production (Fig. 8). We also observed a similar phenomenon in mouse alveolar macrophages (33). Dectin-1 mediates early release of AA from mouse alveolar macrophages primed with GM-CSF, which increases expression of Dectin-1, but does not mediate C. albicans-stimulated COX2 expression or PGE2 production. However, Dectin-1 does contribute to production of TNFα stimulated by C. albicans in GM-CSF-primed alveolar macrophages (33). We also observed that Dectin-1−/− peritoneal macrophages produce less TNFα in response to C. albicans than wild type macrophages (data not shown). Consequently, there is not a generalized attenuation of Dectin-1 signaling in peritoneal macrophages stimulated with C. albicans but preferential use of alternative pathways for sustained cPLA2α activation and expression of COX2.
We also investigated the role of Dectin-1 in mediating C. albicans-stimulated activation of MAPKs, which regulate cPLA2α activation and release of AA as observed previously using zymosan (54). Surprisingly, the activation of MAPKs and phosphorylation of cPLA2α on Ser-505 in response to C. albicans is similar in wild type and Dectin-1−/− peritoneal macrophages, despite results showing that Dectin-1 can mediate activation of ERKs in peritoneal macrophages when stimulated with particulate β-glucan. Therefore, there is preferential use of alternative signaling pathways for activation of both early responses such as MAPK activation and later responses such as COX2 expression in peritoneal macrophages stimulated with C. albicans (Fig. 8). In contrast to peritoneal macrophages, we found that the activation of ERKs by C. albicans is mediated by Dectin-1 in GM-CSF-primed alveolar macrophages and in RAW264.7 cells overexpressing Dectin-1 (32, 33). Therefore, the role for Dectin-1 in mediating macrophage responses is context-dependent and may reflect differences in relative levels of expression of pattern recognition receptors and signaling responses. The early activation of cPLA2α by Dectin-1 in peritoneal macrophages stimulated with C. albicans or zymosan may involve the ability of Dectin-1 to promote increases in [Ca2+]i as shown using RAW264.7 macrophages overexpressing Dectin-1 (Fig. 8). We were unable to accurately quantify differences in calcium mobilization by live cell imaging in wild type and Dectin-1−/− peritoneal macrophages due to the cell-to-cell heterogeneity of calcium responses using the particulate agonist, and because AA release is only partially reduced in Dectin-1−/− macrophages.
Our results demonstrate that MyD88 plays a role in regulating activation of MAPKs (ERKs and p38) and phosphorylation of cPLA2α on Ser-505 that may be the basis for the partially reduced AA release in MyD88−/− macrophages early after stimulation with C. albicans. MyD88 also regulates COX2 expression and production of eicosanoids in peritoneal macrophages measured 6 h after C. albicans infection (Fig. 8). TLR2 and TLR4 do not regulate C. albicans-stimulated activation of ERKs (data not shown) or AA release, but TLR2 partially mediates COX2 expression and prostanoid production (32). Because C. albicans DNA induces TLR9-dependent cytokine production, and TLR9 can mediate cPLA2α activation (62, 63), we compared peritoneal macrophages from wild type and TLR9 knock-out mice but found no role for TLR9 in regulating AA release, ERK activation, COX2 expression, or eicosanoid production in response to C. albicans (data not shown). Another possibility is that C. albicans stimulates production of IL-1α, IL-1β, or IL-18 that act in an autocrine manner through the IL-1 receptor to regulate responses through MyD88.
Our results demonstrate that recognition of C. albicans by peritoneal macrophages involves binding to cell wall mannans and β-glucans because it is blocked by soluble glucan-P, mannose, Candida mannans, and a Dectin-2-specific monoclonal antibody (Fig. 8). Both Dectin-1 and -2 have been reported to mediate binding of zymosan to mouse dendritic cells (53). The ability of Dectin-1−/− macrophages to bind and internalize C. albicans to a similar extent as wild type macrophages suggests that other receptors may compensate in the absence of Dectin-1. C. albicans binding to Dectin-1−/− macrophages is blocked by mannose/mannans but also by soluble glucan-P suggesting the presence of other β-glucan-binding proteins. However, particulate β-glucan does not stimulate AA release in Dectin-1−/− macrophages suggesting that other β-glucan-binding proteins do not signal for cPLA2α activation. Despite similar levels of C. albicans recognition in wild type and Dectin-1−/− macrophages, Dectin-1−/− macrophages release significantly less AA than wild type macrophages supporting a role for this β-glucan receptor in promoting signals for cPLA2α activation. In addition, our data suggest that Dectin-2 may signal for activation of cPLA2α. It has been shown that Dectin-2 mediates production of cysteinyl leukotrienes in dendritic cells stimulated with dust mites (64). Dectin-2, which is highly expressed in tissue macrophages, recognizes high mannose structures (16). The binding of zymosan, C. albicans, and other fungi to Dectin-2 requires calcium and is blocked by mannose (16, 65). We previously reported that chelating extracellular calcium reduces the binding and uptake of zymosan by resident peritoneal macrophages consistent with a role for Dectin-2 (47). Dectin-2 transduces intracellular signals through association with FcRγ chain (65). A recent report found that both Dectin-1 and -2 mediate the Syk-Card-9-dependent responses of dendritic cells to fungi (53). Although we found that soluble glucan-P, mannose, and Dectin-2 antibody block C. albicans recognition to a similar extent, mannose and the Dectin-2 antibody preferentially block ERK activation suggesting that Dectin-2 can provide signals for cPLA2α activation (Fig. 8). A role for Dectin-2 in mediating activation of MAPKs in dendritic cells has been reported (53). The results suggest a role for Dectin-2 and a MyD88-dependent pathway, and not Dectin-1, in promoting ERK activation in C. albicans-stimulated resident peritoneal macrophages (Fig. 8). Dectin-1 may signal for cPLA2α activation by promoting calcium mobilization as demonstrated using RAW264.7 macrophages expressing Dectin-1. The ability of Dectin-2 to provide signals for cPLA2α activation is also supported by results using RAW264.7 macrophages overexpressing Dectin-2, which exhibit enhanced calcium mobilization and the release of AA in response to fungal agents.
In summary, our study demonstrates that fungal agents engage multiple receptors on resident peritoneal macrophages that differentially regulate signals for the activation of cPLA2α and the production of eicosanoids that are important modulators of inflammation.
Supplementary Material
Acknowledgments
We thank Dr. Robert Barkley and Charis Uhlson for mass spectrometry analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants HL34303 (to R. C. M. and C. C. L.), DK54741 (to J. V. B.), and GM53522 (to D. L. W.). This work was also supported by a United Kingdom Medical Research Council Senior Fellowship G0601617 (to P. R. T.) and the Wellcome Trust (to G. D. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1 and 2.
- cPLA2α
- cytosolic phospholipase A2
- AA
- arachidonic acid
- m.o.i.
- multiplicity of infection
- P-βG
- particulate-β-glucan
- RT
- retention time
- PG
- prostaglandin
- LT
- leukotriene
- HETE
- hydroxyeicosatetraenoic acid.
REFERENCES
- 1.Beutler B. (2004) Mol. Immunol. 40, 845–859 [DOI] [PubMed] [Google Scholar]
- 2.Taylor P. R., Martinez-Pomares L., Stacey M., Lin H. H., Brown G. D., Gordon S. (2005) Annu. Rev. Immunol. 23, 901–944 [DOI] [PubMed] [Google Scholar]
- 3.Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 4.Underhill D. M. (2007) Immunol. Rev. 219, 75–87 [DOI] [PubMed] [Google Scholar]
- 5.Trinchieri G., Sher A. (2007) Nat. Rev. Immunol. 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 6.Lee M. S., Kim Y. J. (2007) Annu. Rev. Biochem. 76, 447–480 [DOI] [PubMed] [Google Scholar]
- 7.Pfaller M. A., Diekema D. J. (2007) Clin. Microbiol. Rev. 20, 133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willment J. A., Brown G. D. (2008) Trends Microbiol. 16, 27–32 [DOI] [PubMed] [Google Scholar]
- 9.Poulain D., Jouault T. (2004) Curr. Opin. Microbiol. 7, 342–349 [DOI] [PubMed] [Google Scholar]
- 10.Netea M. G., Brown G. D., Kullberg B. J., Gow N. A. (2008) Nat. Rev. Microbiol. 6, 67–78 [DOI] [PubMed] [Google Scholar]
- 11.Klis F. M., de Groot P., Hellingwerf K. (2001) Med. Mycol. 39, 1–8 [PubMed] [Google Scholar]
- 12.Masuoka J. (2004) Clin. Microbiol. Rev. 17, 281–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinsbroek S. E., Taylor P. R., Martinez F. O., Martinez-Pomares L., Brown G. D., Gordon S. (2008) PLoS Pathog. 4, e1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Veerdonk F. L., Marijnissen R. J., Kullberg B. J., Koenen H. J., Cheng S. C., Joosten I., van den Berg W. B., Williams D. L., van der Meer J. W., Joosten L. A., Netea M. G. (2009) Cell Host Microbe 5, 329–340 [DOI] [PubMed] [Google Scholar]
- 15.Taylor P. R., Reid D. M., Heinsbroek S. E., Brown G. D., Gordon S., Wong S. Y. (2005) Eur. J. Immunol. 35, 2163–2174 [DOI] [PubMed] [Google Scholar]
- 16.McGreal E. P., Rosas M., Brown G. D., Zamze S., Wong S. Y., Gordon S., Martinez-Pomares L., Taylor P. R. (2006) Glycobiology 16, 422–430 [DOI] [PubMed] [Google Scholar]
- 17.Wells C. A., Salvage-Jones J. A., Li X., Hitchens K., Butcher S., Murray R. Z., Beckhouse A. G., Lo Y. L., Manzanero S., Cobbold C., Schroder K., Ma B., Orr S., Stewart L., Lebus D., Sobieszczuk P., Hume D. A., Stow J., Blanchard H., Ashman R. B. (2008) J. Immunol. 180, 7404–7413 [DOI] [PubMed] [Google Scholar]
- 18.Xia Y., Vetvicka V., Yan J., Hanikýrová M., Mayadas T., Ross G. D. (1999) J. Immunol. 162, 2281–2290 [PubMed] [Google Scholar]
- 19.Rice P. J., Kelley J. L., Kogan G., Ensley H. E., Kalbfleisch J. H., Browder I. W., Williams D. L. (2002) J. Leukocyte Biol. 72, 140–146 [PubMed] [Google Scholar]
- 20.Means T. K., Mylonakis E., Tampakakis E., Colvin R. A., Seung E., Puckett L., Tai M. F., Stewart C. R., Pukkila-Worley R., Hickman S. E., Moore K. J., Calderwood S. B., Hacohen N., Luster A. D., El Khoury J. (2009) J. Exp. Med. 206, 637–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reid D. M., Gow N. A., Brown G. D. (2009) Curr. Opin. Immunol. 21, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gantner B. N., Simmons R. M., Canavera S. J., Akira S., Underhill D. M. (2003) J. Exp. Med. 197, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gantner B. N., Simmons R. M., Underhill D. M. (2005) EMBO J. 24, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown G. D., Taylor P. R., Reid D. M., Willment J. A., Williams D. L., Martinez-Pomares L., Wong S. Y., Gordon S. (2002) J. Exp. Med. 196, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmerman J. W., Lindermuth J., Fish P. A., Palace G. P., Stevenson T. T., DeMong D. E. (1998) J. Biol. Chem. 273, 22014–22020 [DOI] [PubMed] [Google Scholar]
- 26.Brown G. D. (2006) Nat. Rev. Immunol. 6, 33–43 [DOI] [PubMed] [Google Scholar]
- 27.Herre J., Marshall A. S., Caron E., Edwards A. D., Williams D. L., Schweighoffer E., Tybulewicz V., Reis e Sousa C., Gordon S., Brown G. D. (2004) Blood 104, 4038–4045 [DOI] [PubMed] [Google Scholar]
- 28.Taylor P. R., Tsoni S. V., Willment J. A., Dennehy K. M., Rosas M., Findon H., Haynes K., Steele C., Botto M., Gordon S., Brown G. D. (2007) Nat. Immunol. 8, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plantinga T. S., van der Velden W. J., Ferwerda B., van Spriel A. B., Adema G., Feuth T., Donnelly J. P., Brown G. D., Kullberg B. J., Blijlevens N. M., Netea M. G. (2009) Clin. Infect. Dis. 49, 724–732 [DOI] [PubMed] [Google Scholar]
- 30.Werner J. L., Metz A. E., Horn D., Schoeb T. R., Hewitt M. M., Schwiebert L. M., Faro-Trindade I., Brown G. D., Steele C. (2009) J. Immunol. 182, 4938–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross O., Gewies A., Finger K., Schäfer M., Sparwasser T., Peschel C., Förster I., Ruland J. (2006) Nature 442, 651–656 [DOI] [PubMed] [Google Scholar]
- 32.Suram S., Brown G. D., Ghosh M., Gordon S., Loper R., Taylor P. R., Akira S., Uematsu S., Williams D. L., Leslie C. C. (2006) J. Biol. Chem. 9, 5506–5514 [DOI] [PubMed] [Google Scholar]
- 33.Parti R. P., Loper R., Brown G. D., Gordon S., Taylor P. R., Bonventre J. V., Murphy R. C., Williams D. L., Leslie C. C. (2010) Am. J. Respir. Cell Mol. Biol. 42, 415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funk C. D. (2001) Science 294, 1871–18755 [DOI] [PubMed] [Google Scholar]
- 35.Ghosh M., Tucker D. E., Burchett S. A., Leslie C. C. (2006) Prog. Lipid Res. 45, 487–510 [DOI] [PubMed] [Google Scholar]
- 36.Evans J. H., Spencer D. M., Zweifach A., Leslie C. C. (2001) J. Biol. Chem. 276, 30150–30160 [DOI] [PubMed] [Google Scholar]
- 37.Perisic O., Paterson H. F., Mosedale G., Lara-González S., Williams R. L. (1999) J. Biol. Chem. 274, 14979–14987 [DOI] [PubMed] [Google Scholar]
- 38.Nalefski E. A., Sultzman L. A., Martin D. M., Kriz R. W., Towler P. S., Knopf J. L., Clark J. D. (1994) J. Biol. Chem. 269, 18239–18249 [PubMed] [Google Scholar]
- 39.Tucker D. E., Ghosh M., Ghomashchi F., Loper R., Suram S., John B. S., Girotti M., Bollinger J. G., Gelb M. H., Leslie C. C. (2009) J. Biol. Chem. 284, 9596–9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin L. L., Wartmann M., Lin A. Y., Knopf J. L., Seth A., Davis R. J. (1993) Cell 72, 269–278 [DOI] [PubMed] [Google Scholar]
- 41.Ensley H. E., Tobias B., Pretus H. A., McNamee R. B., Jones E. L., Browder I. W., Williams D. L. (1994) Carbohydr. Res. 258, 307–311 [DOI] [PubMed] [Google Scholar]
- 42.Williams D. L., McNamee R. B., Jones E. L., Pretus H. A., Ensley H. E., Browder I. W., Di Luzio N. R. (1991) Carbohydr. Res. 219, 203–213 [DOI] [PubMed] [Google Scholar]
- 43.de Carvalho M. S., McCormack F. X., Leslie C. C. (1993) Arch. Biochem. Biophys. 306, 534–540 [DOI] [PubMed] [Google Scholar]
- 44.Bonventre J. V., Huang Z., Taheri M. R., O'Leary E., Li E., Moskowitz M. A., Sapirstein A. (1997) Nature 390, 622–625 [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 46.Rosas M., Liddiard K., Kimberg M., Faro-Trindade I., McDonald J. U., Williams D. L., Brown G. D., Taylor P. R. (2008) J. Immunol. 181, 3549–3557 [DOI] [PubMed] [Google Scholar]
- 47.Girotti M., Evans J. H., Burke D., Leslie C. C. (2004) J. Biol. Chem. 279, 19113–19121 [DOI] [PubMed] [Google Scholar]
- 48.Gijón M. A., Zarini S., Murphy R. C. (2007) J. Lipid Res. 48, 716–725 [DOI] [PubMed] [Google Scholar]
- 49.Hall L. M., Murphy R. C. (1998) J. Am. Soc. Mass Spectrom. 9, 527–532 [DOI] [PubMed] [Google Scholar]
- 50.Taylor P. R., Brown G. D., Reid D. M., Willment J. A., Martinez-Pomares L., Gordon S., Wong S. Y. (2002) J. Immunol. 169, 3876–3882 [DOI] [PubMed] [Google Scholar]
- 51.Kohatsu L., Hsu D. K., Jegalian A. G., Liu F. T., Baum L. G. (2006) J. Immunol. 177, 4718–4726 [DOI] [PubMed] [Google Scholar]
- 52.Jouault T., El Abed-El Behi M., Martínez-Esparza M., Breuilh L., Trinel P. A., Chamaillard M., Trottein F., Poulain D. (2006) J. Immunol. 177, 4679–4687 [DOI] [PubMed] [Google Scholar]
- 53.Robinson M. J., Osorio F., Rosas M., Freitas R. P., Schweighoffer E., Gross O., Verbeek J. S., Ruland J., Tybulewicz V., Brown G. D., Moita L. F., Taylor P. R., Reis e Sousa C. (2009) J. Exp. Med. 206, 2037–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gijón M. A., Spencer D. M., Siddiqi A. R., Bonventre J. V., Leslie C. C. (2000) J. Biol. Chem. 275, 20146–20156 [DOI] [PubMed] [Google Scholar]
- 55.Graham L. M., Brown G. D. (2009) Cytokine 48, 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor P. R., Brown G. D., Herre J., Williams D. L., Willment J. A., Gordon S. (2004) J. Immunol. 172, 1157–1162 [DOI] [PubMed] [Google Scholar]
- 57.Valera I., Fernández N., Trinidad A. G., Alonso S., Brown G. D., Alonso A., Crespo M. S. (2008) J. Immunol. 180, 5727–5736 [DOI] [PubMed] [Google Scholar]
- 58.Cambi A., Netea M. G., Mora-Montes H. M., Gow N. A., Hato S. V., Lowman D. W., Kullberg B. J., Torensma R., Williams D. L., Figdor C. G. (2008) J. Biol. Chem. 283, 20590–20599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rouzer C. A., Kingsley P. J., Wang H., Zhang H., Morrow J. D., Dey S. K., Marnett L. J. (2004) J. Biol. Chem. 279, 34256–34268 [DOI] [PubMed] [Google Scholar]
- 60.Rouzer C. A., Tranguch S., Wang H., Zhang H., Dey S. K., Marnett L. J. (2006) Biochem. J. 399, 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hernanz-Falcon P., Joffre O., Williams D. L., Reis e Sousa C. (2009) Eur. J. Immunol. 39, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee S. H., Lee J. G., Kim J. R., Baek S. H. (2007) Biochem. Biophys. Res. Commun. 364, 996–1001 [DOI] [PubMed] [Google Scholar]
- 63.Miyazato A., Nakamura K., Yamamoto N., Mora-Montes H. M., Tanaka M., Abe Y., Tanno D., Inden K., Gang X., Ishii K., Takeda K., Akira S., Saijo S., Iwakura Y., Adachi Y., Ohno N., Mitsutake K., Gow N. A., Kaku M., Kawakami K. (2009) Infect. Immun. 77, 3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrett N. A., Maekawa A., Rahman O. M., Austen K. F., Kanaoka Y. (2009) J. Immunol. 182, 1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sato K., Yang X. L., Yudate T., Chung J. S., Wu J., Luby-Phelps K., Kimberly R. P., Underhill D., Cruz P. D., Jr., Ariizumi K. (2006) J. Biol. Chem. 281, 38854–38866 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.