Abstract
The serpin mechanism of protease inhibition involves the rapid and stable incorporation of the reactive center loop (RCL) into central β-sheet A. Serpins therefore require a folding mechanism that bypasses the most stable “loop-inserted” conformation to trap the RCL in an exposed and metastable state. This unusual feature of serpins renders them highly susceptible to point mutations that lead to the accumulation of hyperstable misfolded polymers in the endoplasmic reticulum of secretory cells. The ordered and stable protomer-protomer association in serpin polymers has led to the acceptance of the “loop-sheet” hypothesis of polymerization, where a portion of the RCL of one protomer incorporates in register into sheet A of another. Although this mechanism was proposed 20 years ago, no study has ever been conducted to test its validity. Here, we describe the properties of a variant of α1-antitrypsin with a critical hydrophobic section of the RCL substituted with aspartic acid (P8–P6). In contrast to the control, the variant was unable to polymerize when incubated with small peptides or when cleaved in the middle of the RCL (accepted models of loop-sheet polymerization). However, when induced by guanidine HCl or heat, the variant polymerized in a manner indistinguishable from the control. Importantly, the Asp mutations did not affect the ability of the Z or Siiyama α1-antitrypsin variants to polymerize in COS-7 cells. These results argue strongly against the loop-sheet hypothesis and suggest that, in serpin polymers, the P8–P6 region is only a small part of an extensive domain swap.
Keywords: Crystal Structure, Protease Inhibitor, Protein Conformation, Protein Domains, Serpin, Domain Swap, Inhibitor, Polymerization
Introduction
Members of the serpin family of serine protease inhibitors control the biological pathways critical for life, including blood coagulation, complement activation, fibrinolysis, inflammation, and apoptosis (1, 2). The mechanism that serpins employ to inhibit proteases uniquely involves a rapid conformational change to a hyperstable state (3, 4). Serpin function therefore requires a folding pathway that bypasses the most stable conformation in favor of a metastable native state. Exactly how serpins fold is unclear, but the pathway is apparently highly sensitive to mutations, with several missense mutations leading to defects in secretion and subsequent disease (5). The best described case involves the Z mutation (6) in archetypal serpin α1-antitrypsin (α1AT).2 In homozygotes, this mutation leads to a 90% decrease in circulating levels due to the accumulation of hyperstable “polymers” of the protein in the endoplasmic reticulum of hepatocytes. The Z mutation has minimal effect on the activity or stability of the correctly folded native protein (7), so the defect in secretion is due entirely to a perturbation of the folding pathway (8). Polymers isolated from tissue revealed an ordered “beads-on-a-string” morphology by electron microscopy (9) and laddering of bands on native PAGE. Similar morphology and laddering can be obtained in vitro by partial unfolding of serpins into the so-called M* state with either heat or denaturants (10). This phenomenon has become known as serpin polymerization.
The first crystallographic structure of a serpin (11) was of α1AT that had been proteolytically nicked between the P1 and P1′ bond (the scissile bond, using the nomenclature for substrates of Schechter and Berger (12)). It revealed a mixed α/β-structure composed of an N-terminal helical domain and a C-terminal β-barrel domain bridged by a central six-stranded β-sheet (sheet A). Surprisingly, the P1 and P1′ residues were 70 Å apart due to the incorporation of the reactive center loop (RCL) as the fourth strand in sheet A (Fig. 1A). Subsequent structures of native serpins showed that the RCL is indeed a solvent-exposed loop on the “top” of the molecule (shown in the classic orientation) and that strands 3 and 5 of sheet A are hydrogen-bonded to one another as typical parallel β-strands (Fig. 1B). The conformational change from exposed to inserted RCL (known as “loop insertion”) is associated with a significant thermodynamic stabilization so that the resulting six-stranded state is hyperstable. Similar stabilization can be achieved by incorporation of the RCL in the absence of cleavage to make the so-called “latent” conformation or by the incorporation of exogenous peptides with sequences similar to the RCL (4). The observation that peptides can incorporate into β-sheet A led to the hypothesis that the RCL itself could insert into the β-sheet of another monomer in trans to form polymeric hyperstable structures (13). This was the origin of the “loop-sheet” hypothesis. It has subsequently been adjusted to take into account the observation that short peptides related to the portion of the RCL that inserts into the top of sheet A (P14–P9) promote polymerization (14), presumably by opening up the bottom of sheet A to allow the incorporation of the P8–P3 portion of the RCL from another monomer (Fig. 2A). Such a mechanism can also occur through cleavage in the center of the RCL (15), resulting in the incorporation of the N-terminal portion of the RCL in cis and the C-terminal portion in trans (Fig. 2C). The loop-sheet hypothesis in its current manifestation suggests that partial self-insertion (P15–P10) in the context of an intact serpin can similarly open up the bottom of sheet A to allow the intermolecular incorporation of the P8–P3 region (16, 17). However, it is acknowledged that, for polymerization to continue, the self-insertion must be reversed so that a loop-sheet dimer would resemble the model published in 1997 (Fig. 2A) (14).
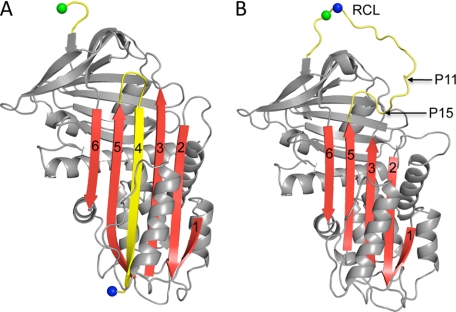
FIGURE 1.
Ribbon diagrams of cleaved and native α1AT. A, the structure of α1AT is composed of a lower N-terminal helical domain and an upper C-terminal β-barrel domain (when in this “classic” orientation). The main features are the yellow RCL and β-sheet A (red, with strands numbered). In this structure, the RCL has been cleaved between the P1 (blue sphere) and P1′ (green sphere) residues and is incorporated as strand 4 in β-sheet A. B, a ribbon depiction of native α1AT (colored as described above) shows the RCL as an exposed loop and a five-stranded β-sheet A. The P1 and P1′ residues are indicated as previously. The first residue to incorporate into β-sheet A upon cleavage is P15 (indicated). Specific cleavage at the P11 residue (indicated) results in the cleaved polymer shown in Fig. 2C.
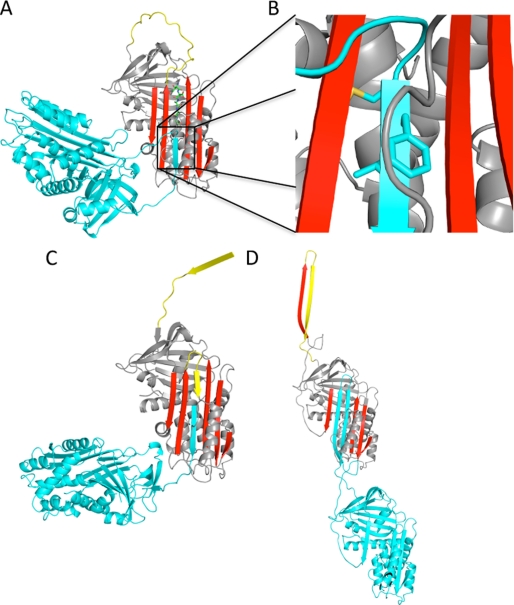
FIGURE 2.
Models of serpin polymers. A, the ribbon diagram shows the peptide-induced loop-sheet polymer. The P14–P9 peptide inserts in register (green rods) to open a gap at the bottom of the sheet between strands 3 and 5A (colored as before). This gap is presumably filled by the in-register insertion of the P8–P3 portion of the RCL from another protomer (cyan). B, a close-up of the P8–P6 region (side chains shown as rods) illustrates the internal orientation of the P8 (Met) and P6 (Leu) side chains and how P7 (Phe) is buried under the helix F loop (gray coil). C, the ribbon diagram of the cleaved serpin polymer, colored as before, reveals how proteolytic nicking of the RCL (here at the P11–P10 bond) leads to self-insertion from P15–P11, followed by insertion of the P10–P3 region from another protomer. D, the model of the open dimer based on the structure of the closed antithrombin dimer shows how strand 5A and the RCL swap from one protomer into another to complete β-sheet A, thereby creating a hyperstable linkage. This mechanism would be less dependent on the composition of the P8–P6 region relative to the peptide and cleavage-induced mechanisms illustrated in A–C.
The stability of the polymeric linkage of loop-sheet polymers thus relies entirely on the in-register insertion of the P8–P3 region into sheet A of another monomer. The P8 and P6 side chains are internally oriented, and P7 is buried by the loop following helix F (Fig. 2B). There are several conceptual difficulties with this model that were explored in a recent molecular modeling study (18). However, the mechanism has never been directly tested by experimental methods. We hypothesized that mutating the P8–P6 region in α1AT from conserved hydrophobic residues Met-Phe-Leu to Asp-Asp-Asp would prevent polymerization by the loop-sheet mechanism because these three residues constitute the bulk of the proposed intermolecular contact for loop-sheet polymers. However, if the P8–P6 region were only a small part of a larger domain swap, as proposed recently (Fig. 2D) (19), then the mutations should have little or no effect on the formation of polymers in vitro (when induced by denaturants or heat) or in vivo (in conjunction with a polymerigenic mutation).
We found that the P8–P6 Asp variant was unable to polymerize by the loop-sheet mechanism whether induced by small peptides or by cleavage in the center of the RCL. In contrast, the Asp mutations had no effect on the formation of polymers in vitro in the presence of low concentrations of guanidine HCl (GdnHCl), or upon heating at 50 °C or in COS-7 cells when coupled with the Z or Siiyama mutation. Further substitutions at P10 (to Pro) and P9 (to Asp) were still unable to prevent polymerization in vitro or in cells. These results argue strongly against the loop-sheet hypothesis and suggest that the P8–P6 region of the RCL is only a small part of a large domain swap, perhaps including strand 5A as seen recently for another serpin (19).
EXPERIMENTAL PROCEDURES
Materials
The P14–P9 peptide of antithrombin (Ac-SEAAAS) was synthesized and purified by SynBioSci Corp. (Livermore, CA). Trypsin and V8 protease were purchased from Sigma, and thrombin was purchased from Haematologic Technologies (Essex Junction, VT).
Mutation, Expression, and Purification of α1AT
For in vitro studies, the template plasmid (pQE30/α1AT) was a double variant with a P1 residue mutation of Met358 to Arg and Cys232 mutated to Ala. For crystallographic studies, α1AT was N-terminally truncated by the addition of a restriction enzyme cleavage site immediately before residue 22 and subsequent cloning into the pET15b vector (Novagen) using NdeI and XhoI sites. Mutagenesis was conducted using the QuikChangeTM kit and associated protocol (Stratagene, La Jolla, CA). For the P8–P6 Asp variant, the residues from P8 to P6 in the RCL (Met351-Phe352-Leu353) were substituted with Asp. For the P10–P6 variant, the P10-to-Pro and P9-to-Asp mutations were introduced into the P8–P6 Asp variant. Recombinant α1AT was produced from Escherichia coli as reported previously (20).
COS-7 Cell Study
For the cell study, α1AT was cloned from pQE30 into the pCEP4 vector (Invitrogen) using NotI and KpnI sites and extended to add the signal sequence derived from α1AT. The Z (E342K), Siiyama (S53F), P8–P6 Asp, and P10 Pro/P9–P6 Asp mutations were then introduced. COS-7 cells were maintained in DMEM (Invitrogen) supplemented with 10% (v/v) of FBS at 37 °C and 5% CO2 in a humidified incubator. α1AT DNA was transfected into cells at 90% confluency in 6-well plates using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. After a 48-h incubation in Opti-MEM culture medium (Invitrogen) at 37 °C, the cell lysate and culture medium were harvested from each well. The cell pellet was lysed in 50 μl of 50 mm Tris (pH 7.4), 150 mm NaCl, 1% Triton X-100, 5 mm EDTA, and 0.5 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) at 4 °C for 1 h. After removal of the insoluble pellet by centrifugation, the supernatant was frozen at −80 °C. The culture medium was transferred via 0.45-μm syringe filter to a Vivaspin 2 concentrator with a 10,000-Da cut-off membrane (Sartorius Stedim Biotech S. A.), concentrated 40 times at 4 °C, and then frozen at −80 °C. Samples of cell lysate and concentrated culture medium were analyzed by 10% nonreducing SDS-PAGE (NuPAGE BisTris gel, Invitrogen) or 8% native PAGE. Gels were analyzed by Western blotting using rabbit anti-α1AT polyclonal primary antibody (AbD Serotec, Oxford, UK) and goat anti-rabbit secondary antibody (Sigma). For the P10–P6 study, we used a mouse monoclonal primary antibody (Abcam Inc., Cambridge, MA) and goat anti-mouse secondary antibody (Thermo Fisher Scientific Inc., Waltham, MA).
CD Spectra and Melts
The structure and thermal unfolding of the intact and cleaved α1AT variants were assessed by CD on a Jasco J-810 spectropolarimeter. Cleaved control α1AT and variants were prepared by incubation with V8 protease and thrombin, respectively. The far-UV spectra from 180 to 260 nm were collected three times and averaged from samples at 0.5 to 1 mg/ml in PBS in a cuvette with a light path of 0.1 mm. Stability was assessed by monitoring change in signal at 222 nm from the same samples while increasing the temperature from 20 to 95 °C at 1 °C/min.
Peptide Polymerization
Control α1AT and variants were prepared at 0.5 mg/ml in PBS and incubated with 0-, 1-, 2-, and 5-fold molar excesses of P14–P9 peptide at 37 °C for 16 h. Polymer formation was assessed by 8% native PAGE.
Protease-induced Polymerization
A novel cleavage site for trypsin was introduced at the P11 residue of the RCL by mutating Ala348 to Arg. The original Arg residue at P1 was mutated to Asp to ensure that the only cleavage site would be at P11. The P11 Arg/P1 Asp variants with and without the P8–P6 Asp mutations were prepared at 0.5 mg/ml in PBS and cleaved by 1:50 (weight) ratio of bovine trypsin at 25 °C for 1 h (the reaction was stopped by the addition of AEBSF). Polymerization was evaluated by 8% native PAGE. To confirm the results, the experiment was run again at a higher concentration of the P8–P6 Asp variant (1 mg/ml) and a longer incubation time (48 h) at 25 °C (trypsin was inhibited by AEBSF after 1 h). Cleaved polymers were still not observed for the variant (data not shown).
GdnHCl and Heat Polymerization
Control α1AT and variants were prepared at 0.1 mg/ml in PBS, and polymerization was induced by incubation with 0, 0.5, 0.75, and 1.0 m GdnHCl at 37 °C for 3 h or by incubation at 50 °C for 0, 1, 4, and 16 h.
Inhibition Assay
The stoichiometry of inhibition was estimated by incubating 0.05 mg/ml thrombin in PBS for 5 min at room temperature with 0.25-, 0.5-, 0.75-, 1-, 2-, and 5-fold molar ratios of control α1AT and with 10-, 20-, 50-, 100-, 200-, and 300-fold molar excesses of the α1AT variants. Reactions were stopped by AEBSF, and final complex formation was evaluated by 10% SDS-PAGE.
Crystallization, Data Collection, and Refinement
The cleaved (N-terminally truncated) P8–P6 Asp and P10 Pro/P9–P6 Asp variants were prepared by incubation with a 1:10 molar ratio of bovine thrombin at 37 °C for 1 h. Thrombin was inhibited by AEBSF and then removed by Q-Sepharose chromatography. The cleaved P8–P6 Asp variant was concentrated to 10 mg/ml in 10 mm Tris (pH 7.4) and 50 mm NaCl and crystallized in 13.3% PEG 550 monomethyl ether, 6.7% PEG 20,000, 0.1 m MES (pH 6.5), and 0.12 m MorpheusTM alcohol mixture. The crystal was flash-cooled to 100 K after cryoprotecting in the original condition plus 20% glycerol, and a data set was collected in-house using a Rigaku rotating copper anode source. The cleaved P10–P6 variant was concentrated to 10 mg/ml in 10 mm Tris (pH 7.4) and 50 mm NaCl and crystallized in 23% PEG 400 and 0.1 m MES-NaOH (pH 6.0). The crystal was flash-cooled to 100 K in 30% PEG 400 and 0.1 m MES-NaOH (pH 6.0), and data were collected on beamline I02 at the Diamond Light Source (Didcot, UK). Diffraction data were processed using Mosflm, Scala, and Truncate (21). Initial phases were determined by molecular replacement with a cleaved α1AT model (Protein Data Bank code 7API) using Phaser (22). The structures were refined using CNS (23) and Refmac5 (24) and rebuilt using XtalView (25) and Coot (26). Structural figures were made using PyMOL (27).
RESULTS
Functional and Structural Characterization of the P8–P6 Asp α1AT Variant
The control and P8–P6 Asp variants were expressed in E. coli on the C232A and M358R (P1) background to remove the exposed Cys residue and to convert the specificity to trypsin-like serine proteases. Correct folding into the native state was confirmed by CD (supplemental Fig. S1) and by thrombin cleavage at P1 (supplemental Fig. S2). The control was an efficient inhibitor of thrombin, but unsurprisingly, the P8–P6 Asp mutations rendered the variant noninhibitory. However, using a massive excess of the variant, we were able to observe some complex by SDS-PAGE, suggestive of a stoichiometry of inhibition of ∼900 (supplemental Fig. S2). The ability of the variant to inhibit thrombin, albeit to a small extent, suggests the full incorporation of the RCL after cleavage at the P1–P1′ bond but at a significantly slowed rate. Consistent with this, CD unfolding showed that the RCL-cleaved control and P8–P6 variants were both hyperstable, with no melting transition up to 95 °C (supplemental Fig. S1). To see how the Asp residues were accommodated in β-sheet A, we solved the structure of the RCL-cleaved P8–P6 Asp variant and found that the Asp residues were incorporated in the normal fashion with the help of a buried water molecule and likely side chain protonation (supplemental Fig. S3A and Table S1). We conclude from these studies that the P8–P6 Asp variant folds into the correct native conformation and that the mutations significantly disfavor or slow insertion of the C-terminal portion of the RCL, as designed.
P8–P6 Asp Mutations Prevent Peptide-induced Polymerization
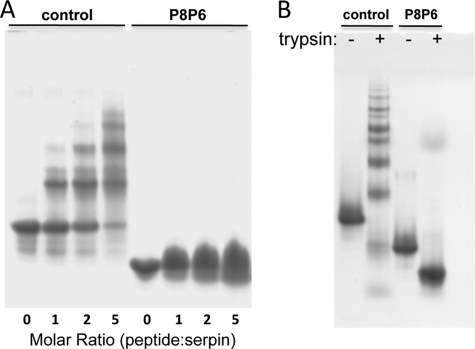
One of the most commonly used models for loop-sheet polymerization is the incubation of antithrombin with a peptide corresponding to the very N-terminal portion of the RCL, namely the P14–P9 peptide (14, 28). We found that the antithrombin-derived peptide (SEAAAS) is also effective at promoting α1AT polymerization. We incubated the SEAAAS peptide with control α1AT and with the P8–P6 Asp variant for 16 h at 37 °C with the ratio of peptide to protein increasing from 1:1 to 5:1 and tested for polymer formation by native PAGE (Fig. 3A). The peptide induced polymer formation in the control in a concentration-dependent manner, as seen by the increased laddering on the native gel and the disappearance of the monomer. In contrast, the P8–P6 variant did not form ladders when incubated with the SEAAAS peptide, even at the highest concentration used. This provides an indication that the Asp residues at P8–P6 prevent the incorporation of the P8–P3 region after the incorporation of the P14–P9 peptide. However, it is possible that the peptide is effectively competing with the P8–P6 Asp RCL to fill the gap created by the initial in-register peptide insertion. This was observed previously in a crystal structure of PAI-1 where two copies of a P14–P10 peptide inserted in the proper position and in the P6–P2 position (29). The increasingly diffuse nature of the monomer band for the peptide-incubated variant might be indicative of single and double insertions. This study demonstrates that the P8–P6 Asp mutation inhibits peptide-induced polymerization of α1AT, but we cannot conclude that it blocks it entirely due to the possible competition from the peptide.
FIGURE 3.
Native PAGE of loop-sheet polymers induced by peptides or cleavage. A, small peptides corresponding to the P14–P9 region induce polymerization of control α1AT (molar ratio indicated) as evidenced by the formation of laddered bands on a native gel. In contrast, the peptide does not induce polymerization of the P8–P6 Asp variant (P8P6) even at the highest peptide concentration. B, the control and P8–P6 Asp variant of α1AT were mutated to create an exclusive trypsin cleavage site at the P11 position in the RCL. Treatment with trypsin converted the control to polymers (with only a small amount of cleaved monomer evident), whereas the P8–P6 Asp variant was completely resistant to polymer formation.
P8–P6 Asp Mutations Prevent Protease-induced Polymerization
A more robust method for inducing serpins to form loop-sheet polymers is to proteolytically nick the RCL a few residues N-terminal of the P1–P1′ bond (i.e. P11–P10 as illustrated in Fig. 2C). This causes the rapid incorporation of the N-terminal portion of the RCL into sheet A but leaves the C-terminal part exposed on the top and only capable of insertion in trans. Polymers were observed by native PAGE and electron microscopy, even at vanishingly low concentrations (15), illustrating the favorability of polymer formation by this method. The loop-sheet linkage was subsequently verified by two independent crystal structures (30, 31). To test if the P8–P6 Asp mutations prevent polymerization by this mechanism, we mutated the P1 residue to Asp and placed an Arg at P11. Incubation with trypsin caused the rapid and full polymerization of control α1AT, but the P8–P6 Asp variant remained monomeric after cleavage at P11 (Fig. 3B). We can conclude from this study that Asp at P8–P6 prevents polymerization by the loop-sheet mechanism.
P8–P6 Asp Mutations Do Not Prevent GdnHCl- or Heat-induced Polymerization
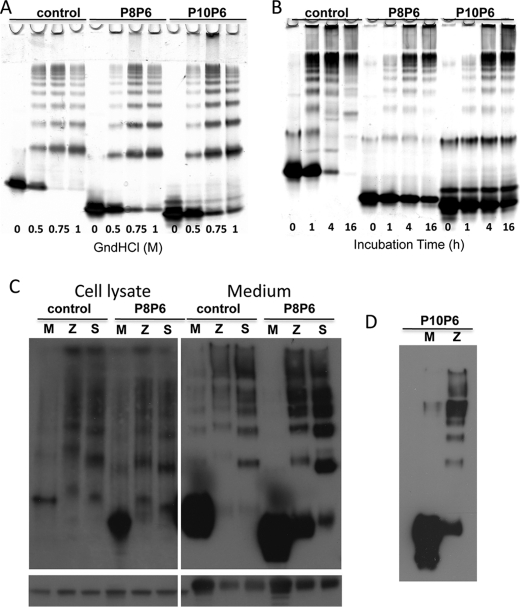
The standard method for producing serpin polymers in vitro is to incubate at low concentration of denaturant or at a temperature 5–10 °C below the melting point. To determine whether the P8–P6 Asp mutations affect the ability of α1AT to polymerize under these commonly used in vitro conditions, we incubated it and a control for 3 h at 37 °C in GdnHCl concentrations ranging from 0 to 1 m or at 50 °C for up to 16 h. The reactions were run on native polyacrylamide gel, and the results are shown in Fig. 4 (A and B). It is evident from the appearance of laddered bands on the native gels that the P8–P6 variant has little effect on the ability of α1AT to polymerize under either condition. The persistence of the monomer band for the variant at the highest GdnHCl concentration and at the longest incubation time is consistent with the ∼5 °C increase in melting temperature for the native variant relative to the control (supplemental Fig. S1). The P8–P6 Asp mutations clearly do not prevent polymerization by whatever mechanism is induced by either GdnHCl or heat.
FIGURE 4.
In vitro and in vivo polymer formation is unaffected by the RCL mutations. A, incubation at 37 °C with increasing amounts of GdnHCl leads to polymer bands on native PAGE for control, the P8–P6 Asp variant (P8P6) and the P10 Pro/P9–P6 Asp variant (P10P6). B, a native gel of the control and variants after heating at 50 °C for the times indicated shows the formation of polymers over time. The apparent resistance of the variants to polymerization is likely due to the increased stability of the native variants relative to the control (∼5 °C). C, shown are Western blots of COS-7 cell lysates and media when the control and P8–P6 Asp variant are without (M) or coupled with the Z (Z) or Siiyama (S) mutations. The native gels (upper) show that polymers are formed for the control and variant when on the Z or Siiyama background whether samples were taken from cell lysate or secreted medium. Western blots of SDS gels are given below to indicate the amount of α1AT loaded. D, shown are cell media samples for the control (M) and Z versions of the P10 Pro/P9–P6 Asp variant on a native gel.
P8–P6 Asp Mutations Do Not Affect Polymerization in COS-7 Cells
We cannot assume that polymers formed in vitro upon partial denaturation are identical to those formed in cells from a folding intermediate. So, to test the effect of the P8–P6 Asp mutations on polymerization in cells, we created the Z and Siiyama variants on the control or P8–P6 Asp backgrounds and transiently transfected COS-7 cells. Samples from cell lysates and from the media were run on native polyacrylamide gel to assess the amount of monomeric and polymeric species (Fig. 4C). In the absence of the polymerigenic mutations, both the control and P8–P6 Asp were secreted predominantly as monomers. However, when coupled with either the Z or Siiyama mutation, the level of secretion was decreased, and the material was predominantly polymeric. Similar results were obtained from cell lysates (Fig. 4C). Although there is some indication that the P8–P6 Asp mutations rescue folding of the Z variant to some degree (larger monomer band than that with control Z), there is no rescue for the Siiyama variant when loading is taken into account. Nevertheless, it is overwhelmingly clear from these results that the P8–P6 Asp variant is capable of polymerizing in cells when coupled with polymer-inducing mutations.
Addition of P10 Pro and P9 Asp to the P8–P6 Asp Mutations Does Not Prevent Polymerization in Vitro or in Cells
We were somewhat surprised to find that aspartic acids at P8–P6 did not prevent the insertion of the RCL upon cleavage at P1 by thrombin and that the cleaved form was still stable up to 95 °C (supplemental Fig. S1B). In an attempt to obtain a variant incapable of loop insertion in the critical P8–P3 region, we added two further mutations to the P8–P6 Asp variant, namely, P10 Pro and P9 Asp. In contrast to the P8–P6 variant, the P10–P6 variant displayed a complete absence of inhibitory activity (supplemental Fig. S2), and when cleaved at the P1 residue by thrombin, its stability was increased from 61 to only 81 °C (supplemental Fig. S1). However, when we solved the crystal structure of the cleaved P10–P6 variant, the RCL was found to be fully inserted (supplemental Fig. S3B and Table S1). Whether the RCL is stably inserted from P10–P3 in solution is unclear, but melting data prove that the additional mutations have had the desired effect of further destabilizing the RCL interactions within sheet A. Despite the destabilizing effect of the P10–P6 mutations, the variant was still capable of polymerizing in vitro with GdnHCl or heat (Fig. 4, A and B) and in COS-7 cells when coupled with the Z mutation (Fig. 4D).
DISCUSSION
Serpin polymerization underlies a large number of loss-of-function diseases, including emphysema (α1AT, α1-antichymotrypsin), angioedema (C1 inhibitor), and thrombosis (antithrombin). In addition, the accumulation of polymers within cells occasionally leads to gain-of-function toxicity, resulting in liver disease (α1AT) and dementia (neuroserpin). It has long been acknowledged that the design of treatments for these disorders requires insight into the conformation of the polymerigenic folding intermediate (M*) and into the structural basis of the polymeric linkage. Over the last 30 years, we and others have solved a multitude of crystal structures of serpins in various conformations, illustrating the flexibility of the fold and the promiscuity with which peptides and protein loops can insert into β-sheet A. The structure of a cleaved serpin polymer was even solved by two groups independently. However, the structure of an intact serpin polymer has eluded the field until recently, when we solved the structure of a self-terminating antithrombin dimer (19). The structure surprisingly showed a domain swap including the entirely of both strands 4 and 5A and immediately explained why partial unfolding is necessary to obtain antithrombin polymers in vitro (i.e. to release strand 5A). Importantly, this structure provided a plausible alternative to the loop-sheet hypothesis and stimulated renewed interest into the mechanism of serpin polymerization. This study was designed to determine whether the P8–P6 region of the RCL provides the majority of the polymeric contact or if it instead merely constitutes a small part of an extensive domain swap.
We made and characterized two α1AT variants that were designed to disfavor incorporation of the critical P8–P6 region into β-sheet A. The P8–P6 Asp mutations had the desired effect of slowing RCL incorporation after cleavage at P1, as indicated by the high stoichiometry of inhibition, but they did not prevent full loop insertion or significantly destabilize the cleaved form relative to the control. Further mutations of P10 Pro and P9 Asp did significantly destabilize the cleaved form, with the Tm decreasing from >95 °C to 81 °C, yet the variant was still capable of full RCL incorporation as determined by the crystal structure. These variants were thus well suited to test the loop-sheet hypothesis because they did not prevent insertion of the P8–P6 region if part of a large domain swap (i.e. at least the whole RCL from P15 to P3) but would still be expected to slow or block insertion of the P8–P3 region in isolation. This is indeed what we found whether by adding a P14–P9 peptide to induce polymerization or by specific cleavage at the P11–P10 bond. The latter study is compelling evidence that the mutations prevent the intermolecular insertion of the isolated P10–P3 region if the P8–P6 residues are mutated from hydrophobic to acidic amino acids. It is, of course, possible that the mutations just slow polymerization by this method and that the time of incubation was insufficiently long or that the concentration was insufficiently high to observe polymers. This was tested by increasing the length of incubation and the protein concentration, yet polymers were still not observed by native PAGE (data not shown). In contrast, when normal in vitro methods were used (i.e. denaturant or heat), the apparent rates of polymerization for the variants were similar to those of the control, despite the increase in thermal stability for the native variants (∼5 °C increase in Tm). We can thus conclude that the P8–P6 Asp variant is incapable of forming polymers by the two established models of the loop-sheet mechanism yet polymerizes readily under normal in vitro conditions. Therefore, α1AT does not polymerize via the loop-sheet mechanism in vitro.
Although it has long been assumed that polymers formed in vitro are the same as those formed in vivo, it is unclear whether the family of partially unfolded states obtained using heat or GdnHCl resembles the polymerigenic folding intermediate. We therefore made the P8–P6 Asp mutations on the polymer-inducing Z and Siiyama α1AT backgrounds and tested polymer formation in cells. Similar to what was observed in vitro, we found that the P8–P6 Asp mutations had minimal effect on polymerization in vivo. This suggests that the loop-sheet mechanism is not in operation for polymerigenic mutants of α1AT in cells.
The P8–P6 Asp mutations were designed to test the loop-sheet hypothesis, in particular, that the intermolecular insertion of the P8–P3 region of the RCL is the principal polymer contact. If this were the case, then mutation of hydrophobic residues to Asp would prevent or at least disfavor the loop-sheet mechanism. But what if the loop-sheet mechanism actually utilizes more of the RCL, say P14–P3. This would potentially explain why the mutations prevented peptide- and cleavage-induced polymerization but not normal in vitro or in vivo polymerization. We recently undertook a modeling study to ascertain the degree of possible RCL incorporation in trans and found that the maximal extent physically possible is P12–P3 (18). However, as a necessary consequence of increased RCL incorporation, the polymer will compress and no longer resemble beads on a string but rather a rigid rod. In addition, our further mutations of P10 to Pro and P9 to Asp had no effect on in vitro polymerization, contrary to what would be expected if the insertion from P12–P9 could somehow overcome the defect caused by the P8–P6 Asp mutations. We thus conclude that partial intermolecular RCL insertion is not the polymeric linkage for α1AT.
This leaves two other possibilities: either the RCL is part of an extensive domain swap, as seen in our crystal structure of the antithrombin dimer (19), or the RCL is not involved. The evidence that the RCL must be involved is strong. The only known mechanism of generating a hyperstable serpin is to expand sheet A to the six-stranded form, and it has been demonstrated that the polymeric linkage is hyperstable, implying involvement of the RCL. Studies have also shown that the RCL is protected from proteolysis in serpin polymers (32).
In summary, we have demonstrated that RCL mutations that slow or disfavor incorporation into β-sheet A abrogate loop-sheet polymerization of α1AT by the two commonly used models (peptides and proteolysis) but have little or no effect on polymerization in vitro by heat or GdnHCl or in vivo when coupled to polymer-inducing mutations. We conclude that the RCL, in particular the P10–P6 region, constitutes only a small part of an extensive domain swap that likely includes the preceding strand 5 from β-sheet A. In light of these findings and the recent crystal structure of the domain-swapped antithrombin dimer, the loop-sheet hypothesis should be reconsidered.
Supplementary Material
This work was supported by a Medical Research Council senior nonclinical fellowship (J. A. H.) and a Medical Research Council project grant. This work was carried out with the support of the Diamond Light Source.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (codes 3NDD and 3NDF) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- α1AT
- α1-antitrypsin
- RCL
- reactive center loop
- GdnHCl
- guanidine HCl
- AEBSF
- 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., Pemberton P. A., Remold-O'Donnell E., Salvesen G. S., Travis J., Whisstock J. C. (2001) J. Biol. Chem. 276, 33293–33296 [DOI] [PubMed] [Google Scholar]
- 2.Gettins P. G. (2002) Chem. Rev. 102, 4751–4804 [DOI] [PubMed] [Google Scholar]
- 3.Huntington J. A., Read R. J., Carrell R. W. (2000) Nature 407, 923–926 [DOI] [PubMed] [Google Scholar]
- 4.Huntington J. A. (2006) Trends Biochem. Sci. 31, 427–435 [DOI] [PubMed] [Google Scholar]
- 5.Belorgey D., Hägglöf P., Karlsson-Li S., Lomas D. A. (2007) Prion 1, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomas D. A., Evans D. L., Finch J. T., Carrell R. W. (1992) Nature 357, 605–607 [DOI] [PubMed] [Google Scholar]
- 7.Knaupp A. S., Levina V., Robertson A. L., Pearce M. C., Bottomley S. P.J. Mol. Biol. 396, 375–383 [DOI] [PubMed] [Google Scholar]
- 8.Yu M. H., Lee K. N., Kim J. (1995) Nat. Struct. Biol. 2, 363–367 [DOI] [PubMed] [Google Scholar]
- 9.Lomas D. A., Finch J. T., Seyama K., Nukiwa T., Carrell R. W. (1993) J. Biol. Chem. 268, 15333–15335 [PubMed] [Google Scholar]
- 10.Dafforn T. R., Mahadeva R., Elliott P. R., Sivasothy P., Lomas D. A. (1999) J. Biol. Chem. 274, 9548–9555 [DOI] [PubMed] [Google Scholar]
- 11.Loebermann H., Tokuoka R., Deisenhofer J., Huber R. (1984) J. Mol. Biol. 177, 531–557 [PubMed] [Google Scholar]
- 12.Schechter I., Berger A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 13.Schulze A. J., Baumann U., Knof S., Jaeger E., Huber R., Laurell C. B. (1990) Eur. J. Biochem. 194, 51–56 [DOI] [PubMed] [Google Scholar]
- 14.Chang W. S., Whisstock J., Hopkins P. C., Lesk A. M., Carrell R. W., Wardell M. R. (1997) Protein Sci. 6, 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mast A. E., Enghild J. J., Salvesen G. (1992) Biochemistry 31, 2720–2728 [DOI] [PubMed] [Google Scholar]
- 16.Gooptu B., Lomas D. A. (2009) Annu. Rev. Biochem. 78, 147–176 [DOI] [PubMed] [Google Scholar]
- 17.Miranda E., Pèrez J., Ekeowa U. I., Hadzic N., Kalsheker N., Gooptu B., Portmann B., Belorgey D., Hill M., Chambers S., Teckman J., Alexander G. J., Marciniak S. J., Lomas D. A. (2010) Hepatology, in press [DOI] [PubMed] [Google Scholar]
- 18.Huntington J. A., Whisstock J. (2010) Biol. Chem., in press [DOI] [PubMed] [Google Scholar]
- 19.Yamasaki M., Li W., Johnson D. J., Huntington J. A. (2008) Nature 455, 1255–1258 [DOI] [PubMed] [Google Scholar]
- 20.Zhou A., Carrell R. W., Huntington J. A. (2001) J. Biol. Chem. 276, 27541–27547 [DOI] [PubMed] [Google Scholar]
- 21.Leslie A. G. W. (1992) Joint CCP4 and EACMB Newsletter on Protein Crystallography, No. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 22.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 23.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.McRee D. E. (1992) J. Mol. Graphics 10, 44–46 [Google Scholar]
- 26.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 28.Zhou A., Stein P. E., Huntington J. A., Sivasothy P., Lomas D. A., Carrell R. W. (2004) J. Mol. Biol. 342, 931–941 [DOI] [PubMed] [Google Scholar]
- 29.Xue Y., Björquist P., Inghardt T., Linschoten M., Musil D., Sjölin L., Deinum J. (1998) Structure 6, 627–636 [DOI] [PubMed] [Google Scholar]
- 30.Huntington J. A., Pannu N. S., Hazes B., Read R. J., Lomas D. A., Carrell R. W. (1999) J. Mol. Biol. 293, 449–455 [DOI] [PubMed] [Google Scholar]
- 31.Dunstone M. A., Dai W., Whisstock J. C., Rossjohn J., Pike R. N., Feil S. C., Le Bonniec B. F., Parker M. W., Bottomley S. P. (2000) Protein Sci. 9, 417–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedersen K. E., Einholm A. P., Christensen A., Schack L., Wind T., Kenney J. M., Andreasen P. A. (2003) Biochem. J. 372, 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.