Abstract
Synaptotagmins (Syt) are a large family of proteins that regulate membrane traffic in neurons and other cell types. One isoform that has received considerable attention is SYT4, with apparently contradictory reports concerning the function of this isoform in fruit flies and mice. SYT4 was reported to function as a negative regulator of neurotrophin secretion in mouse neurons and as a positive regulator of secretion of a yet to be identified growth factor from muscle cells in flies. Here, we have directly compared the biochemical and functional properties of rat and fly SYT4. We report that rat SYT4 inhibited SNARE-catalyzed membrane fusion in both the absence and presence of Ca2+. In marked contrast, fly SYT4 stimulated SNARE-mediated membrane fusion in response to Ca2+. Analysis of chimeric molecules, isolated C2 domains, and point mutants revealed that the C2B domain of the fly protein senses Ca2+ and is sufficient to stimulate fusion. Rat SYT4 was able to stimulate fusion in response to Ca2+ when the conserved Asp-to-Ser Ca2+ ligand substitution in its C2A domain was reversed. In summary, rat SYT4 serves as an inhibitory isoform, whereas fly SYT4 is a bona fide Ca2+ sensor capable of coupling Ca2+ to membrane fusion.
Keywords: Calcium, Exocytosis, Fusion Protein, Membrane Fusion, Membrane Reconstitution, SNARE, Synaptotagmin 4, Synaptotagmin IV, Syt IV
Introduction
Neurons harbor two distinct major classes of secretory vesicles: small synaptic vesicles (SV),2 which mediate rapid synaptic transmission, and large dense core vesicles (LDCV), which secrete neuropeptide growth factors as well as other hormones (1). Significant progress has been made in the identification and function of positive and negative regulators of SV exocytosis (2), but at present less is known concerning the regulation of LDCV exocytosis.
Early studies indicate that a member of the synaptotagmin family of proteins, SYT4 (3), is a component of SVs (4), whereas other reports rule out the presence of SYT4 on SVs (5, 6). More recent studies, carried out using fruit flies and mouse neurons, suggest that SYT4 might play a direct role in the regulation of secretion from LDCVs in both organisms (7–9). More specifically, SYT4 was shown to be expressed in muscle cells at the Drosophila neuromuscular junction (7). Loss of postsynaptic SYT4 results in defects in presynaptic nerve terminal growth and plasticity, and it is proposed that SYT4 functions as a positive regulator of an unidentified retrograde messenger (7).
A different view arose from similar studies carried out using cultured mouse neurons (8). In this case, SYT4 was shown to co-localize with brain-derived neurotrophic factor (BDNF) in LDCVs that are targeted to both pre- and postsynaptic compartments. Knock-out and overexpression experiments revealed that SYT4 serves as a negative regulator of BDNF secretion at both loci. SYT4 is rapidly up-regulated in response to activity (10–12), and it was proposed that up-regulation of SYT4 serves to down-regulate synaptic function via inhibition of BDNF (8). Indeed, SYT4 serves to limit the magnitude of long-term potentiation via its ability to inhibit the release of BDNF (8).
Collectively, it appears that the positive role played by SYT4 at the fly neuromuscular junction and the negative role of SYT4 in the release of BDNF from mouse neurons are contradictory findings. The focus of the current study is to use direct approaches to resolve this apparent controversy. Before delving further, it will be useful to review a few key findings regarding the Syt family.
All of the Syt isoforms have similar overall structures: a short luminal domain, a single transmembrane domain, and a cytoplasmic domain comprising tandem C2 domains called C2A and C2B (13). In SYT1, each C2 domain binds two to three Ca2+ ions via four to five acidic residues in two flexible loops (14–16); upon binding Ca2+ these loops partially insert into membranes that harbor acidic phospholipids (17, 18). The insertion step drives localized bending of the target membrane to facilitate fusion (19, 20). Syt has also been shown to mediate vesicle aggregation; this could juxtapose SNAREs to facilitate fusion (21, 22). In addition, SYT1 binds to target membrane SNARE proteins in a Ca2+ promoted manner (23, 24); it is thought that the Ca2+-independent component of this interaction serves to clamp or inhibit the fusion complex (25). Then, in response to Ca2+, SYT1 can influence the folding and assembly of SNAREs to facilitate fusion (26).
In contrast to SYT1, SYT4 harbors a Asp-to-Ser substitution of one of the Ca2+ ligands in its C2A domain (3). Interestingly, fly SYT4 harbors the same Asp-to-Ser mutation within its C2A domain, but in contrast to the rat protein, fly SYT4 appears to retain some degree of Ca2+, and Ca2+-dependent effector, binding activity (27, 28). This observation is consistent with the notion that fly SYT4 might serve to facilitate SNARE-mediated membrane fusion reactions, whereas rat SYT4 inhibits these reactions. Here, we directly test this idea by assaying both proteins in a reconstituted membrane fusion system. Our results confirmed that rat SYT4 is a negative regulator of SNARE catalyzed membrane fusion and also directly revealed, for the first time, that fly 4 serves as a positive regulator of fusion. Hence, despite their ∼50% sequence identity and the conserved substitution of a Ca2+ ligand in their C2A domains, mouse and fly SYT4 are not functional orthologs. Further experiments provided insights into the molecular basis for these functional differences.
EXPERIMENTAL PROCEDURES
DNA Constructs
A plasmid for the expression of mouse synaptobrevin 2 in Escherichia coli was provided by J. E. Rothman (Yale University, New Haven, CT) (29); full-length t-SNARE heterodimers were generated as described previously by subcloning cDNA encoding full-length rat SNAP-25B and rat syntaxin 1A into the pRSFDuet-1 vector (Novagen) (25). cDNA encoding fly SYT1, fly SYT4, fly n-syb, fly syntaxin 1A, and fly SNAP-25 were provided by J. T. Littleton (Massachusetts Institute of Technology, Boston). For expression in bacteria, the cytoplasmic domains of fly SYT1-(148–474) and fly SYT4-(134–474) were subcloned into pGEX-4T vectors. Point mutations were generated by QuikChange mutagenesis (Stratagene). Rat chimeras were subcloned into pGEX-4T vectors as follows: rat1A 4B, residues 96–263 of SYT1 and residues 279–425 of SYT 4; rat4A 1B, residues 108–278 of SYT4 and residues 264–421 of SYT1. Full-length fly n-syb was subcloned into the pET-28a vector using the EcoRI and BamHI sites. To generate fly t-SNARE heterodimers, cDNA encoding full-length fly SNAP-25 was subcloned into pRSFDuet-1 (Novagen) using the EcoRI and NotI sites; full-length fly syntaxin was subcloned into a downstream site via BgIII and KpnI sites. Rat and fly SYT4 chimeras were subcloned into pGEX-4T vectors as follows: rat4A fly4B, residues 108–278 of rat SYT4 and residues 322–474 of fly SYT4; fly4A rat4B, residues 134–321 of fly SYT4 and residues 279–425 of rat SYT4. The cytoplasmic domain of SYT11 was subcloned into pGEX-4T vectors.
Protein Expression and Purification
Recombinant proteins were generated as described previously (30).
Liposome Preparation
Proteoliposomes were prepared as described (31). Briefly, lipids were dried under a stream of nitrogen and resuspended in elution buffer (25 mm HEPES, 400 mm KCl, 10% glycerol, 1 mm dithiothreitol, 1% n-octylglucoside) containing SNARE proteins. Mixtures were diluted with dialysis buffer (25 mm HEPES, 100 mm KCl, 10% glycerol, 1 mm dithiothreitol) and centrifuged for 5 h at 41,000 rpm in an Accudenz gradient. Liposomes were collected (1.2 ml) from the 0 and 30% Accudenz interface.
For preparation of protein free liposomes, lipids (15% phosphatidylserine (PS), 30% phosphatidylethanolamine (PE), 55% phosphatidylcholine (PC)) were dried under a stream of nitrogen and resuspended in dialysis buffer. The mixtures were then extruded through polycarbonate membranes (100 nm, GE Healthcare) to form unilamellar liposomes.
In Vitro Fusion Assays
Fusion assays were performed as described (30) but using 10-fold lower amounts of vesicles and proteins. Under these conditions, less Syt is needed to drive fusion at rates comparable with our earlier studies. Briefly, 75-μl fusion reactions were prepared, including 4.5 μl of t-SNARE vesicles or protein-free vesicles, 0.5 μl of 7-nitro-2-1,3-benzoxadiazol-4-yl (NBD)-rhodamine-labeled v-SNARE vesicles and 1 μm Syt. The mixtures were preincubated at 37 °C for 20 min in the presence of 0.2 mm EGTA followed by injection of Ca2+ (1 mm); fusion was monitored for an additional hour. At the end of each run, 20 μl of the detergent n-dodecyl-β-d-maltoside was added to each reaction to yield the maximum fluorescence signals at “infinite” dilution of the fluorescence resonance energy transfer (FRET) donor-acceptor pair. NBD dequenching was monitored using a BioTek Synergy HT plate reader.
Co-sedimentation Assays
Syt proteins (4 μm) were incubated with increasing concentrations of liposomes (15% PS, 55% PC, and 30% PE) for 15 min at room temperature in a final reaction volume of 100 μl. The mixtures were then centrifuged at 70,000 rpm for 1 h. The supernatant of each sample was collected, mixed with 25 μl of 3× SDS loading buffer, and boiled for 5 min. Samples were subjected to SDS-PAGE and stained with Coomassie Blue.
Co-flotation Assays
45 μl of PS-free t-SNARE vesicles were mixed with the indicated Syt proteins (10 μm final concentration) in the presence of either 1 mm Ca2+ or 0.2 mm EGTA in a total reaction volume of 100 μl at room temperature for 30 min. Samples were mixed with 100 μl of 80% Accudenz and transferred to centrifuge tubes (Beckman Instruments). 35, 30, and 0% Accudenz were sequentially added to form step gradients. Samples were centrifuged at 55,000 rpm for 105 min. 40 μl of each sample was collected at the interface between the 30 and 0% Accudenz layers and analyzed by SDS-PAGE and staining with Coomassie Blue.
Immunoprecipitation
Syt proteins (2 μm) were mixed with t-SNARE heterodimers (2 μm) in TBS buffer (20 mm Tris, 150 mm NaCl, pH 7.4) plus 0.5% Triton X-100 in a total volume of 150 μl for 1 h at 4 °C in the presence of 2 mm EGTA or 1 mm Ca2+. t-SNARE heterodimers were immunoprecipitated with 2 μl of an anti-syntaxin antibody (HPC-1) for 2 h followed by the addition of 40 μl of a 50% slurry of protein G-Sepharose Fast Flow beads (GE Healthcare). The mixtures were incubated for 1 h, and beads were collected by centrifugation. Pellets were washed with TBS buffer four times, boiled in sample buffer, subjected to SDS-PAGE, and stained with Coomassie Blue.
RESULTS
Opposite Effects of Rat and Fly SYT4 in Reconstituted Membrane Fusion Reactions
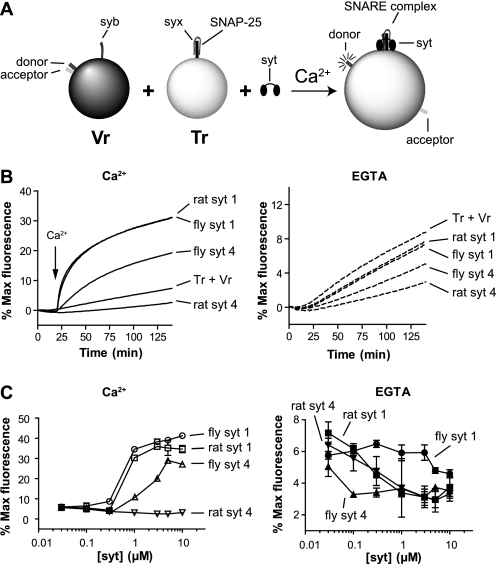
Rat and fly SYT4 exhibit ∼50% sequence identity within their cytoplasmic domains and, as their names indicate, are thought to be encoded by orthologous genes. Compared with SYT1, they both bear an Asp-to-Ser substitution of one of the acidic Ca2+ ligands within the C2A domain. However, recent studies suggest that rat and fly SYT4 have distinct physiological functions (7–9, 32). These differences in function may be because of the distinct biochemical properties of these two proteins. Here, we addressed this question by carrying out direct comparisons of rat and fly SYT4 in a well characterized reconstituted membrane fusion assay (25, 30, 31). SNAREs, from either rat or fly, were reconstituted into PS/PC/PE (15/55/30%) liposomes. v-SNARE vesicles bear a fluorescence resonance energy transfer donor-acceptor pair (NBD-rhodamine) attached to the head group of PE. Fusion of v-SNARE-bearing vesicles with unlabeled t-SNARE vesicles serves to dilute the donor-acceptor pair, resulting in dequenching of the NBD signal, which is monitored using a plate reader (Fig. 1A).
FIGURE 1.
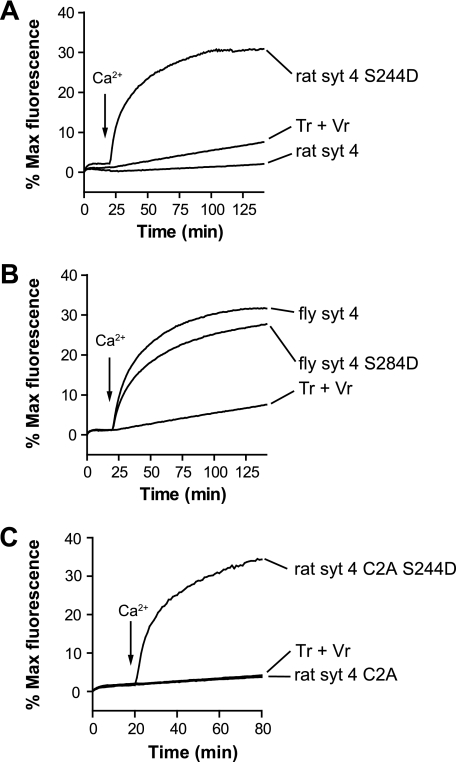
Effect of rat and fly SYT4 on reconstituted SNARE-mediated membrane fusion reactions. A, schematic diagram of the in vitro fusion assay. Tr, t-SNARE vesicle; Vr, v-SNARE vesicle. B, left, the cytoplasmic domain of rat or fly SYT4 (1 μm) was added to SNARE-bearing liposome fusion reactions, and samples were incubated for 20 min prior to the addition of Ca2+. After injection of Ca2+ (1 mm [final], indicated by arrows), fusion was monitored for another 120 min at 37°C. As controls, rat and fly SYT1 were assayed under identical conditions. NBD dequenching signals were normalized to the maximum fluorescence signal, obtained by adding detergent, and plotted as a function of time. Right, experiments were also carried out in the continued presence of 0.2 mm EGTA. C, the final extent of fusion, regulated by SYT1 or SYT4, was plotted against protein concentration in the presence of 1 mm Ca2+ (left panel) or 0.2 mm EGTA (right panel) (n = 3).
Rat and fly SYT4 were preincubated with SNARE-bearing vesicles for 20 min. Ca2+ was then added to yield a final free concentration of 1 mm, and fusion was monitored for another 120 min. Rat and fly SYT4 exhibited marked functional differences in this fusion assay (Fig. 1, B and C). Rat SYT4 inhibited fusion in the absence and presence of Ca2+, whereas fly SYT4 promoted robust fusion in response to Ca2+; fusion was not observed using protein-free vesicles (data not shown). As a positive control we included rat and fly SYT1 in parallel fusion assays. As expected, rat SYT1 gave rise to efficient Ca2+-promoted fusion activity. Interestingly, similar results were obtained using the fly ortholog of SYT1. Because Syt proteins operate, in part, by engaging SNARE proteins (25, 26, 30, 33, 34), these findings further suggested that the determinants that underlie Syt-SNARE interactions are conserved across species. These results also helped to validate the use of vertebrate SNAREs to study invertebrate Syt proteins. However, to ensure that these results were not specific for rat SNAREs, we repeated these experiments with fly SNARE-bearing vesicles, and similar results were obtained. Rat SYT4 inhibited fly SNARE-mediated fusion, whereas fly SYT4 stimulated fly SNARE-mediated fusion (supplemental Fig. S1). Both proteins were able to clamp fusion in EGTA.
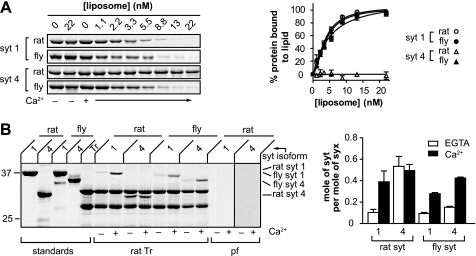
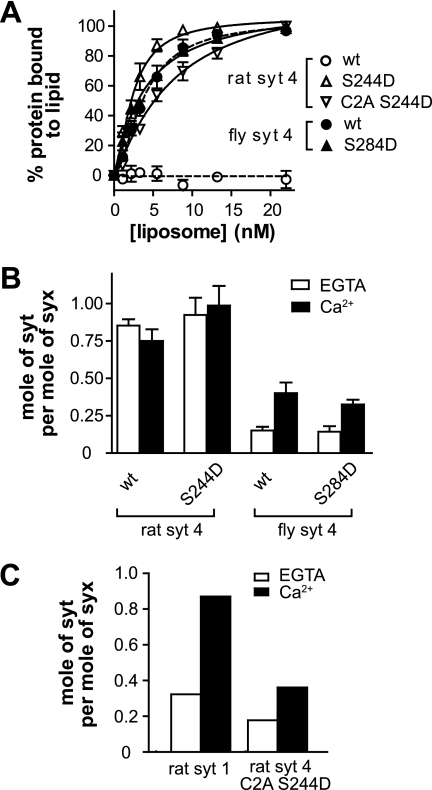
As alluded to above, Syt proteins have been proposed to regulate fusion by interacting with PS and t-SNAREs (35). We therefore compared the PS and t-SNARE binding activities of fly and rat SYT1 and SYT4 to determine whether these effector interactions were indeed correlated with Ca2+-regulated fusion activity. PS binding activity was monitored using a co-sedimentation assay in which Syt was incubated with increasing amounts of PS-bearing liposomes. Liposomes were then pelleted via centrifugation. To avoid the loss of material in the pellet due to washing steps, we monitored the depletion of Syt from the supernatant and used this information to calculate the amount of bound material. Using this approach we found that rat SYT4 failed to bind to PS in either the absence or presence of Ca2+. In contrast, fly SYT4, as well as rat and fly SYT1, bound to PS in a Ca2+-dependent manner (Fig. 2A). These data are consistent with previous reports indicating that fly, but not rat, SYT4 binds to PS in response to Ca2+ (28, 36).
FIGURE 2.
PS and t-SNARE binding activities of rat and fly SYT4. A, left, representative gel of the co-sedimentation assay used to monitor Syt-membrane interactions, in the presence of 0.2 mm EGTA or 1 mm Ca2+. Syt was incubated with liposomes (15% PS, 30% PE, 55% PC) for 20 min. Bound and free proteins were then separated via sedimentation as described under “Experimental Procedures.” The supernatant fraction, which was depleted of Syt protein upon binding liposomes, was subjected to SDS-PAGE and stained with Coomassie Blue. Rat SYT4 failed to bind PS-harboring liposomes; in contrast, fly SYT4, rat SYT1, and fly SYT1 all bound to PS-harboring vesicles in a Ca2+-dependent manner. Right, the amount of bound protein was calculated and plotted against [liposome] (n = 3). B, left, the t-SNARE binding activities of SYT4 and SYT1 were monitored using a co-flotation assay as described under “Experimental Procedures.” Syt proteins were incubated with PS-free t-SNARE vesicles (Tr) (30% PE, 70% PC) in 0.2 mm EGTA and then floated through a density gradient with or without added Ca2+ (1 mm). Syt that was associated with vesicles (via interactions with t-SNARE) was collected from the top layer of the gradient, subjected to SDS-PAGE, and stained with Coomassie Blue. A representative gel is shown. Rat SYT4 exhibited strong t-SNARE binding activity in EGTA; this binding was unaffected by Ca2+. In contrast, rat SYT1, fly SYT1, and fly SYT4 all exhibited weak t-SNARE binding activity in the absence of Ca2+ and much stronger binding in the presence of Ca2+. Neither rat SYT1 nor rat SYT4 bound to protein-free (pf) liposomes that lacked PS. Right, the molar ratio of Syt to syntaxin (syx) in each sample was plotted.
Next, we examined the t-SNARE binding activities of rat and fly SYT4 using a co-flotation assay in which t-SNAREs are reconstituted into liposomes that lack PS. Soluble Syt fragments were incubated with the liposomes and centrifuged through an Accudenz step gradient. Syt that was bound to vesicles via interactions with t-SNAREs co-floated with the proteoliposomes to the top of the 0–30% Accudenz interface. Vesicles were collected from this interface, subjected to SDS-PAGE, and stained with Coomassie Blue. Rat SYT4 strongly bound to t-SNARE heterodimers in the absence Ca2+; binding was not influenced by the addition of Ca2+. In contrast, fly SYT4, as well as rat and fly SYT1, bound to t-SNAREs weakly in the absence of Ca2+; the addition of Ca2+ resulted in a significant increase in t-SNARE binding activity (Fig. 2B). Together with the PS binding data detailed above, we propose that rat SYT4 is unable to stimulate fusion because it fails to interact with either PS or t-SNARE heterodimers in response to Ca2+.
Rat SYT1 and SYT4 Chimeras Failed to Stimulate Fusion
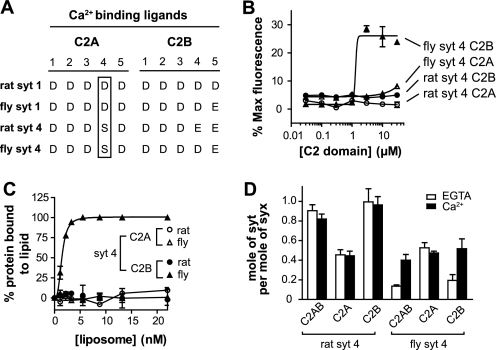
Both rat and fly SYT4 bear the same aspartate to serine substitution in one of the Ca2+ ligands in C2A (Fig. 4A), yet one protein is an active Ca2+ sensor and the other is not. To gain greater insight into this critical difference, we made use of chimeric proteins that harbored tethered C2 domains from different Syt isoforms.
FIGURE 4.
The C2B domain of fly SYT4 directly promotes fusion in response to Ca2+. A, alignment of Ca2+ ligands in rat and fly Syt. The conserved Asp-to-Ser Ca2+ ligand substitution is indicated by an open rectangular box. B, fusion reactions were carried out as described in the legend for Fig. 1B, using isolated fly and rat SYT4 C2A or C2B domains. Normalized fluorescence signals were plotted against [C2 domain]. Fly SYT4 C2B triggered fusion in response to Ca2+; rat SYT4 C2A, C2B and fly SYT4 C2A all failed to stimulate fusion. C, the ability of isolated C2 domains from rat and fly SYT4 to bind PS-bearing liposomes was monitored using a co-sedimentation assay. The amount of bound protein was determined and plotted against [liposome]. Fly SYT4 C2B bound to PS-bearing liposomes in response to Ca2+; the other three C2 domains failed to exhibit significant levels of binding. D, the t-SNARE binding activities of the isolated C2 domains from rat and fly SYT4 were monitored using a co-flotation assay as described in the legend for Fig. 2B. The molar ratio of Syt to syntaxin in each sample was plotted. Both C2 domains of rat SYT4, as well as fly SYT4 C2A, bound to t-SNAREs in a Ca2+-independent manner. In contrast, fly SYT4 C2B bound to t-SNARE in a Ca2+-dependent manner (n = 3). For representative gels of these experiments, please see supplemental Fig. S3.
First, we noted that in the case of rat SYT1, the isolated C2B domain alone is able to stimulate fusion in the reconstituted fusion assay (30), and thus differences in the C2B domain between rat and fly SYT4 could potentially contribute to the observed differences in our fusion assays. However, it is also possible that an inactive C2A domain in rat SYT4 might be able to down-regulate the activity of its adjacent C2B domain (e.g. as shown in supplemental Fig. S2, the D230S mutation in the C2A domain of the cytoplasmic domain of rat SYT1 significantly reduced the ability of adjacent C2B domain to stimulate fusion).
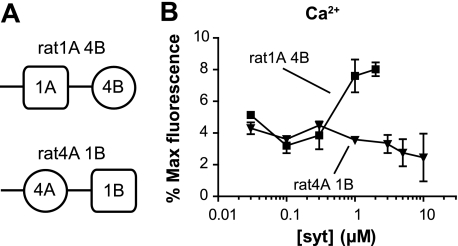
To begin to discern between these possibilities, we built two chimeras between rat SYT1 and rat SYT4: the C2A domain from SYT1 was fused with the C2B domain from rat SYT4 (designated rat1A 4B). This chimera was used to address the question of whether the C2A domain of SYT1 could activate the C2B domain of SYT4 (37). The second chimera, in which the C2A domain from SYT4 was fused with the C2B domain from SYT1 (designated rat4A 1B), was used to determine whether an active C2B domain could still regulate fusion when tethered to a “dead” C2A domain (Fig. 3A).
FIGURE 3.
Rat SYT1 and SYT4 chimeras have modest effects on reconstituted membrane fusion reactions. A, schematic diagram summarizing the structural organization of rat SYT1 and SYT4 chimeras. B, the extent of fusion regulated by two rat chimeric proteins, rat1A 4B and rat4A 1B in the presence of 1 mm Ca2+, was plotted against [Syt]. Neither chimera stimulated significant levels of fusion (n = 3).
We tested these two chimeras in the fusion assay and found that neither was able to stimulate fusion to any significant degree, although there was a minor increase in fusion when using relatively high concentrations (1–2 μm) of rat1A 4B (Fig. 3B). These experiments suggested that both C2 domains from rat SYT4 are defective in terms of stimulating membrane fusion in response to Ca2+.
Functional Comparison of the Isolated C2 Domains from Rat and Fly SYT4
As shown above, rat and fly SYT4 have distinct effects in membrane fusion reactions. To further elucidate the molecular basis for this difference, we purified the isolated C2 domains from each protein and analyzed them in the fusion assay. Interestingly, the isolated C2B from fly SYT4 was able to stimulate fusion in response to Ca2+. In contrast, rat SYT4 C2B, as well as rat and fly SYT4 C2A, failed to stimulate fusion (Fig. 4B). These results confirm that rat SYT4 fails to stimulate membrane fusion because of a lack of activity in both of its C2 domains.
We then tested the PS binding activities of the isolated SYT4 C2 domains using the supernatant depletion liposome co-sedimentation assay (Fig. 4C and supplemental Fig. S3A). Fly SYT4 C2B bound to PS in response to Ca2+. In contrast, rat SYT4 C2B, as well as rat and fly SYT4 C2A, failed to bind to PS in either the presence or absence of Ca2+.
We also tested the t-SNARE binding activities of each C2 domain using the reconstituted t-SNARE liposome co-flotation assay. Analogous to the PS binding activity of each C2 domain described above, fly SYT4 C2B bound to t-SNARE heterodimers in a Ca2+-promoted manner. In contrast, rat SYT4 C2B exhibited robust Ca2+-independent t-SNARE binding activity. The rat and fly SYT4 C2A domains bound to t-SNARE heterodimers weakly in the absence and presence of Ca2+ (Fig. 4D and supplemental Fig. S3B).
In light of these findings, using isolated C2 domains, we built and tested another chimeric protein: the C2A domain of rat SYT4 was fused to the C2B domain of fly SYT4 (designated rat4A fly4B) (supplemental Fig. S4A). We found that this chimera was not able to stimulate fusion when tested at concentrations up to 1 μm (supplemental Fig. S4B). These findings indicate that rat SYT4 C2A is able to “shut off” a functional adjacent C2B domain. These data are consistent with our findings in Fig. 3B demonstrating that rat SYT4 C2A is also able to inhibit the function of an adjacent SYT1 C2B domain. For completeness, we tested an additional chimera: the C2A domain of fly SYT4 was fused to the C2B domain of rat SYT4 (designated fly4A rat4B). This chimera also failed to stimulate fusion, in agreement with the observation that each of the isolated C2 domains used to build the construct was without activity in the reconstituted fusion assay (supplemental Fig. S4).
Reversal of the Ser-244 Mutation Endows Rat SYT4 with the Ability to Function as a Ca2+ Sensor for Fusion
As detailed above, SYT4, in both fly and rat, harbors a Asp-to-Ser substitution of one of the Ca2+ ligands within its C2A domain. Previous studies have examined the functional consequences of this mutation in a neuroendocrine cell line. First, amperometric recordings of LDCV exocytosis in PC12 cells overexpressing wild type rat SYT4 (which harbors Ser-244) revealed an increase in stand-alone foot (SAF) signals as compared with cells overexpressing SYT1. SAF represent events in which fusion pores open and close again without dilating. The S244D mutation in rat SYT4 reduced the duration of SAF to that in cells overexpressing SYT1, but the frequency of SAF remained as high as that in cells overexpressing wild type SYT4 (38). These data suggest that the S244D mutation converts, at least partially, the biological activity of SYT4 so that it is more similar to SYT1. We therefore tested whether the S244D mutation might be able to rescue the ability of rat SYT4 to stimulate membrane fusion in vitro. As shown in Fig. 5A, we found that the S244D mutant form of rat SYT4 was in fact able to stimulate membrane fusion. This gain of function suggests that rat SYT4 evolved to inhibit membrane fusion. For comparison, we generated and tested a fly SYT4 mutant that carries an analogous mutation (fly SYT4 S284D). The mutant fly SYT4 stimulated fusion in response to Ca2+ to levels similar to wild type fly SYT4, and no apparent gain of function was observed (Fig. 5B).
FIGURE 5.
The S244D mutation in rat SYT4 results in robust Ca2+ sensor function during reconstituted membrane fusion. A, fusion assays were carried out as described in the legend for Fig. 1B but with the indicated Syt constructs. In contrast to wild type rat SYT4, the S244D mutant stimulated membrane fusion in the presence of Ca2+. B, an analogous mutation was also analyzed in fly SYT4 (S284D); this mutation was largely without effect. C, the S244D mutation endowed the isolated C2A domain of SYT4 with the ability to stimulate fusion in response to Ca2+. Tr, t-SNARE vesicle; Vr, v-SNARE vesicle.
To further understand how the S244D mutation “activates” rat SYT4, we examined its PS and t-SNARE binding activity, again using the co-sedimentation and co-flotation assays detailed above. These experiments revealed that the point mutation gave rise to Ca2+-regulated binding of the intact cytoplasmic domain of rat SYT4 to both of these effectors (Fig. 6, A and B, and supplemental Fig. S5, A and B).
FIGURE 6.
PS and t-SNARE binding activities of rat and fly SYT4 Ser-to-Asp mutants. A, liposome binding assays were carried out by co-sedimentation and analyzed as described in the legend for Fig. 2A. Rat SYT4 failed to bind. Rat SYT4 S244D, wild type fly SYT4, and fly SYT4 S284D all bound to PS-bearing liposomes in response to Ca2+. B, the molar ratio of Syt to syntaxin, as determined from co-flotation assays (described in the legend for Fig. 2B), was plotted (n = 3). Wild type and S244D mutant rat SYT4 bound to t-SNAREs in a Ca2+-independent manner. In contrast, wild type fly SYT4 and the S284D mutant exhibited Ca2+-promoted t-SNARE binding activity. C, co-immunoprecipitation of Syt with t-SNARE heterodimers composed of full-length SNAP-25B and syntaxin 1A. The isolated C2A domain of rat SYT4 S244D bound to t-SNAREs in a Ca2+-dependent manner. Rat SYT1 was analyzed in parallel and served as a control. For representative gels of these experiments, please see supplemental Fig. S5.
The gain of function brought about by the Ser-to-Asp mutation could be restricted to the C2A domain of rat SYT4. Alternatively, and as noted above, this mutation could act by altering the ability of C2A to influence the adjacent C2B domain (supplemental Fig. S2). To address this question, we analyzed the isolated C2A domain, which harbored this mutation, in the fusion assay and found that it stimulated fusion in response to Ca2+ (Fig. 5C). Hence, the S244D mutation endows the C2A domain of rat SYT4 with the ability to function as a Ca2+ sensor capable of regulating SNARE-catalyzed fusion reactions.
Next, we examined the PS and t-SNARE binding activity of the isolated C2A domain that bore the S244D mutation using co-sedimentation and co-immunoprecipitation assays. Isolated S244D SYT4 C2A bound to PS-bearing liposomes in response to Ca2+ (Fig. 6A and supplemental Fig. S5A), consistent with a previous report (39). Given that the t-SNARE binding activity of the rat SYT4 C2A S244D mutant was too weak to be quantified using co-flotation assays (where low affinity interactions can result in significant levels of dissociation during centrifugation), we used a co-immunoprecipitation approach to monitor binding. Isolated S244D rat SYT4 C2A was incubated with full-length t-SNARE heterodimers in detergent and immunoprecipitated using an anti-syntaxin antibody. Bound material was subjected to SDS-PAGE and the gels stained with Coomassie Blue. Rat SYT4 S244D C2A bound to t-SNARE heterodimers weakly in the absence of Ca2+; binding was significantly increased by the addition of Ca2+. SYT1 served as a control and exhibited robust Ca2+-dependent t-SNARE binding activity (Fig. 6C and supplemental Fig. S5C).
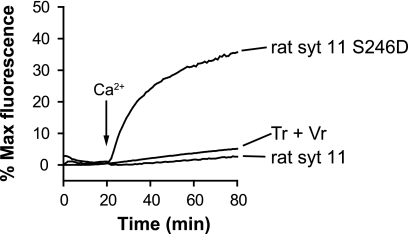
Among all of the Syt isoforms, SYT11 has the greatest degree of homology to rat SYT4, and harbors the Asp-to-Ser substitution of the same Ca2+ ligand as SYT4 (40). Therefore, in the final series of experiments, we compared rat SYT4 and SYT11 in the reconstituted fusion assay. Analogous to SYT4, wild type SYT11 failed stimulate fusion, whereas reversal of the Ca2+ ligand mutation in the C2A domain of SYT11 back to an aspartic acid residue endowed the protein with the ability to stimulate membrane fusion in response to Ca2+ (Fig. 7).
FIGURE 7.
Analysis of wild type and mutant SYT11 in reconstituted membrane fusion assays. Wild type rat SYT11 failed to stimulate membrane fusion. The S246D mutation, which restored a Ca2+ ligand within the C2A domain, endowed the protein with the ability to stimulate fusion in response to Ca2+. Tr, t-SNARE vesicle; Vr, v-SNARE vesicle.
DISCUSSION
SYT4 is an interesting member of the Syt family because its expression is induced by seizures and activity (10). Indeed, SYT4 KO mice exhibit defects in memory and learning tasks, and SYT4 has emerged as a critical regulator of synaptic plasticity in mice (8, 41–43). SYT4 has been studied in detail in fruit flies, where it also affects aspects of synaptic plasticity and neuronal growth (7, 32). However, studies based on mice and flies have resulted in apparently contradictory findings. Namely, studies of mammalian SYT4 indicate that it plays an inhibitory role in secretion. For instance, overexpression of SYT4 reduces the frequency of LDCV fusion events in PC12 cells, shortens the time from fusion pore opening to dilation (44), increases the frequency and duration of kiss-and-run events (38), modulates LDCV exocytosis in peptidergic nerve terminals of the neurohypophysis (9), and inhibits pre- and postsynaptic BDNF release in neurons (8). In contrast, studies using fruit flies as a model system suggest that fly SYT4 is a positive regulator of secretion. SYT4 in muscle cells at the fly neuromuscular junction is required for normal presynaptic growth and for aspects of synaptic plasticity. It therefore has been proposed that fly SYT4 serves to promote the release of a retrograde factor from muscle cells to influence presynaptic structure and function (7). The goal of the current study was to investigate the seemingly disparate functions reported for fly and rat SYT4.
Using a reconstituted system, we directly compared the effects of fly and rat SYT4 on SNARE-mediated membrane fusion reactions. First, we confirmed that rat SYT4 failed to stimulate fusion in response to Ca2+ (45). Strikingly, and in marked contrast to the rat protein, fly SYT4 promoted robust Ca2+ stimulated fusion in vitro (Fig. 1B). These results helped to resolve the disparate findings regarding the function of SYT4; the fly and rat proteins are not functional orthologs of one another. We noted that the cytoplasmic domain of fly SYT4 is homologous to rat SYT11 (49% identity) (40). However, given that rat SYT11 is an inhibitory isoform (45), as in the case of rat SYT4, it too does not appear to serve as a functional ortholog of fly SYT4.
Previous studies have established that rat SYT1 plays a dual role in regulating membrane fusion reactions in vitro and at synapses (25, 30, 35, 46–49). Although rat SYT1 promotes fusion in the presence of Ca2+, this isoform also serves to clamp or inhibit fusion in the absence of Ca2+. This clamping activity appears to be mediated by the Ca2+-independent component of Syt-t-SNARE interactions (25). Although rat SYT4 does not stimulate fusion in response to Ca2+, it strongly clamps fusion in the absence of Ca2+ (Fig. 1B, right panel) (45). Consistent with the strong clamping function of SYT4, its Ca2+-independent t-SNARE binding activity is also very robust (Fig. 2B) (38).
Although one of the acidic Ca2+ ligands within the C2A domain of SYT4 has been replaced over the course of evolution with a serine residue, all of the acidic Ca2+ ligands are conserved within the C2B domain. Thus, the apparently obvious explanation for the functional difference between rat SYT1 and SYT4 is the Ser-to-Asp mutation within C2A. However, our in vitro fusion data, using chimeras between SYT1 and SYT4, indicate that both C2 domains of rat SYT4 exhibit functional deficiencies (Fig. 3). This notion was confirmed by analysis of the isolated C2 domains. In contrast to SYT1, in which the isolated C2B domain is able to regulate fusion (30), neither isolated C2A nor C2B from rat SYT4 was able to stimulate fusion.
When we examined the isolated C2 domains of fly SYT4 in the fusion assay, both fly and rat C2A failed to stimulate fusion (owing to the Ser-to-Asp mutation), but interestingly, the isolated C2B of fly SYT4 was able to stimulate fusion, whereas rat C2B did not. The finding that the C2B domains of rat and fly SYT4 have such divergent properties in reconstituted fusion reactions was highly unexpected; these proteins exhibit 56% sequence identity and their isolated C2B domains have perfectly conserved acidic Ca2+ ligands. Our biochemical analysis revealed that the C2B domain of rat SYT4 does not possess Ca2+ dependent PS or t-SNARE binding activity. In contrast, the C2B domain from fly SYT4 exhibits Ca2+-promoted interactions with both of these effectors (Fig. 4, C and D). Moreover, a gain-of-function mutation, in which a Ca2+ ligand was restored by replacing the serine at position 244 with an aspartate residue in rat SYT4, endowed the protein with the ability to stimulate membrane fusion in response to Ca2+. Further analysis showed that this functional “rescue” was due to the gain of Ca2+-regulated PS and t-SNARE binding activity exhibited by the C2A domain.
Expression levels of rat SYT4 are controlled by neuronal activity; increases in activity result in up-regulation, thus allowing SYT4 to compete with positive regulators of LDCV secretion to down-regulate exocytosis (9, 44). In neurons, this down-regulation results in a reduction in BDNF secretion to dampen neuronal activity and to place an upper limit on long term potentiation (8). Rat SYT4 therefore appears to have evolved as a homeostatic regulator of synaptic activity. In contrast, fly SYT4 appears to serve as a positive regulator of secretion. Whether flies express inhibitory isoforms of Syt remains an open issue. In this light it is notable that 17 isoforms of Syt have been identified in vertebrates, but only seven isoforms are expressed in flies. Future studies, in which each isoform of fly Syt is screened in fusion reactions using a variety of fly SNARE proteins, should reveal the inhibitory isoforms of Syt (45).
In summary, we have directly addressed the abilities of rat and fly SYT4 to regulate SNARE-catalyzed membrane fusion reactions. The major finding here is that fly SYT4 was able to respond to Ca2+ to drive rapid and robust membrane fusion. This contrasts sharply with the inability of rat SYT4 to couple Ca2+ to fusion. However, both rat and fly SYT4 were able to clamp fusion to some extent in the absence of Ca2+, and our biochemical data indicate this is due to relatively efficient binding of these proteins to t-SNAREs to shut them off. Finally, although most attention has been focused on the conserved Asp-to-Ser substitution of a Ca2+ ligand in the C2A domain of fly and rat SYT4, the data reported here indicate that functional differences between these proteins extend to their C2B domains.
Supplementary Material
Acknowledgments
We thank Dr. C. Dean, Dr. J. D. Gaffaney, and Dr. E. Hui for helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant MH 61876 (to E. R. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- SV
- synaptic vesicle
- LDCV
- large dense core vesicle
- SNARE
- soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- BDNF
- brain-derived neurotrophic growth factor
- Syt
- synapatotagmin
- SYT1
- full-length synapatotagmin 1
- C2A
- membrane proximal C2 domain of SYT
- C2B
- membrane distal C2 domain of SYT
- PS
- 1,2-dioleoyl-sn-glycero-3-[phospho-l-serine]
- PC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- NBD
- 7-nitro-2-1,3-benzoxadiazol-4-yl
- SAF
- stand-alone foot.
REFERENCES
- 1.De Camilli P., Jahn R. (1990) Annu. Rev. Physiol. 52, 625–645 [DOI] [PubMed] [Google Scholar]
- 2.Lin R. C., Scheller R. H. (2000) Annu. Rev. Cell Dev. Biol. 16, 19–49 [DOI] [PubMed] [Google Scholar]
- 3.Hilbush B. S., Morgan J. I. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8195–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osborne S. L., Herreros J., Bastiaens P. I., Schiavo G. (1999) J. Biol. Chem. 274, 59–66 [DOI] [PubMed] [Google Scholar]
- 5.Ibata K., Hashikawa T., Tsuboi T., Terakawa S., Liang F., Mizutani A., Fukuda M., Mikoshiba K. (2002) Neurosci. Res. 43, 401–406 [DOI] [PubMed] [Google Scholar]
- 6.Berton F., Cornet V., Iborra C., Garrido J., Dargent B., Fukuda M., Seagar M., Marquèze B. (2000) Eur. J. Neurosci. 12, 1294–1302 [DOI] [PubMed] [Google Scholar]
- 7.Yoshihara M., Adolfsen B., Galle K. T., Littleton J. T. (2005) Science 310, 858–863 [DOI] [PubMed] [Google Scholar]
- 8.Dean C., Liu H., Dunning F. M., Chang P. Y., Jackson M. B., Chapman E. R. (2009) Nat. Neurosci. 12, 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z., Bhalla A., Dean C., Chapman E. R., Jackson M. B. (2009) Nat. Neurosci. 12, 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vician L., Lim I. K., Ferguson G., Tocco G., Baudry M., Herschman H. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2164–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babity J. M., Armstrong J. N., Plumier J. C., Currie R. W., Robertson H. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2638–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson G. D., Vician L., Herschman H. R. (2001) Mol. Neurobiol. 23, 173–185 [DOI] [PubMed] [Google Scholar]
- 13.Perin M. S., Johnston P. A., Ozcelik T., Jahn R., Francke U., Südhof T. C. (1991) J. Biol. Chem. 266, 615–622 [PubMed] [Google Scholar]
- 14.Sutton R. B., Davletov B. A., Berghuis A. M., Südhof T. C., Sprang S. R. (1995) Cell 80, 929–938 [DOI] [PubMed] [Google Scholar]
- 15.Shao X., Fernandez I., Südhof T. C., Rizo J. (1998) Biochemistry 37, 16106–16115 [DOI] [PubMed] [Google Scholar]
- 16.Ubach J., Zhang X., Shao X., Sudhof T. C., Rizo J. (1998) EMBO J. 17, 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman E. R., Davis A. F. (1998) J. Biol. Chem. 273, 13995–14001 [DOI] [PubMed] [Google Scholar]
- 18.Bai J., Tucker W. C., Chapman E. R. (2004) Nat. Struct. Mol. Biol. 11, 36–44 [DOI] [PubMed] [Google Scholar]
- 19.Hui E., Johnson C. P., Yao J., Dunning F. M., Chapman E. R. (2009) Cell 138, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens S., Kozlov M. M., McMahon H. T. (2007) Science 316, 1205–1208 [DOI] [PubMed] [Google Scholar]
- 21.Popoli M., Mengano A. (1988) Neurochem. Res. 13, 63–67 [DOI] [PubMed] [Google Scholar]
- 22.Popoli M., Paternò R. (1992) Neuroreport 3, 177–180 [DOI] [PubMed] [Google Scholar]
- 23.Chapman E. R., Hanson P. I., An S., Jahn R. (1995) J. Biol. Chem. 270, 23667–23671 [DOI] [PubMed] [Google Scholar]
- 24.Schiavo G., Stenbeck G., Rothman J. E., Söllner T. H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chicka M. C., Hui E., Liu H., Chapman E. R. (2008) Nat. Struct. Mol. Biol. 15, 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhalla A., Chicka M. C., Tucker W. C., Chapman E. R. (2006) Nat. Struct. Mol. Biol. 13, 323–330 [DOI] [PubMed] [Google Scholar]
- 27.Littleton J. T., Serano T. L., Rubin G. M., Ganetzky B., Chapman E. R. (1999) Nature 400, 757–760 [DOI] [PubMed] [Google Scholar]
- 28.Dai H., Shin O. H., Machius M., Tomchick D. R., Südhof T. C., Rizo J. (2004) Nat. Struct. Mol. Biol. 11, 844–849 [DOI] [PubMed] [Google Scholar]
- 29.Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., Rothman J. E. (1998) Cell 92, 759–772 [DOI] [PubMed] [Google Scholar]
- 30.Gaffaney J. D., Dunning F. M., Wang Z., Hui E., Chapman E. R. (2008) J. Biol. Chem. 283, 31763–31775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucker W. C., Weber T., Chapman E. R. (2004) Science 304, 435–438 [DOI] [PubMed] [Google Scholar]
- 32.Barber C. F., Jorquera R. A., Melom J. E., Littleton J. T. (2009) J. Cell Biol. 187, 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Kim-Miller M. J., Fukuda M., Kowalchyk J. A., Martin T. F. (2002) Neuron 34, 599–611 [DOI] [PubMed] [Google Scholar]
- 34.Bai J., Wang C. T., Richards D. A., Jackson M. B., Chapman E. R. (2004) Neuron 41, 929–942 [DOI] [PubMed] [Google Scholar]
- 35.Chapman E. R. (2008) Annu. Rev. Biochem. 77, 615–641 [DOI] [PubMed] [Google Scholar]
- 36.Chapman E. R., Desai R. C., Davis A. F., Tornehl C. K. (1998) J. Biol. Chem. 273, 32966–32972 [DOI] [PubMed] [Google Scholar]
- 37.Bai J., Wang P., Chapman E. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1665–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C. T., Lu J. C., Bai J., Chang P. Y., Martin T. F., Chapman E. R., Jackson M. B. (2003) Nature 424, 943–947 [DOI] [PubMed] [Google Scholar]
- 39.von Poser C., Ichtchenko K., Shao X., Rizo J., Südhof T. C. (1997) J. Biol. Chem. 272, 14314–14319 [DOI] [PubMed] [Google Scholar]
- 40.Craxton M. (2004) BMC Genomics 5, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferguson G. D., Anagnostaras S. G., Silva A. J., Herschman H. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5598–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson G. D., Wang H., Herschman H. R., Storm D. R. (2004) Hippocampus 14, 964–974 [DOI] [PubMed] [Google Scholar]
- 43.Ferguson G. D., Herschman H. R., Storm D. R. (2004) Neuropharmacology 47, 604–611 [DOI] [PubMed] [Google Scholar]
- 44.Wang C. T., Grishanin R., Earles C. A., Chang P. Y., Martin T. F., Chapman E. R., Jackson M. B. (2001) Science 294, 1111–1115 [DOI] [PubMed] [Google Scholar]
- 45.Bhalla A., Chicka M. C., Chapman E. R. (2008) J. Biol. Chem. 283, 21799–21807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geppert M., Goda Y., Hammer R. E., Li C., Rosahl T. W., Stevens C. F., Südhof T. C. (1994) Cell 79, 717–727 [DOI] [PubMed] [Google Scholar]
- 47.Littleton J. T., Stern M., Perin M., Bellen H. J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10888–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiAntonio A., Schwarz T. L. (1994) Neuron 12, 909–920 [DOI] [PubMed] [Google Scholar]
- 49.Mackler J. M., Drummond J. A., Loewen C. A., Robinson I. M., Reist N. E. (2002) Nature 418, 340–344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.