Abstract
Bone marrow-derived mesenchymal stem cells (BM-MSCs) are often recruited to solid tumors, integrate into the tumor stroma, and contribute to tumor development. TNFα is a major inflammatory cytokine present in the tumor microenvironment and has a profound influence on the progression of tumor development. This study was aimed to investigate the role of BM-MSCs in tumor promotion in response to TNFα. Quantitative real-time PCR arrays show that diverse cytokines/chemokines were induced in TNFα-treated BM-MSCs; in particular, CXCR3 ligand chemokines, including CXCL9, CXCL10, and CXCL11, were potently induced. A serial and site-directed mutation analysis in the CXCL9, CXCL10, and CXCL11 promoters revealed that NF-κB binding elements were responsible for TNFα-induced promoter activation of CXCR3 ligand chemokines. TNFα stimulated NF-κB activity, and ectopic expression of NF-κB enhanced TNFα-induced promoter activities of the CXCR3 ligand chemokines. Gel shift and supershift assays showed that NF-κB was associated with CXCR3 ligand chemokine promoters in response to TNFα treatment. All three CXCR3 ligand chemokines enhanced the migration and invasive motility of MDA-MB-231 breast cancer cells expressing CXCR3. Treatment of MDA-MB-231 cells with CXCL10 activated small GTPase of Rho family proteins, such as RhoA and Cdc42. CXCL9-, CXCL10-, or CXCL11-induced invasive capability of MDA-MB-231 cells was completely abrogated in the presence of a neutralizing anti-CXCR3 antibody in the culture medium. Moreover, CXCL9, CXCL10, and CXCL11 stimulated the expression of MMP-9, but not MMP-2, in MDA-MB-231 cells. These results suggest that BM-MSCs promote the locomotion of breast cancer cells through CXCR3 ligand-mediated actin rearrangement by TNFα in the tumor microenvironment.
Keywords: Chemokines, NF-κB Transcription Factor, Transcription Promoter, Tumor Metastases, Tumor Necrosis Factor (TNF), CXCR3 Ligand Chemokine
Introduction
Genetic alterations of epithelial cells can lead to the initiation and progression of carcinomas. However, the metastatic potency of tumor cells is largely acquired from paracrine signals from tumor-associated stromal cells (1, 2). Epithelial tumor cells stimulate adjacent stromal cells, which play an important role in the recruitment of immune cells essential for the generation of a tumor microenvironment (1, 3–8). Activated tumor-associated fibroblasts (myofibroblasts) and infiltrating immune cells within the tumor microenvironment secrete diverse cytokines, chemokines, and growth factors to promote tumor growth and potentiate cancer progression, including angiogenesis, invasion, and metastasis (2, 4, 6–9). The role of tumor-associated fibroblasts gains further support from a study showing that stromal fibroblasts within invasive breast carcinoma promote tumor growth and angiogenesis through the secretion of chemokine stromal-derived factor-1 (SDF1)/CXCL12 and CXCL14 (2, 3).
Bone marrow-derived mesenchymal stem cells (BM-MSCs)2 support the growth of hematopoietic progenitor cells and contribute to the regeneration of mesenchymal tissues. BM-MSC can differentiate into a variety of cell types, including osteoblasts, chondrocytes, myocytes, adipocytes, and many other types of cells (10). BM-MSCs are recruited to tumor stroma (11) and can differentiate into myofibroblast-like carcinoma-associated fibroblasts and become part of the tumor microenvironment that supports tumor growth (12–15). Karnoub et al. (1) demonstrated that human breast carcinoma cells have the ability to attract BM-MSCs and to stimulate the secretion of the chemokine CCL5/RANTES from BM-MSCs. Secreted CCL5/RANTES acts in a paracrine fashion on the cancer cells to enhance cancer motility, invasion, and metastasis. Notably, they suggest that the acquisition and maintenance of increased metastatic potency depend on continuing contact with stromal cells and need continual tumorigenic signals from their stromal microenvironment (1). As MSCs are described as co-conspirators of tumor development in tumor microenvironments, additional studies of factors are essential in understanding the dynamic interplay between MSC and tumor cells.
To better understand the functional contributions of the BM-MSCs in tumor progression, we sought to elucidate the properties of BM-MSCs within the tumor microenvironment. TNFα is an inflammatory cytokine mainly produced by tumor stromal cells (16). TNFα induces diverse inflammatory cytokines and chemokines, modulates a broad range of inflammatory and immunological processes, and plays crucial roles in tumor progression including tumor invasion and metastasis within tumor microenvironments (17). In the present study we show that BM-MSCs produce many kinds of cytokine/chemokines in response to TNFα stimulation. This study focuses on the role of CXCR3 ligands, including CXCL9 (18), CXCL10 (IP-10), and CXCL11 (I-TAC), in tumor progression in response to TNFα stimulation as they are not well studied. We observed that these CXCR3 ligand chemokines enhanced the migration and invasion motility of MDA-MB-231 breast carcinoma cells through activation of the small GTPases of Rho family, RhoA, and Cdc42. We also show that the NF-κB pathway is involved in TNFα-induced transcriptional activation of these CXCR3 ligand chemokines. Our findings further support the functional role of BM-MSCs in the promotion of motility and invasiveness of breast cancer cells within the tumor microenvironment, particularly through secretion of CXCR3 ligands chemokines by TNFα stimulation.
EXPERIMENTAL PROCEDURES
Cell Culture
The human BM-MSCs were obtained from the Cell Bank of RIKEN BioResource Center (Ibaraki, Japan) and maintained in MSC GS medium (ScienceCell, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT). MDA-MB-231 breast cancer cells were from American Type Culture Collection (Manassas, VA) and maintained in the ATCC-formulated Leibovitz's L-15 medium containing 10% FBS at 37 °C in humidified 100% air, as described in ATCC product catalogue.
SYBR Green Real-time PCR Array
BM-MSCs were stimulated with 10 ng/ml TNFα for 24 h, and total RNA was extracted using a Trizol RNA extraction kit (Invitrogen). The first-strand cDNA was synthesized from 500 ng of total RNA using an iScript cDNA synthesis kit (Bio-Rad). SYBR® Green real-time PCR arrays were performed using the commercially available Human RT2 ProfilerTM PCR Array system (Cat. No. PAHS-011; SuperArray Bioscience; Frederick, MD) profiling the expression of 84 genes encoding C-C and C-X-C chemokines and their receptors as well as other related genes for the gene expression analysis. Real-time PCR was performed with the iCycler iQTM (Bio-Rad) according to the manufacturer's instructions. The expression profile of 84 genes was analyzed with the aid of a software program provided by the manufacturer (Bio-Rad) using the mean threshold cycle number (Ct) of five housekeeping genes as reference endogenous controls for each array plate.
Quantitative Real-time PCR Analysis
Relative expression levels of mRNAs were measured by quantitative real-time PCR with a TaqMan-iQTM supermix kit (Bio-Rad) using the Bio-Rad iCycler iQTM according to the manufacturer's instruction. The TaqManTM fluorogenic probes and PCR primers were designed by Metabion, Int. (Martinsried, Germany). The sequences of primers for quantitative real-time PCR were as follows: forward CXCL9, 5′-actatccacctacaatccttgaaagac-3′; reverse CXCL9, 5′-tcacatctgctgaatctgggtttag-3′; CXCL9 TaqMan probe, 5′-FAM-tgccccaagcccttcctgcgagaaaa-BHQ-3′; forward CXCL10, 5′-agcaaggaaaggtctaaaagatctcc-3′; reverse CXCL10, 5′-ggcttgacatatactccatgtaggg-3′; CXCL10 TaqMan probe, 5′-FAM-aggcagcctctgtgtggtccatcctt-BHQ-3′; forward CXCL11, 5′-tgctacagttgttcaaggcttcc-3′; reverse CXCL11, 5′-ggtacattatggaggctttctcaatatc-3′; CXCL11 TaqMan probe, 5′-FAM-ccccagggcctatgcaaagacagcgt-BHQ-3′; forward MMP-2, 5′-ggaatgccatccccgataacc-3′; reverse MMP-2, 5′-ggttctccagcttcaggtaatagg-3′; MMP-2 TaqMan probe, 5′-FAM-cagggcggcggtcacagctacttctt-BHQ-1–3′; forward MMP-9, 5′-cctcgccctgaacctgagc-3′; reverse MMP-9, 5′-gctctgaggggtggacagtg-3′; MMP-9 TaqMan probe, 5′-FAM-cctccaaccaccaccacaccgcagc-BHQ-1–3′; forward GAPDH, 5′-tcgacagtcagccgcatcttc-3′; reverse GAPDH, 5′-cgcccaatacgaccacctccg-3′; GAPDH TaqMan probe, 5′-Yakima Yellow TM-cgtcgccagcccagccacgc-BHQ-1–3′ (FAM, 6-carboxyfluorescein; BHQ, tert-butylhydroquinone). Expression values were normalized to GAPDH mRNA (ΔCt) by subtracting the cycle threshold (Ct) value of GAPDH mRNA from the Ct value of the experimental value using the software program provided by the manufacturer. The relative -fold changes were normalized for GAPDH mRNA in the same samples.
Northern Blot Analysis
For each sample 10 μg of total RNA were electrophoresed on a formaldehyde/agarose gel and transferred to a Hybond N+ nylon membrane (Amersham Biosciences). Northern blotting was performed with [α-32P]dCTP-labeled CXCL9, CXCL10, or CXCL11 cDNA probes followed by hybridization with a GAPDH cDNA probe.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was done using 10 μg of nuclear extracts prepared from BM-MSCs either untreated or treated with TNFα (10 ng/ml) and 32P-labeled oligonucleotide probes as previously described (19). The sequences of oligonucleotide probes and competitors were as follows: CXCL9 NF-κB, 5′-cagaaattcccttggatctgagactagggtttccccca-3′ (−172/−135); CXCL9 mt κB, 5′-cagaaataaaattggatctgagactagggttaaaacca-3′ (−172/−135); CXCL10 NF-κB#1, 5′-gcagagggaaattccgtaactt-3′ (−182/−161); CXCL10 mt κB#1, 5′-gcagagggaaaaaaagtaactt-3′ (−182/−161); CXCL10 NF-κB#2, 5′-caacatgggacttccccaggaac-3′ (−128/−106); CXCL11 NF-κB, 5′-aagagggaaattcctgtgcc-3′ (−70/−51) (“mt” indicates mutant oligonucleotides carrying mutations within the core sequence of the NF-κB binding site at the underlined nucleotides). The samples were electrophoresed in nondenaturing 6% polyacrylamide gels and visualized by autoradiography.
Construction and Mutagenesis of the Promoter-Reporter Constructs for CXCL9, CXCL10, and CXCL11
Amplification of the 5′-flanking region of the CXCL9, CXCL10, and CXCL11 genes was performed by PCR using the high-fidelity LabopassTM SP-TaqDNA polymerase (COSMO Genetech Co., Seoul, Korea) and human leukocyte genomic DNA (Promega, Madison, WI). The nucleotide upstream of the transcription start site was numbered −1. For the CXCL9 promoter reporter, PCR products were obtained with forward primer, 5′-ttatggccattcttgcaggt-3′ (−971/−951), and reverse primer, 5′-tggagttccaagtcactcctgt-3′ (+8/+29), of the CXCL9 gene and subcloned into the pGL3-basic vector (Promega), yielding pCXCL9-Luc(−971/+29). A serial deletion construct was generated by PCR using forward primers 5′-ttgtcaggtatatagattgtg-3′ (−750/−730) and 5′-ccaaaggctatcagtgttgct-3′ (−250/−230) with +8/+29 reverse primer, yielding pCXCL9-Luc(−750/+29) and pCXCL9-Luc(−250/+29), respectively. For the CXCL10 promoter reporter, PCR products were obtained with forward primer 5′-tttgccacgattcatcatcc-3′ (−993/−974) and reverse primer 5′-aatgtctcagaaaacgtgggg-3′ (−13/+8) of the CXCL10 gene and subcloned into the pGL3-basic vector (Promega), yielding pCXCL10-Luc(−993/+8). A serial deletion construct was generated by PCR using forward primers 5′-cctaggatagctatgaatcct-3′ (−750/−730) and 5′-ctttttttcaagaaacagttc-3′ (−250/−230) with −13/+8 reverse primer, yielding pCXCL10-Luc(−750/+8) and pCXCL10-Luc(−250/+8), respectively. For the CXCL11 promoter reporter, PCR products were obtained with forward primer 5′-tagctgtgggcctcaacaag-3′ (−965/−946) and reverse primer 5′-ggtgctgctgctgctacttc-3′ (+51/+70) of the CXCL11 gene and subcloned into the pGL3-basic vector (Promega), yielding pCXCL11-Luc(−965/+70). A serial deletion construct was generated by PCR using forward primers 5′-gggaaacttctttaggcact-3′ (−500/−481) and 5′-ttaccatctgggttttcacag-3′ (−200/−180) with +51/+70 reverse primer, yielding pCXCL11-Luc(−500/+70) and pCXCL10-Luc(−200/+70), respectively. Point mutations for the NF-κB-binding motif (ttcc to aaaa) in the pCXCL9-Luc(−250/+29), (pCXCL10-Luc(−250/+8) and pCXCL9-Luc(−200/+70) were generated using the technique of overlapping two-step PCR reactions, yielding pCXCL9-Luc(−250/+29mtNFκB), pCXCL10-Luc(−250/+8mtNFκB), and pCXCL9-Luc(−200/+70mtNFκB). All the resultant constructs were verified by DNA sequencing and by restriction enzyme digests.
Promoter Reporter Assay
BM-MSCs were seeded onto 12-well plates and transfected with 0.5 μg of the CXCL9, CXCL10, or CXCL11 promoter construct using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions. To monitor transfection efficiency, 50 ng of the pRL-null plasmid encoding Renilla luciferase was included in all samples. Where indicated, 0.2 μg of mammalian expression vectors was also included. At 24 h post-transfection, the levels of firefly and Renilla luciferase activity were measured sequentially from a single sample using the Dual-GloTM Luciferase Assay System (Promega). The firefly luciferase activity was normalized to the Renilla activity, and the relative amount of luciferase activity in the untreated cells was designated as 1. The luminescence was measured with a luminometer (Centro LB960; Berthold Tech, Bad Wildbad, Germany).
Western Blot Analysis
Cells were lysed in 20 mm HEPES (pH 7.2) containing 1% Triton X-100, 10% glycerol, 150 mm NaCl, 10 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride. Primary antibodies used in these experiments were anti-glyceraldehyde phosphate-3-dehydrogenase (GAPDH; 1:2000), anti-α-tubulin (1:1000, anti-p65 NF-κB, or anti-lamin B (1:1000), all from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) or anti-IκB (Cell Signaling Technology, Denver, MA).
Cell Migration
Migration of MDA-MB-231 cells was measured using the in vitro cell scratch assay. After cells grown in six-well plates had reached confluence, a scratch was made with a pipette tip followed by extensive washing with serum-free medium to remove cell debris. CXCL9 (100 ng/ml), CXCL10 (20 ng/ml), CXCL11 (50 ng/ml), or 0.1% bovine serum albumin as vehicle control was then added. Cells were allowed to migrate into the scrapped area for up to 12 h at 37 °C. At indicated time points, cells were photographed with a Nikon E500 camera (Nikon Corporation, Tokyo, Japan).
Evaluation of Actin Reorganization by Confocal Laser Scanning Microscopy
MDA-MB-231 cells grown on glass coverslips were treated with 20 ng/ml CXCL10 for 24 h and fixed in 4% paraformaldehyde and then permeabilized in 0.3% Triton X-100. Actin rearrangement was determined using the rhodamine-phalloidin-based F-Actin Visualization Biochem KitTM (Cytoskeleton, Inc.; Denver, CO) according to the manufacturer's instructions. Polymerized F-actin was analyzed with a confocal-laser scanning microscope (FV-1000; Olympus, Tokyo, Japan).
Pulldown Assay for RhoA, Rac1, or Cdc42 Activation
The assay of RhoA, Rac1, or Cdc42 activation was performed using commercially available kits, the RhoA or Rac1/Cdc42 activation assay Biochem KitTM (Cytoskeleton, Inc.; Denver, CO), according to the manufacturer's instructions. Briefly, active GTP-bound Rho, Rac, or Cdc42 was precipitated with GST-bait beads. Active RhoA was captured using GST-rhotekin Rho-binding domain (GST-RBD), whereas active Rac1/2/3 and Cdc42 were captured using GST-PAK1 p21-binding domain (GST-PBD). Precipitated Rho, Rac, or Cdc42 was detected using Western blotting.
Transwell Invasion Assay
The cancer cell Transwell migration assay was performed utilizing the CytoSelectTM 24-well cell migration and invasion assay in a colorimetric format (8-μm pore size; Cell Biolabs, Inc., San Diego, CA). The assay was performed exactly as specified by the manufacturer's protocol. In brief, cells were serum-starved for 24 h, and a cell suspension containing 5 × 105 cells/ml in serum-free medium containing CXCL9 (100 ng/ml), CXCL10 (20 ng/ml), or CXCL11 (50 ng/ml) was prepared and seeded on the upper chamber onto a polycarbonate filter coated with Matrigel-based basement membrane proteins. Cells were allowed to invade for 24 h. Cells that had invaded to the bottom of the membrane were stained, extracted, and quantified at A560 nm as described in the assay protocol.
RESULTS
Various Cytokines and Chemokines Are Expressed by BM-MSC in Response to TNFα Stimulation
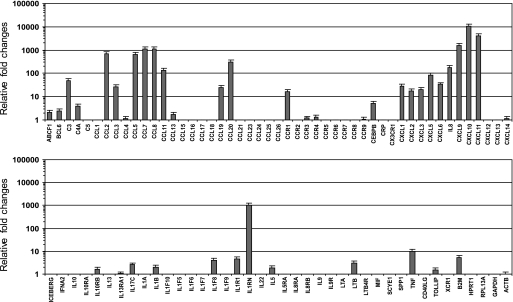
BM-MSCs interacting with breast cancer cells secrete CCL5 (RANTES), which then acts in a paracrine fashion on cancer cells to enhance their motility, invasion, and metastasis (1). To further understand the cross-talk between BM-MSC and cancer cells in the tumor microenvironment, BM-MSCs were treated with TNFα, a major inflammatory cytokine in the tumor microenvironment, and then changes in expression of chemokines and their receptors were screened using a SYBR Green real-time PCR array system. Notably, the mRNA levels of C-C and C-X-C chemokines were highly accumulated, whereas those of other cytokines, such as IL1, IL5, IL6, and IL13, were relatively low (Fig. 1). Particularly, all three CXCR3 ligand chemokines, CXCL9, CXCL10, and CXCL11, were the most highly accumulated. We chose to focus further analysis on CXCR3 ligand chemokines (CXCL9, CXCL10, and CXCL11) because BM-MSC-produced CXCR3 ligands have not been well characterized.
FIGURE 1.
Relative expression of various cytokines and chemokines in response to TNFα stimulation in BM-MSCs. BM-MSCs were either left untreated or treated with 10 ng/ml TNFα for 24 h. Relative -fold changes of mRNA levels between control and TNFα-treated cells were evaluated using SYBR Green quantitative real-time RT-PCR array system (RT2 ProfilerTM).
Production of CXCR3 Ligands from TNFα-treated BM-MSCs
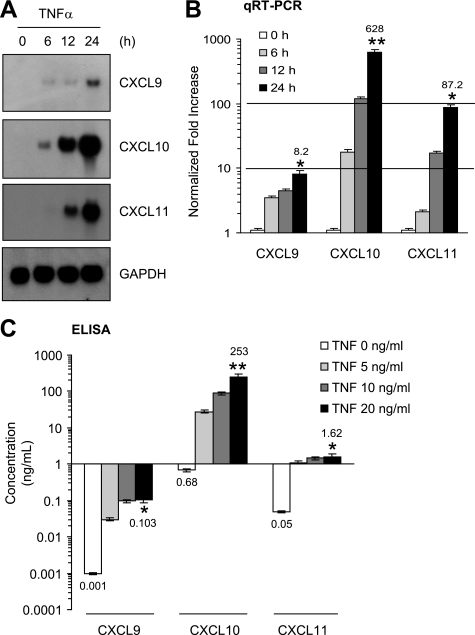
To confirm the quantitative real-time PCR array results, Northern blot analysis was performed. Resting BM-MSCs did not express detectable levels of CXCL9, CXCL10, or CXCL11 transcripts; however, TNFα-treated BM-MSCs exhibited a progressive accumulation of all these CXCR3 ligand mRNAs for more than 24 h (Fig. 2A). To more precisely measure the induction fold of CXCR3 ligand transcripts, we conducted quantitative real-time PCR (Fig. 2B). Consistent with Northern blot data, the increases of all these CXCR3 ligand transcripts were detectable as early as 6 h after TNFα treatment. After 24 h, the mRNA levels of CXCL9, CXCL10, or CXCL11 had accumulated by 8.2-, 628-, or 87.2-fold higher than basal levels, respectively. To further evaluate whether BM-MSCs could produce CXCR3 ligand proteins in response to TNFα, ELISA was used to detect proteins present in the conditioned medium of TNFα-treated BM-MSCs (Fig. 2C). Dose-dependent extracellular releases of all three CXCL9, CXCL10, or CXCL11 proteins were observed after treatment with TNFα. Among them, the secreted amount of CXCL10 was highest: CXCL9 (0.1 ng/ml), CXCL10 (253 ng/ml), and CXCL11 (1.6 ng/ml) per 2 × 105 cells treated with 20 ng/ml TNFα for 24 h. Thus, BM-MSCs produce CXCR3 ligand chemokines in response to TNFα stimulation. In particular, CXCL10 was most highly produced.
FIGURE 2.
Up-regulation of CXCR3 ligand chemokines by TNFα in BM-MSCs. A, BM-MSCs were stimulated with TNFα (10 ng/ml) for various lengths of time (6–24 h). Total RNAs were prepared, and Northern blotting was performed. GAPDH expression was used as an internal control. Each blot shown is representative of at least three separate experiments. B, quantification of mRNA levels was performed by real time-PCR (qRT-PCR) using samples prepared in A. Values were normalized for GAPDH levels. The data shown represent the mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01; both compared with unstimulated control. C, BM-MSCs cultured in 24-well plates were incubated with different concentrations of TNFα. After 24 h, CXCL9, CXCL10, or CXCL11 protein in the conditioned medium was measured using ELISA. The data shown represent the mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01; both compared with unstimulated conditioned medium. qRT-PCR, quantitative real-time PCR.
NF-κB Mediates TNFα-induced CXCR3 Ligands Expression in BM-MSCs
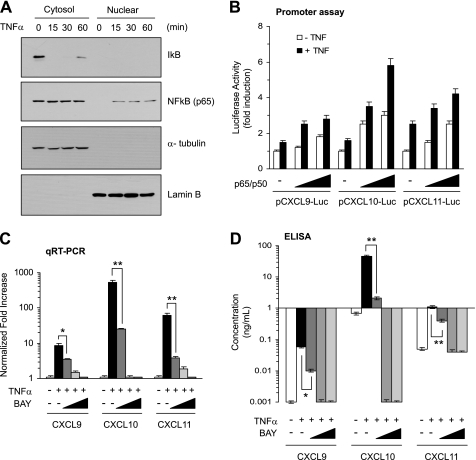
NFκB functions as a tumor promoter and plays a critical role in the production of diverse inflammatory cytokines in the tumor microenvironment (20, 21). Previous reports have demonstrated that NF-κB is activated by TNFα stimulation in diverse cell types (16, 21). We confirmed the activation of NF-κB in BM-MSCs (Fig. 3A). As proximal regions of all three CXCR3 ligand chemokine genes contain p65 NF-κB-binding sites, we determined whether NF-κB could trans-activate the CXCL9, CXCL10, or CXCL11 promoters. NF-κB expression plasmids (combined p65 and p50) were co-transfected into BM-MSCs with their promoter reporter constructs, pCXCL9-Luc(−980/+29), pCXCL10-Luc(−1049/+8), or pCXCL11-Luc(−980/+70), and the luciferase reporter activity was measured. Exogenous expression of p65/p50 NF-κB was able to enhance all three CXCR3 ligand promoter activities in a dose-dependent manner (Fig. 3B). Analysis of mRNA expression by quantitative real-time PCR demonstrated that pretreatment with the NF-κB inhibitor, BAY 11-7082, dose-dependently inhibited TNFα inducibility of CXCR3 ligands expression (Fig. 3C). Furthermore, pretreatment with BAY 11-7082 dose-dependently abrogated production of CXCL9, CXCL10, or CXCL11 proteins present in the conditioned medium of TNFα-treated BM-MSCs (Fig. 3D).
FIGURE 3.
Involvement of NF-κB in TNFα-induced CXCR3 ligand chemokines expression in BM-MSCs. A, BM-MSCs were treated with 10 ng/ml TNFα for the indicated times. Cytosol and nuclear fractions were isolated, and the amounts of IκB and p65 NF-κB were measured by Western blotting. α-Tubulin and lamin B were used as a loading control for the cytosol and nuclear fractions, respectively. B, BM-MSCs were transiently cotransfected with 0.2 μg of promoter reporter plasmids, CXCL9-Luc(−971/+29), CXCL10-Luc(−993/+8), or CXCL11-Luc(−965/+70) and different concentrations (50 and 100 ng) of expression plasmids for NF-κB (p65 plus p50) along with 50 ng of the expression plasmid for Renilla luciferase, pRL-null, for normalization of transfection efficiency. Forty-eight hours later the cells were lysed, and dual luciferase activity was measured. The data shown represent the means ± S.D. of three independent experiments performed in triplicate. C, BM-MSCs were pretreated with different concentrations of BAY 11–702 (5, 10, and 20 μm) before 30 min of TNFα (10 ng/ml) stimulation. Total RNAs were prepared, and quantification of mRNA levels was performed by real time-PCR analysis. Values were normalized to GAPDH levels. The data shown represent the mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01. D, BM-MSCs were pretreated with BAY 11–702 as in C. After 24 h, ELISA was performed to measure CXCL9, CXCL10, or CXCL11 protein in the conditioned medium. The data shown represent the mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01. qRT-PCR, quantitative real-time PCR.
NF-κB Binding Site Is Essential for TNFα-induced CXCL9, CXCL10, or CXCL11 Expression in BM-MSCs
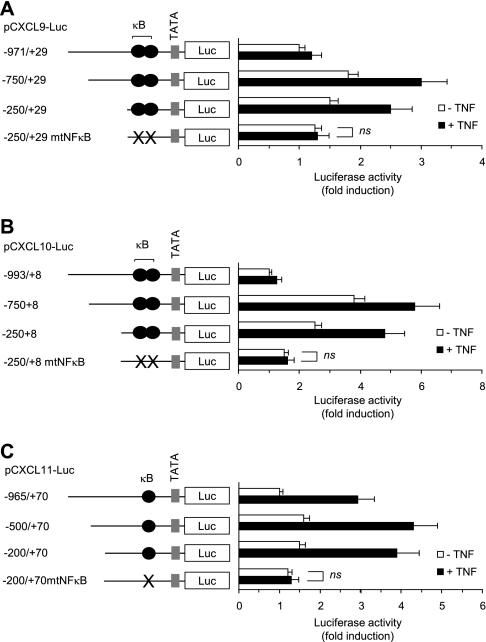
To determine whether NF-κB binding elements within the CXCR3 ligand chemokine promoters are required for TNFα-induced promoter activation in BM-MSCs, a serial deletion and site-directed mutagenesis were carried out. We found that 5′-deletion constructs, which contain NF-κB binding sites, did not significantly affect basal and inducible promoter activities by TNFα. Targeted mutation of the p65 NF-κB binding sites within the proximal promoter regions of the CXCL9 (Fig. 4A), CXCL10 (Fig. 4B), or CXCL11 (Fig. 4C) gene completely abolished TNFα inducibility. These results suggest that the NF-κB sites within the minimal promoter region of the CXCL9, CXCL10, or CXCL11 gene are essential for TNFα-induced transcriptional activation in BM-MSCs.
FIGURE 4.
NF-κB binding site is essential for TNFα-induced promoter activation of CXCR3 ligand chemokines. BM-MSCs were transiently transfected with 0.2 μg of serial deletion or NF-κB site mutant (mtNFκB) constructs of CXCL9 (A), CXCL10 (B), or CXCL11 (C) along with 50 ng of the pRL-null for normalization of transfection efficiency. After 24 h, cells were either unstimulated or stimulated with 10 ng/ml TNFα for an additional 24 h. The cells were lysed, and dual luciferase activity was measured. The schematics of the reporter constructs used in the transfection assay show serial deletion or targeted mutation of the conserved NF-κB-binding motif (κB). The data shown represent the means ± S.D. of three independent experiments performed in triplicate. ns, not significant (p > 0.05).
NF-κB Directly Binds to Its Consensus Site within the CXCL9, CXCL10, or CXCL11 Promoter
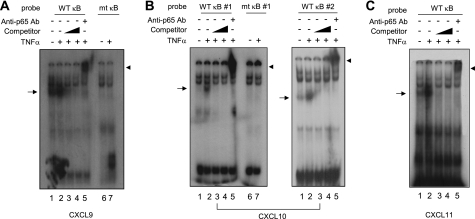
To investigate whether NF-κB binds directly to the putative NF-κB binding sites in the CXCL9, CXCL10, or CXCL11 promoter in response to TNFα stimulation, nuclear extracts from BM-MSCs were prepared. EMSA was performed with the p65 NF-κB binding sequences within their promoter regions. TNFα stimulation of BM-MSCs resulted in the formation of a DNA-protein complex with p65 NF-κB sites within the CXCL9 (Fig. 5A), CXCL10 (Fig. 5B), or CXCL11 (Fig. 5C) promoter (lane 2). These complexes were completely competed with a 10- or 100-times molar excess of unlabeled NF-κB-binding oligonucleotides (lanes 3 and 4), establishing binding specificity. When an antibody against p65 NF-κB was added, TNFα-induced DNA-protein complexes were supershifted (lane 5), indicating that the protein-DNA complex contains p65 NF-κB. In addition, when radiolabeled oligonucleotides containing mutations within the core sequence of the NF-κB binding site in the CXCL9 (Fig. 5A) or CXCL10 (Fig. 5B) promoter were used, this DNA-protein complex was not observed (lane 7). Collectively, these results demonstrate that NF-κB specifically interacts with the consensus NF-κB-binding sites within the CXCL9, CXCL10, and CXCL11 promoters, forming DNA-protein complexes in response to TNFα stimulation in BM-MSCs.
FIGURE 5.
Association of NF-κB to CXCR3 ligand chemokine promoters. BM-MSCs were either untreated or treated with 10 ng/ml TNFα for 4 h, and nuclear extracts were prepared. EMSA was done with 32P-labeled oligonucleotide probes corresponding to positions −172 to −135 of the CXCL9 promoter containing wild-type (WT κB) or mutant NF-κB sites (mt κB) (A), −182 to −161 (left) or −128/−106 (33) of the CXCL10 promoter containing wild-type (WT κB) or mutant NF-κB sites (mt κB) (B), or −70 to −51 of the CXCL11 promoter containing wild-type NF-κB site (WT κB) (C). For the competition assay, unlabeled oligonucleotides (Competitor) were added at 10- or 100-fold molar excess before the addition of labeled probes. The supershift assay was performed by the addition of an anti-NFκB (p65) antibody. Arrows indicate DNA-NF-κB complexes. Arrowheads indicate supershifted DNA-NF-κB complexes.
CXCR3 Ligand Chemokines Promote Motility of MDA-MB-231 Cells
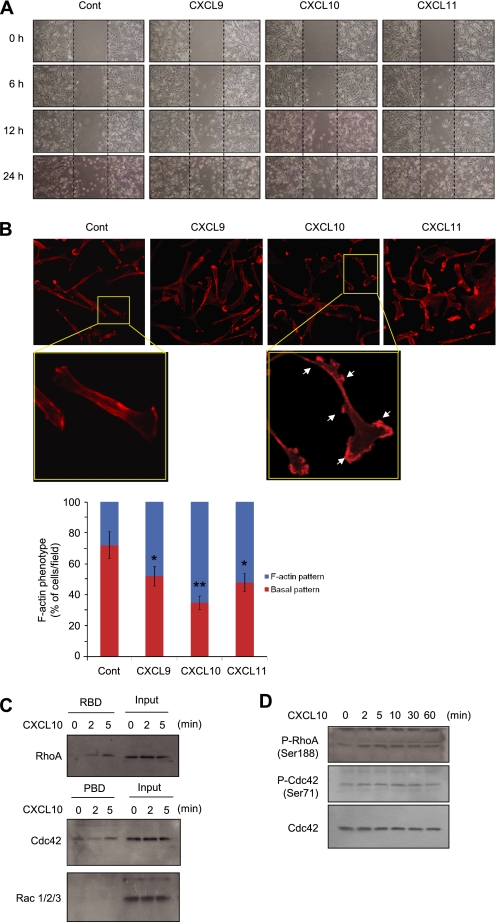
CXCR3 is abundantly expressed in diverse cancer cells, including breast carcinoma (22–24). We also observed that CXCR3 was highly expressed in MCF7 and MDA-MB-231 breast cancer cells and moderately expressed in HeLa and HCT116 carcinoma cells (supplemental Fig. S1). Cell migration is an essential feature in tumor invasion and metastasis. To understand whether CXCR3 ligands produced from BM-MSCs play roles in the promotion of cell motility, we tested their effect on the migration of MDA-MB-231 cells using the cell scratch assay. After confluent cell monolayers were disrupted by scraping with a pipette tip; the scraped closure was followed for up to 24 h. Cells treated with CXCL9, CXCL10, or CXCL11 for 24 h displayed more efficiently enhanced closure than did control cells (Fig. 6A). Under these conditions, cell growth rates were not altered within 12 h of the application of CXCR3 ligands (supplemental Fig. S2). Thus, we suggest that CXCR3 ligands must be applied for longer than 12 h to increase cell mobility.
FIGURE 6.
CXCR3 ligand chemokines promote motility of MDA-MB-231 cells. A, migration of MDA-MB-231 cells treated with CXCL9 (100 ng/ml), CXCL10 (20 ng/ml), or CXCL11 (50 ng/ml) was analyzed using a cell scratch assay. Dotted lines indicate the scraped boundaries at the beginning of the experiment. Cont, control. B, MDA-MB-231 cells treated with CXCL9 (100 ng/ml), CXCL10 (20 ng/ml), or CXCL11 (50 ng/ml) for 12 h were stained with rhodamine-phalloidin (1:100) and analyzed actin rearrangement. Arrows indicate polarized F-actin (top panels). The percentage of cells in at least three fields in each of two independent experiments exhibit a polarized F-actin phenotype (bottom graph). Bars represent the mean ± S.D. *, p < 0.05; **, p < 0.01 (Student's t test) compared with control samples. C, MDA-MB-231 cells were treated with 20 ng/ml CXCL10 for indicated time periods, and whole cell lysates were precipitated with GST-RBD or GST-PBD beads as indicated. The proteins bound to the beads were separated on SDS-PAGE, and the amounts of RhoA, Rac1/2/3 or Cdc42 were measured by Western blotting. D, MDA-MB-231 cells treated with 20 ng/ml CXCL10 for indicated time periods were harvested, and whole cell lysates were analyzed the phosphorylation status of RhoA (Ser-188) or Cdc42 (Ser-71) by Western blotting. The whole amount of Cdc42 was used as a loading control.
Cell migration is largely dependent on rearrangement of actin cytoskeleton. Monomeric G-actin can polymerize into microfilaments (F-actin). To determine whether BM-MSC-secreted CXCR3 ligand chemokines can regulate actin rearrangement, the formation of F-actin in MDA-MB-231 cells were analyzed by rhodamine-phalloidin staining. Treatment with CXCL9, CXCL10, or CXCL11 increased numbers of cells displaying a polarized F-actin pattern ∼2–3-fold over control cultures, with F-actin primarily detected at the cell edges (Fig. 6B). Activation of the small GTPase of Rho family plays pivotal roles in regulating cell polarity and motility (25). It has been demonstrated that RhoA activation is critical in regulating the motility and invasion of MDA-MB-231 cells (26–28). RBD of the Rho effector protein, rhotekin, binds to activated GTP-bound RhoA and RhoC (29), whereas p21-activated kinase/PBD binds to activated GTP-bound Rac1 and Cdc42 (30). To determine whether CXCR3 ligands can activate the Rho family of GTPases, a pulldown assay was subjected using GST-fused RBD or PBD. Treatment of MDA-MB-231 cells with CXCL10 caused a time-dependent increase in the amount of RhoA and Cdcd42 precipitated by GST-RBD and GST-PBD-agarose beads, respectively, whereas Rac1/2/3 was not pulled down by GST-PBD beads (Fig. 6C). Furthermore, in accord with pulldown assay, active phosphorylation of RhoA (Ser-88) and Cdc42 (Ser-1) was also increased after CXCL10 treatment (Fig. 6D). These data suggest that RhoA and Cdc42, at least in part, might be involved in facilitation of MDA-MB-231 cell motility through actin rearrangement in response to CXCR3 ligand chemokines produced by BM-MSCs.
CXCR3 Ligand Chemokines Promote Invasive Motility of MDA-MB-231 Cells
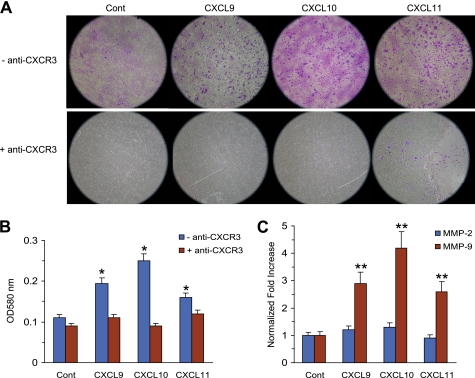
The addition of conditioned medium from BM-MSCs treated with TNFα (MSC-CM) increased the expression of MMP-2 and MMP-9 mRNA in MDA-MB-231 cells (supplemental Fig. S3A) and stimulated the invasion activity of MDA-MB-231 cells, responses that were abrogated by exposure to anti-CXCR3 antibody (supplemental Fig. S3B). These observations suggest that BM-MSCs recruited to cancer cells promote cancer cell invasion at least in part through CXCR3. To determine the effects of CXCR3 ligand chemokines on the invasion activity of MDA-MB-231 cells, Matrigel-based Transwell migration assays were performed. Numbers of invasive MDA-MB-231 cells present on the lower surfaces of basement membrane-coated filters were significantly increased by treatment with CXCL9, CXCL10, or CXCL11 (Fig. 7A, top panel). To further investigate whether enhanced invasive motility of MDA-MB-231 cells was mediated through CXCR3 receptor, anti-CXCR3 antibody was added before treatment with CXCL9, CXCL10, or CXCL11. Preincubation with an anti-CXCR3 antibody almost completely blocked the invading activity of MDA-MB-231 cells (Fig. 7, bottom panels, and B), suggesting that the observed CXCL9-, CXCL10-, or CXCL11-enhanced invasive motility depends on the CXCR3-mediated response. To test whether tumor cell lines expressing CXCR3 respond similarly, we conducted similar experiment using breast (MCF7, T47D) and lung (A549) carcinoma cells. We found that treatment with CXCR3 ligands promoted invasive motility of all cell lines tested (supplemental Fig. S4). Expression of gelatinases such as MMP-2 and MMP-9 from cancer cells is important for tumor invasion and metastasis (31). To assess whether CXCR3 ligands would regulate gelatinase expression on cancer cells, MDA-MB-231 cells were treated with CXCR3 ligand chemokines, and MMP-2 and MMP-9 mRNA levels were measured using quantitative real-time PCR analysis. The amount of MMP-9 mRNA was elevated ∼2.9-, 4.2- or 2.6-fold after CXCL9, CXCL10, or CXCL11 stimulation, respectively, as compared with that of an unstimulated control, whereas MMP-2 mRNA was not significantly induced (Fig. 7C).
FIGURE 7.
CXCR3 ligand chemokines promote invasion ability of MDA-MB-231 cells through induction of MMP-9 expression. A and B, a cell suspension containing MDA-MB-231 cells in serum-free medium containing CXCL9 (100 ng/ml), CXCL10 (20 ng/ml), or CXCL11 (50 ng/ml) in the absence or presence of anti-CXCR3 antibody (5 μg/ml) was seeded onto a basement membrane-coated filter and allowed to invade for 24 h. Invasive cells on the bottom of the insert were stained (A) and extracted for measurement of the OD at 560 nm in a plate reader (B). Cont, control. C, MDA-MB-231 cells were stimulated with CXCL9 (100 ng/ml), CXCL10 (20 ng/ml), or CXCL11 (50 ng/ml) for 24 h. Total RNA was isolated and assessed for MMP-2 or MMP-9 mRNA expression by real time-PCR. Values were normalized for GAPDH mRNA levels. The data shown represent the mean ± S.D. of three independent experiments performed in triplicate. ns, not significant; **, p < 0.01 compared with unstimulated control.
Collectively, these results suggest that each CXCR3 ligand produced by BM-MSCs can individually facilitate migration and invasive motility of CXCR3-expressed breast cancer cells, probably through a mechanism involving the up-regulation of MMP-9 expression at least in part.
DISCUSSION
In this study we demonstrate that BM-MSCs produce all three CXCR3 ligand chemokines, CXCL9, CXCL10, and CXCL11, in response to TNFα through an NF-κB-mediated pathway. All three CXCR3 ligands have the ability to promote the migration and invasion activity of CXCR3-expressing MDA-MB-231 breast cancer cells. We suggest that BM-MSCs recruited to tumor sites contribute to tumor motility and invasiveness through production of CXCR3 ligand chemokines in response to TNFα stimulation in the tumor microenvironment.
It is now widely accepted that a variety of stromal cells, including fibroblasts, endothelial cells, and immune cells, are recruited to tumors and play an important role in enhancing tumor growth, angiogenesis, invasion, and metastasis through secretion of diverse chemokines (9, 32). BM-MSCs are also recruited to tumor stroma and can functionally differentiate into carcinoma-associated fibroblasts-like cells (1, 2, 9, 11, 14, 15, 33). TNFα is a major inflammatory cytokine mainly produced by stromal cells, and it plays a pivotal role in the creation of the tumor microenvironment through the induction of extracellular matrix-degrading enzymes and diverse inflammatory cytokines and chemokines (16, 32). Although chemokines classically represent the chemotactic activity of leukocytes, accumulating evidence demonstrates that diverse chemokines can contribute to tumor growth, angiogenesis, and metastasis (9, 22, 32, 34–38). However, little is known about whether BM-MSCs recruited to tumor sites could be activated by TNFα, a major inflammatory cytokine in the tumor microenvironment, to promote tumor progression. To address this issue, we first screened chemokines expressed in BM-MSCs by TNFα using a quantitative real-time PCR array system. We found that BM-MSCs express diverse CC chemokines, including CCL2 (MCP-1), CCL5 (RANTES), CCL7 (MCP-3), CCL8 (MCP-2), and CCL20 (LARC) as well as CXC chemokines, including CXCL8 (IL-8), CXCL9 (18), (IP-10), and CXCL11 (I-TAC), in response to TNFα stimulation. This finding supports the notion that BM-MSCs could contribute to the facilitation of a local tumor microenvironment as a component of tumor stromal cell response to TNFα.
Among the cytokines induced by TNFα in BM-MSCs, three C-X-C type chemokines (CXCL9, CXCL10, and CXCL11), which share a common receptor (CXCR3), were highly expressed. Classically, it is well known that these CXCR3 ligand chemokines are induced by IFNγ in diverse cell types, including neutrophils, monocytes, fibroblasts, keratinocytes, endothelial cells, and astrocytes, and they act as selective chemotactic factors for monocytes (39), dendritic cells (40), natural killer cells (41), and Th1 cells (42, 43). The effect of these CXCR3 ligands is known to exert angiostatic and antitumor activities on microvascular endothelial cells and activated T lymphocytes (44–46). In contrast, some studies have demonstrated that CXCR3 and their ligands are overexpressed in some cancer cells, including renal cell carcinoma (47), melanoma (48), breast cancer (23, 24), ovarian carcinoma (49), and thyroid carcinoma (50), to promote tumor progression and metastasis in an autocrine fashion. Our data show that all three CXCR3 ligands promote the migration and invasion activity of MDA-MB-231 cells, which were almost completely blocked by the addition of a neutralizing anti-CXCR3 antibody to the culture medium. In addition, we found that CXCR3 ligands promoted the invasive migration of MCF7, T47D, and A549 cells, all of which express CXCR3, suggesting that the effects of CXCR3 ligands on tumor cell migration may also apply to other cancer cells. Furthermore, these CXCR3 ligands cause the activation of small GTPase Rho A and up-regulation of MMP-9 mRNA expression in MDA-MB-231 cells, which probably contribute to the migration and invasion activity of tumor cells. Thus, BM-MSCs stimulated by TNFα could support tumor motility and invasion through the production of CXCR3 ligands, particularly CXCL10, in the tumor microenvironment.
A previous study has reported that BM-MSCs comingled with breast carcinoma cells, but not BM-MSC alone, and accelerate the tumor growth and metastasis through the secretion of CCL5, demonstrating the importance of the interaction between breast cancer cells and BM-MSCs for tumor development in the tumor microenvironment (1). Our results show a similar contribution of BM-MSCs in the progression of breast cancer cell motility and invasiveness through the secretion of CXCR3 ligand chemokines but point to an important difference. Here, we found that the production of CXCR3 ligands from BM-MSCs is not required for direct interaction between BM-MSCs and cancer cells but is stimulated by the soluble factor, TNFα, which is enriched in the tumor microenvironment. In fact, we found that treatment of BM-MSCs with conditioned media from various breast cancer cell lines, including MDA-MB-231, MCF7, and T47D cells, up-regulated the expression of CXCR3 ligands (supplemental Fig. S5), supporting the idea that BM-MSCs are stimulated by soluble factors secreted from cancer cells. Because the major source of TNFα is from tumor-associated stromal cells, it is likely that BM-MSCs recruited to the tumor site can be activated by tumor-associated stromal cells, such as myofibroblasts and infiltrated immune cells. However, as TNFα can be produced in diverse cancer cells, we cannot rule out a possible interaction between cancer cells and BM-MSCs for CXCR3 ligand expression.
In summary, this study demonstrates the role of BM-MSCs in the promotion of tumor motility and invasiveness in CXCR3-expressing breast cancer cells through the secretion of CXCR3 ligand chemokines (CXCL9, CXCL10, and CXCL11) in response to TNFα stimulation. Control of TNFα production or inhibition of the CXCR3 ligand expression from BM-MSCs may provide a new opportunity for the attenuation of CXCR3-expressing tumor growth.
Supplementary Material
This work was supported by Disease Network Research Program Grant 2009-0084181) and by Basic Science Research Program Grant 2009-0076856 through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology, Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental S1–S5.
- BM-MSC
- bone marrow-derived mesenchymal stem cell
- RBD
- Rho-binding domain
- PBD
- p21-activated kinase/p21-binding domain.
REFERENCES
- 1.Karnoub A. E., Dash A. B., Vo A. P., Sullivan A., Brooks M. W., Bell G. W., Richardson A. L., Polyak K., Tubo R., Weinberg R. A. (2007) Nature 449, 557–563 [DOI] [PubMed] [Google Scholar]
- 2.Allinen M., Beroukhim R., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Schnitt S., Sellers W. R., Polyak K. (2004) Cancer Cell 6, 17–32 [DOI] [PubMed] [Google Scholar]
- 3.Orimo A., Gupta P. B., Sgroi D. C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V. J., Richardson A. L., Weinberg R. A. (2005) Cell 121, 335–348 [DOI] [PubMed] [Google Scholar]
- 4.Bhowmick N. A., Neilson E. G., Moses H. L. (2004) Nature 432, 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camps J. L., Chang S. M., Hsu T. C., Freeman M. R., Hong S. J., Zhau H. E., von Eschenbach A. C., Chung L. W. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tlsty T. D. (2001) Semin. Cancer Biol. 11, 97–104 [DOI] [PubMed] [Google Scholar]
- 7.Bissell M. J., Radisky D. (2001) Nat. Rev. Cancer 1, 46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu M., Polyak K. (2008) Curr. Opin. Genet. Dev. 18, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karnoub A. E., Weinberg R. A. (2006) Breast Dis. 26, 75–85 [DOI] [PubMed] [Google Scholar]
- 10.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. (1999) Science 284, 143–147 [DOI] [PubMed] [Google Scholar]
- 11.Studeny M., Marini F. C., Dembinski J. L., Zompetta C., Cabreira-Hansen M., Bekele B. N., Champlin R. E., Andreeff M. (2004) J. Natl. Cancer Inst. 96, 1593–1603 [DOI] [PubMed] [Google Scholar]
- 12.Spaeth E. L., Dembinski J. L., Sasser A. K., Watson K., Klopp A., Hall B., Andreeff M., Marini F. (2009) PLoS. One 4, e4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emura M., Ochiai A., Horino M., Arndt W., Kamino K., Hirohashi S. (2000) In Vitro Cell Dev. Biol. Anim. 36, 77–80 [DOI] [PubMed] [Google Scholar]
- 14.Mishra P. J., Mishra P. J., Humeniuk R., Medina D. J., Alexe G., Mesirov J. P., Ganesan S., Glod J. W., Banerjee D. (2008) Cancer Res. 68, 4331–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii G., Sangai T., Oda T., Aoyagi Y., Hasebe T., Kanomata N., Endoh Y., Okumura C., Okuhara Y., Magae J., Emura M., Ochiya T., Ochiai A. (2003) Biochem. Biophys. Res. Commun. 309, 232–240 [DOI] [PubMed] [Google Scholar]
- 16.Balkwill F. (2009) Nat. Rev. Cancer 9, 361–371 [DOI] [PubMed] [Google Scholar]
- 17.Fries M., Chauhan H. J., Domingo G. J., Jung H. I., Perham R. N. (2003) Eur. J. Biochem. 270, 861–870 [DOI] [PubMed] [Google Scholar]
- 18.Riccardi C., Bruscoli S., Ayroldi E., Agostini M., Migliorati G. (2001) Adv. Exp. Med. Biol. 495, 31–39 [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y. Y., Guo L., Zhao X. J., Liu H., Lei T., Ma D. J., Gao X. Y. (2009) Exp. Mol. Med. 41, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karin M. (2006) Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 21.Pikarsky E., Porat R. M., Stein I., Abramovitch R., Amit S., Kasem S., Gutkovich-Pyest E., Urieli-Shoval S., Galun E., Ben-Neriah Y. (2004) Nature 431, 461–466 [DOI] [PubMed] [Google Scholar]
- 22.Raman D., Baugher P. J., Thu Y. M., Richmond A. (2007) Cancer Lett. 256, 137–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta D., Flaxenburg J. A., Laxmanan S., Geehan C., Grimm M., Waaga-Gasser A. M., Briscoe D. M., Pal S. (2006) Cancer Res. 66, 9509–9518 [DOI] [PubMed] [Google Scholar]
- 24.Goldberg-Bittman L., Neumark E., Sagi-Assif O., Azenshtein E., Meshel T., Witz I. P., Ben-Baruch A. (2004) Immunol. Lett. 92, 171–178 [DOI] [PubMed] [Google Scholar]
- 25.Raftopoulou M., Hall A. (2004) Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 26.Pillé J. Y., Denoyelle C., Varet J., Bertrand J. R., Soria J., Opolon P., Lu H., Pritchard L. L., Vannier J. P., Malvy C., Soria C., Li H. (2005) Mol. Ther. 11, 267–274 [DOI] [PubMed] [Google Scholar]
- 27.Sahai E., Garcia-Medina R., Pouysségur J., Vial E. (2007) J. Cell Biol. 176, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu D., Asiedu M., Wei Q. (2009) Oncogene 28, 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid T., Furuyashiki T., Ishizaki T., Watanabe G., Watanabe N., Fujisawa K., Morii N., Madaule P., Narumiya S. (1996) J. Biol. Chem. 271, 13556–13560 [DOI] [PubMed] [Google Scholar]
- 30.Benard V., Bohl B. P., Bokoch G. M. (1999) J. Biol. Chem. 274, 13198–13204 [DOI] [PubMed] [Google Scholar]
- 31.Deryugina E. I., Quigley J. P. (2006) Cancer Metastasis Rev. 25, 9–34 [DOI] [PubMed] [Google Scholar]
- 32.Balkwill F. (2004) Nat. Rev. Cancer 4, 540–550 [DOI] [PubMed] [Google Scholar]
- 33.Direkze N. C., Hodivala-Dilke K., Jeffery R., Hunt T., Poulsom R., Oukrif D., Alison M. R., Wright N. A. (2004) Cancer Res. 64, 8492–8495 [DOI] [PubMed] [Google Scholar]
- 34.Tanaka T., Bai Z., Srinoulprasert Y., Yang B. G., Yang B., Hayasaka H., Miyasaka M. (2005) Cancer Sci. 96, 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh S., Sadanandam A., Singh R. K. (2007) Cancer Metastasis Rev. 26, 453–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strieter R. M., Burdick M. D., Mestas J., Gomperts B., Keane M. P., Belperio J. A. (2006) Eur. J. Cancer 42, 768–778 [DOI] [PubMed] [Google Scholar]
- 37.Ben-Baruch A. (2006) Cancer Metastasis Rev. 25, 357–371 [DOI] [PubMed] [Google Scholar]
- 38.Ali S., Lazennec G. (2007) Cancer Metastasis Rev. 26, 401–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janatpour M., Hudak S., Sathe M., Sedgwick J., McEvoy L. (2001) J. Exp. Med. 194, 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penna G., Sozzani S., Adorini L. (2001) J. Immunol. 167, 1862–1866 [DOI] [PubMed] [Google Scholar]
- 41.Loetscher M., Gerber B., Loetscher P., Jones S. A., Piali L., Clark-Lewis I., Baggiolini M., Moser B. (1996) J. Exp. Med. 184, 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonecchi R., Bianchi G., Bordignon P., D'Ambrosio D., Lang R., Borsatti A., Sozzani S., Allavena P., Gray P., Mantovani A. (1998) J. Exp. Med. 187, 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sallusto F., Lenig D., Mackay C. R., Lanzavecchia A. (1998) J. Exp. Med. 187, 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tannenbaum C. S., Tubbs R., Armstrong D., Finke J. H., Bukowski R. M., Hamilton T. A. (1998) J. Immunol. 161, 927–932 [PubMed] [Google Scholar]
- 45.Teichmann M., Meyer B., Beck A., Niedobitek G. (2005) J. Pathol. 206, 68–75 [DOI] [PubMed] [Google Scholar]
- 46.Romagnani P., Annunziato F., Lasagni L., Lazzeri E., Beltrame C., Francalanci M., Uguccioni M., Galli G., Cosmi L., Maurenzig L., Baggiolini M., Maggi E., Romagnani S., Serio M. (2001) J. Clin. Invest. 107, 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suyama T., Furuya M., Nishiyama M., Kasuya Y., Kimura S., Ichikawa T., Ueda T., Nikaido T., Ito H., Ishikura H. (2005) Cancer 103, 258–267 [DOI] [PubMed] [Google Scholar]
- 48.Kawada K., Hosogi H., Sonoshita M., Sakashita H., Manabe T., Shimahara Y., Sakai Y., Takabayashi A., Oshima M., Taketo M. M. (2007) Oncogene 26, 4679–4688 [DOI] [PubMed] [Google Scholar]
- 49.Furuya M., Suyama T., Usui H., Kasuya Y., Nishiyama M., Tanaka N., Ishiwata I., Nagai Y., Shozu M., Kimura S. (2007) Hum. Pathol. 38, 1676–1687 [DOI] [PubMed] [Google Scholar]
- 50.Melillo R. M., Castellone M. D., Guarino V., De Falco V., Cirafici A. M., Salvatore G., Caiazzo F., Basolo F., Giannini R., Kruhoffer M., Orntoft T., Fusco A., Santoro M. (2005) J. Clin. Invest. 115, 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.