Abstract
Cell division and cell wall synthesis are closely linked complex phenomena and play a crucial role in the maintenance and regulation of bacterial virulence. Eukaryotic-type Ser/Thr kinases reported in prokaryotes, including that in group A Streptococcus (GAS) (Streptococcus pyogenes Ser/Thr kinase (SP-STK)), regulate cell division, growth, and virulence. The mechanism of this regulation is, however, unknown. In this study, we demonstrated that SP-STK-controlled cell division is mediated under the positive regulation of secretory protein that possesses a cysteine and histidine-dependent aminohydrolases/peptidases (CHAP) domain with functionally active cell wall hydrolase activity (henceforth named as CdhA (CHAP-domain-containing and chain-forming cell wall hydrolase). Deletion of the CdhA-encoding gene resulted in severe cell division and growth defects in GAS mutants. The mutant expressing the truncated CdhA (devoid of the CHAP domain), although displayed no such defects, it became attenuated for virulence in mice and highly susceptible to cell wall-acting antibiotics, as observed for the mutant lacking CdhA. When CdhA was overexpressed in the wild-type GAS as well as in heterologous strains, Escherichia coli and Staphylococcus aureus, we observed a distinct increase in bacterial chain length. Our data reveal that CdhA is a multifunctional protein with a major function of the N-terminal region as a cell division plane-recognizing domain and that of the C-terminal CHAP domain as a virulence-regulating domain. CdhA is thus an important therapeutic target.
Keywords: Bacterial Protein Kinases, Bacterial Signal Transduction, Cell Division, Cell Surface Protein, Gene Knockout, Multifunctional Enzymes, Phagocytosis, Prokaryotic Protein Kinases, Serine Threonine Protein Kinase, CHAP Domain
Introduction
Group A Streptococcus (GAS3; Streptococcus pyogenes), a human pathogen, is known to cause the broadest range of diseases of any known pathogen (1). These diseases range from mild pharyngitis and impetigo to severe, debilitating, and often fatal toxic shock syndrome and necrotizing fasciitis (2–4). Morbidity and mortality caused by GAS-associated autoimmune sequelae, rheumatic heart disease (5, 6), and glomerulonephritis (1, 7), especially in underdeveloped and developing countries (8), continue to challenge us clinically and scientifically (9, 10). GAS expresses an array of surface-bound and secreted proteins, which either alone or in concert play important roles in bacterial adhesion, invasion, and further proliferation in a variety of constantly fluctuating host environments (11). The expression of these proteins is also bacterial growth-dependent and controlled by the two-component regulatory systems composed of an environmental sensor, histidine kinase, and a response regulator, which serves as a transcription factor (12). In other words, two-component regulatory systems play an important role in regulating genes required for successful colonization and infection by GAS. We, previously identified a novel one-component signaling system that is structurally very similar to eukaryotic-type serine threonine kinase (13). GAS mutants lacking eukaryotic-type serine threonine kinase (SP-STK) are defective in cell division and growth but display increased production of capsule and hemolysin (13), indicating that SP-STK, which cotranscribes with serine threonine phosphatase, positively regulates bacterial growth and cell division while negatively regulating bacterial capsule and hemolysin production (13), which is reminiscent of the CovR-mediated regulation (14).
The phenotypic characteristics of SP-STK mutant mimic those reported for Gram-positive pathogen mutants that lack Streptococcus pneumoniae/Streptococcus agalactiae orthologs (15–17). In GAS, PscB ortholog is SPy0019, a secreted protein of unknown function. The purified SPy0019 has been previously reported to possess a human immunoglobulin binding ability and hence was termed as SibA (immunoglobulin-binding protein of group A Streptococcus) (18). Besides pneumococcal and group B streptococcal PcsB (15–17), SPy0019 is structurally similar to Streptococcal mutans GbpB/SagA (19), Streptococcus thermophilus Cse (20), Enterococcus faecalis/faecium (SalB/SagA) (21, 22), and Listeria monocytogenes p45 protein (23). All of these proteins are involved in some way in the regulation of cell division. However, the exact function of these proteins in bacterial cell division and in particular SPy0019 in GAS cell division and pathogenesis is not known.
Because GAS mutants lacking SP-STK display defective cell division (13), we hypothesized that SP-STK regulates GAS cell division by regulating the expression of SPy0019. Further, protein domain analysis of SPy0019 and its orthologs revealed the presence of a cysteine and histidine-dependent aminohydrolases/peptidases (CHAP) domain at its C-terminal end (24). The ubiquitous presence of this domain and its possible cell wall hydrolase activity suggest that these structurally related proteins may have a specific role in cell wall synthesis. The latter is closely linked with cell division. However, the role of the CHAP domain in cell division, cell wall synthesis, or bacterial pathogenesis has not been proven conclusively, partly because of the fact that these proteins are found to be essential for bacterial survival. Available studies are therefore limited to conditional mutants or mutants with depleted protein expression (15, 16). In this report, we characterized the structure and functions of SPy0019, phenotype of domain-specific GAS mutants, and their possible roles in cell division and pathogenesis of GAS. In this study, we have further demonstrated the role of this protein in bacterial chain formation. Cumulatively, our findings indicated that SPy0019, by serving as a GAS cell division plane recognition protein, plays a critical role in the regulation of early events in cell division process, which in turn help regulate cell wall synthesis and ultimately virulence. In particular, we show that the N-terminal portion of SPy0019 plays a significant role in cell division and bacterial growth, whereas the CHAP domain plays a significant role in the regulation of GAS virulence but not in cell division or cell wall synthesis. We therefore preferred to term SPy0019 henceforth as CdhA (CHAP domain-containing and chain-forming cell wall hydrolase from group A Streptococcus).
EXPERIMENTAL PROCEDURES
Bacterial Strain, Growth Conditions, and Cell Culture
The wild-type GAS strain M1-SF370 (ATCC 700294) (25) and the derived mutants were grown at 37 °C in Todd-Hewitt broth (Difco) supplemented with 0.5% (w/v) yeast extract (THY) or on proteose peptone blood agar plates with or without spectinomycin (500 μg/ml) (13). Staphylococcus aureus RN4220 was grown in trypticase soy broth or agar as described previously (26). Escherichia coli strains XL1-Blue and BL21 (DE3 pLysS), were grown in Luria-Bertani (LB) broth or agar at 37 °C. The N-terminal His-tagged expression vector pET14B (Novagen) was used to make recombinant His6-tagged proteins. Vector pFW5 containing the spectinomycin gene (aad9) was used to transform M1-SF370 as described before (27, 13, 28). Vector pDC123 (29) and pCN40tet (26) were used for the complementation experiments within M1-SF370 and S. aureus RN4220, respectively. Culture plates and media supplemented with ampicillin (100 μg/ml), spectinomycin (500 μg/ml), chloramphenicol (10 μg/ml), and tetracycline (12 μg/ml) were used for the selection of mutants or for construction of complemented mutant strains. Detroit 562 human pharyngeal cell lines (ATCC CCL138) were cultured and maintained in MEM with 10% FBS at 37 °C in a humidified CO2 incubator as described previously (13).
Production of His-tagged Recombinant CdhA Protein and Affinity-purified Anti-CdhA Rabbit Polyclonal Antibodies
Recombinant CdhA and corresponding rabbit polyclonal antibodies were made essentially as described previously (13). Briefly, using M1-SF370 genomic DNA as template, the cdhA gene (encoding 1125 bp excluding the first 72 bp, which encodes for a signal sequence) was amplified by PCR using gene-specific primers (primer 1 (GBSPy19-F) and primer 2 (GBSPy19-R)), sequentially digested with XhoI and BamHI, and cloned into pET14B vector digested with the same enzymes to obtain pETCdhA. E. coli XL1-Blue cells were transformed with pETCdhA, and the recombinant His-tagged CdhA (rCdhA) protein was expressed in E. coli BL-21 (DE3-pLysS) strain after IPTG induction and purified by Ni+2-NTA affinity column chromatography (Qiagen) under denaturing conditions using 8 m urea as per the manufacturer's instructions. The purified protein was then renatured by dialyzing against 50 mm ammonium bicarbonate, pH 8.6, concentrated, and redialyzed against 50 mm Tris/HCl, pH 8.0. Rabbit polyclonal antibody against rCdhA was custom-raised by Lampire Biological Laboratories (Pipersville, PA) and was purified by sequential Protein G and CdhA-DADPA affinity column chromatography as described previously (13). Fab-specific anti-CdhA antibody fragment after pepsin digestion was obtained as described previously (28).
Construction of cdhA Deletion Mutants
From the wild-type M1-SF370 strain, two CdhA-specific isogenic mutant strains were created. To create a mutant lacking the entire CdhA (M1ΔCdhA), an upstream 930-bp PCR product (including the first 61 nucleotides of SPy0019) obtained with primer 3 (GB19Bam-UpNt) and primer 4 (GB19Hin-UpCt) and a downstream 774-bp PCR product (including the last 267 nucleotides of SPy0019) obtained with primer 5 (GB19NcoDnNt) and primer 6 (GB19PstDnCt) were digested with BamHI/HindIII and NcoI/PstI for upstream and downstream fragments, respectively (for all primer sequence, see supplemental Table S1). They were inserted serially into the MCS-I and MCS-II of pFW5 (27) digested with the same enzymes to obtain plasmid pFW5ΔcdhA. The resultant plasmid was then used to transform the wild-type GAS strain M1-SF370 by electroporation to obtain M1ΔcdhA mutant as a result of a double crossover homologous recombination event, which was confirmed by PCR and DNA sequencing using flanking primers of the genomic DNA (primer 7 (upNT-Pr3) and primer 8 (dnCT-Pr6)) and aad9-specific primers (primers 9 and 10) in a primer pair combinations of 7/10 and 9/8.
To elucidate the function of the C-terminal domain of CdhA, a mutant (M1CdhAΔC55) expressing a truncated form of CdhA lacking the last 55 amino acids (the catalytic portion of the CHAP domain (24)) was constructed (supplemental Table S2). An internal fragment of 1,075 bp in size (from nucleotide 157 to 1231 of cdhA) generated by PCR using primer 11 (GB19C55-F) and primer 12 (GB19C55-R) was cloned into BamHI/HindIII sites of MCS-I of pFW5 (supplemental Tables S1 and S2). The resultant plasmid pFW5cdhAΔC55 was then used to transform the wild-type GAS strain, M1-SF370, by electroporation yielding M1CdhAΔC55 mutant (supplemental Table S2). Integrity of the mutants was verified by colony PCR employing primer pairs 7/10 and 9/8 or 9/2 as described above and further confirmed by DNA sequencing.
Construction of Plasmid for Complementation of cdhA and Overexpression of CdhA
The wild-type cdhA gene along with its native promoter (including a 100-bp upstream region) was amplified by PCR using primer 13 (GB19PDCHin-F) and primer 14 (GB19PDCKpn-R) and cloned into pDC123 (29) between HindIII and KpnI sites to obtain pDC123-cdhA. The insert was ligated in the plasmid in the opposite orientation of the cat gene to allow the expression of cdhA by its native promoter. To understand the role of CdhA in SP-STK-mediated regulation of cell division, the latter was used to transcomplement M1ΔSTK (13) as well as M1ΔCdhA to obtain M1ΔSTK::cdhA and M1ΔCdhA::cdhA, respectively, as described before (13). To understand the innate function of CdhA, the wild-type strain M1-SF370 (M1-WT) was similarly transformed with pDC123-cdhA to obtain M1-WT::cdhA. The effect of the overexpression of CdhA on cell division in other species of Gram-positive and Gram-negative bacteria was studied by complementing cdhA in S. aureus RN4220 and E. coli BL21, respectively. For S. aureus, the wild-type cdhA gene along with a 100-bp upstream region containing its native promoter was cloned into pCN40tet between XmaI and KpnI sites as described recently (26) (see supplemental Table S1). The effect of overexpression of CdhA on cell division in E. coli BL21 cells under IPTG induction was measured as described above.
Microscopy
Growth curve patterns of the wild type, mutants, and complemented GAS strains were monitored for 16 h in THY broth. At the late log phase of growth, the GAS strains were harvested, washed, and subjected to light (Gram stain, phase-contrast) and fluorescence microscopy followed by scanning and transmission electron microscopy. For fluorescence microscopy, GAS strains were adsorbed onto the poly-l-lysine-coated glass slides for 30 min, washed, and stained with CdhA-specific IgGFab (1:50 dilution) followed by FITC-labeled goat anti-rabbit IgG (Fab′-specific) (Bio-Rad; 1:50). Stained bacteria were kept in Slow FadeTM equilibrium buffer and stained with DAPI (1 μg/ml) for 5 min and were visualized by fluorescence microscopy (Nikon Eclipse E600). For electron microscopy, the wild-type, mutant, and complemented streptococcal strains were grown until late log phase in THY broth, washed once with phosphate-buffered saline, and fixed in 0.1 m cacodylate buffer containing 2.5% glutaraldehyde and 4% paraformaldehyde for 12–16 h at 4 °C. Samples were then processed for transmission and scanning electron microscopy at the Ohio State University Campus Microscopy and Imaging Facility equipped with a NOVA nanoSEM 400 scanning electron microscope with secondary and low vacuum detectors and cryo-capable digital TEM (Technai G2 Spirit, EFI) as described previously (13).
Human IgG-binding Assay
To determine changes in human immunoglobulin binding activity as a result of the deletion of CdhA/SibA/SPy0019, an hIgG-binding assay was carried out essentially as described previously with minor modifications (30). Briefly, wild-type, mutant, and cdhA-complemented GAS strains were suspended in 0.05 m Tris/HCl buffer, pH 8.0, containing 0.5% BSA to an A600 of 1.00. Equal volumes of the bacterial suspension and a serially diluted hIgG solution (Sigma; starting concentration 100 μg) were mixed in a final volume of 200 μl in 96-deep well plates for 1 h followed by same volume of alkaline phosphatase-conjugated monoclonal antibody against anti-hIgG (Sigma; 1:3000) for 1 h. Bacteria were thoroughly washed with the suspension buffer after every step and finally resuspended in 0.05 m Tris/HCl, pH 10.0, containing 5 mm MgCl2. The contents of each well were then transferred to a new 96-well microtiter plate and centrifuged. Indirect measurement of bacteria-bound hIgG was determined spectrophotometrically after adding 200 μl of ready-to-use phosphatase substrate (Sigma) in each well and incubating further for 1 h using a 96-well plate reader (PolarStar Galaxy, BMG). The binding of bacteria with different concentrations of hIgG was carried out in triplicates in three independent experiments. The values obtained for reactions carried out with only conjugate antibody were treated as background values and were subtracted from the test values. The results were statistically evaluated by non-parametric two-way analysis of variance, and p values were determined using GraphPad Prism 4 software.
Streptococcal Peptidoglycan Preparation
Peptidoglycan from the overnight-grown culture of the wild-type M1-SF370 strain was extracted as described previously with some modifications (26, 31). Briefly, the bacterial pellet obtained from the overnight grown culture was chilled and suspended in 4% SDS, boiled for 30 min, and centrifuged at high speed. The pellet of crude cell wall was then broken using FastPrep20 (MP Biomedical) and subjected to sequential treatment of amylase, DNase/RNase followed by trypsin to remove cell wall-associated carbohydrate, nucleic acid, and surface proteins, respectively. Subsequently, by an additional step of boiling, all of these enzymes were inactivated. Other cell wall-associated lipid/protein components were then released and removed by sequential treatment of 8 m LiCl, EDTA, water, and acetone followed by lyophilization. The acetone powder of the cell wall was then treated with hydrofluoric acid followed by alkaline phosphatase to remove lipoteichoic acid and boiling in a water bath for 5 min. Peptidoglycan thus obtained was dried and stored at 4 °C until further use in subsequent zymogram assays.
Zymogram Assay
A cell wall hydrolase zymogram assay was performed using SDS-PAGE and renaturing SDS-PAGE essentially as described previously (32) with the following modifications. S. pyogenes M1-SF370 peptidoglycan (0.5%, w/v) was included in 10% polyacrylamide gels for detection of cell wall hydrolase activity of CdhA in culture supernatants. Culture supernatants (1.6 ml) from each of the late log phase cultures of wild-type, two CdhA-specific mutants, and cdhA-complemented M1ΔCdhA mutant strains were separately collected and precipitated overnight with 80% of ammonium sulfate saturation at 4 °C. The resulting precipitates were centrifuged (4 °C, 10,000 × g, 40 min) and solubilized in a fixed volume of sample loading buffer. Proteins were then resolved on the polyacrylamide gel containing GAS peptidoglycan. After electrophoresis, the gels were washed twice in distilled water at room temperature with gentle shaking and incubated at 37 °C (with shaking) in 40 ml of renaturing buffer (50 mm MES/NaOH, pH 6, 0.2% (w/v) Triton X-100) for 2 days with periodic buffer replacement after every 5–6 h. Gels were stained with 0.1% (w/v) methylene blue in 0.01%(w/v) KOH for 2 h at room temperature with gentle shaking followed by destaining with distilled water as described previously (32). The clear band on a blue gel was perceived as cell wall hydrolase activity. Stained gels with clear bands were scanned, and the images were inverted to visualize the clear bands as dark bands, using Photoshop software (Adobe).
Bacterial Adherence Assays
GAS adherence to Detroit 562 human pharyngeal cells (ATCC CCL138) was determined essentially as described previously (13). Briefly, adherence assays were performed for a period of 3 h using a confluent culture of Detroit 562 cells (grown in 24-well tissue culture plates), which were infected with the wild-type and mutant strains (multiplicity of infection, 50:1 (bacteria/cell)). Before infecting the confluent cell culture, washed bacteria were water-sonicated gently on ice to disrupt the aggregates. The number of colony-forming units in this preparation was determined prior to performing this experiment. Adherent bacteria were removed by repeated washing and were counted on proteose peptone blood agar plates as colony-forming units (cfu).
Phagocytosis/Bactericidal Assays
The ability of GAS to resist phagocytosis and survive in human blood was analyzed in a bactericidal assay as described previously (13). Briefly, 50 μl (∼150–200 cfu) of GAS grown at the late log phase was added to freshly drawn heparinized blood (1 ml) in a sterile glass tube and incubated at 37 °C for 3 h under constant gentle mixing. The number of cfu in the inoculum and those obtained after incubation in human blood were analyzed by the pour plate method using sheep blood agar plate. M1Δemm1 mutant strain (13) lacking the M protein (SPy2018) was used as a positive control. Bacterial growth (multiplication factor) was calculated as the mean number of cfu after 3 h divided by the initial mean number of cfu (t = 0).
An approved Ohio State University institutional review board protocol was obtained to draw blood from healthy non-smoking individuals/volunteers (18–65 years of age). A written informed consent was provided by study participants.
Estimation of Hyaluronic Acid of GAS Capsule
The amount of hyaluronic acid capsule of the wild-type and mutant GAS strains was measured spectrophotometrically (640 nm) using the standard curve generated with known concentrations of hyaluronic acid and Stain-All chromogenic reagent (Sigma; 1-ethyl-2-[3-(1-ethyl-naptho-[1,2-d]thiazolin-2-ylidene)-2-methylpropenyl]-naptho-[1,2-d]thiazolium bromide) in a final volume of 50 μl as described earlier (13).
Antibiotic Sensitivity Assay
Susceptibility of the wild-type strain and the isogenic M1ΔCdhA and M1CdhAΔC55 to various antibiotics was determined using the E-TestR method (AB BIODISK North America Inc., Piscataway, NJ). For this, antibiotic-specific Etest strips containing a gradient of five representative antibiotics with MIC range as indicated (benzylpenicillin (PG) (MIC 0.002–32 μg/ml), cefotaxime (CT) (MIC 0.016–256 μg/ml), erythromycin (MIC 0.016–256 μg/ml), and clindamycin (MIC 0.016–256 μg/ml), ciprofloxacin (MIC 0.002–32 μg/ml)) were used. Inoculum preparation, media, usage of E-test antibiotic strips, incubation time, and interpretation of the results obtained for bactericidal and bacteriostatic antibiotics were according to the manufacturer's instructions and essentially as described (26).
Bacterial Virulence Assay
In vivo virulence of the wild-type and mutant strains was measured in the mouse peritonitis model. A group of 10 mice (BALB/c, 5 weeks old, 20–22 g, Charles River Laboratories) were injected intraperitoneally with 2 × 108 cfu. Morbidity and mortality in the infected animals were observed twice daily for 10 days. The data presented in the mortality/survival curve were statistically evaluated using the log rank test using GraphPad Prism4 software. The dissemination of GAS in liver, spleen, kidney, and lungs was determined by enumerating the cfu on blood agar plates as β-hemolytic colonies. All animal experiments were performed per the Ohio State University institutional animal care and use committee-approved protocol. The infected animals developing the signs and symptoms of extreme distress (e.g. due to increased bacteremia after I/P infection), such as gasping, asyphyxia, apnea, curved body posture with limited movements, or low heart beat, within 24 h after or during the time of observation after infection were euthanized for further study.
SDS-PAGE and Western Blotting
Procedures of SDS-PAGE of whole bacterial extracts and electrotransfer of resolved proteins on PVDF membranes were performed as described (13). Western blots were developed using Protein-G affinity-purified rabbit polyclonal anti-CdhA and anti-M6 (10B6) mouse monoclonal antibody (33), followed by corresponding alkaline phosphatase-labeled goat anti-rabbit/mouse conjugate antibodies to detect the expression level of CdhA and the M protein in culture supernatants/cell walls of GAS obtained at early, middle, and/or late log phases. Specific-reactive bands were visualized using alkaline phosphatase-specific chromogenic substrates (13).
Microarray Analyses and Quantitative Real-time PCR
The complete/whole M1 genome (25) oligonucleotide-based microarray was designed, synthesized, and made by Illumina Inc. Oligonucleotides (50–60-mer) were designated to hybridize 1,769 ORFs in the genome of S. pyogenes type M1 strain SF370 (25). The oligonucleotides were spotted onto poly l-lysine microarray slides by a robotic microarrayer (Omni GRAD 100 microarrayer) of the Center for Applied Genomics at the Public Health Research Institute, University of Medicine and Dentistry of New Jersey (Newark, NJ).
Microarray analyses were performed to determine relative transcript abundance of M1ΔCdhA and M1CdhAΔC55 with M1-SF370. Bacteria were grown in Todd-Hewitt broth until late log phase, harvested by centrifugation, and washed twice with sterile PBS. Total RNA was isolated from these washed streptococcal strains using a Qiagen RNeasy minikit. High purity and integrity of RNA was confirmed by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Synthesis and labeling of cDNA were carried out using 20 μg of total RNA, 1.5 μg of random hexamer, 0.5 mm dNTPs (except that 0.2 mm dTTP was replaced by the same amount of amino-allyl dUTP to incorporate dUTP into first-strand cDNA) in each of the reverse transcriptase reactions (Superscript II reverse transcriptase, Invitrogen).
Purified cDNA preparations from the wild-type, M1ΔCdhA, and M1CdhAΔC55 mutant strains were labeled with either Alexafluor-555 or Alexafluor-647 depending on the experimental design (i.e. dye swap experiment). Differentially labeled probes were then combined and purified. Using three independently isolated RNA preparations from each mutant (biological replicates), a total of 11 experiments for M1ΔCdhA strain (incorporating dye swaps) and seven experiments for M1CdhAΔC55 strain were performed. Signals of the bound reagents on the microarray spots in terms of relative fluorescence values were measured and quantified by a laser scanner (GenePix 4100) at 10 μm/pixel resolution. The resulting images were processed using Gene Pix Pro software (version 4.0, Axon Instruments).
The web application CARMAweb (comprehensive R-based microarray analysis web service) was used for the normalization and analysis of microarray data (34). All raw data (in the form of *.GPR files) were uploaded to the web application in the data directory. Using an appropriate navigation tree, background correction from the foreground signal was applied and within microarray normalization was achieved using the Lowess method. Genes flagged as bad spots by the scanning software were excluded from the analysis, and all flagged spots were given a weight of zero. The normalized data were then subjected to fold-change analysis and t statistics using the Bioconductor multtest package. Differentially expressed genes were defined based on cut-off value +1 ⩽ Ratio (Log2) ⩽ −1 with p < 0.05. All microarray data (intensity and expression ratio) for 11 M1ΔCdhA experiments and seven M1CdhAΔC55 experiments have been deposited in the GEO data base with accession numbers GSE15546 (M1ΔCdhA) and GSE15598 (M1CdhAΔC55).
The expressed mRNA levels from microarray analysis were confirmed by real-time PCR using 1 μg of total RNA in a reverse transcriptase reaction to synthesize first strand cDNA according to the manufacturer's instruction (Roche Applied Science RT kit). Real-time PCRs for selected genes were performed using Brilliant SYBR Green QPCR Master Mix (Roche Applied Science) and specific primers (supplemental Table S3) and conducted using a LightCycler® 480 (Roche Applied Science) real-time PCR instrument. The copy numbers for all of the genes were normalized with gyrA using at least three biological replicates, and results were analyzed using Exor4 software (Roche Applied Science) (35).
Statistical Analysis
Unless otherwise indicated, statistical analysis was carried out from at least three independent experiments and based on nonparametric two-tailed Student's t test with Welch's correction using GraphPad Prism 4 software.
RESULTS
SP-STK Positively Regulates CdhA, a CHAP Domain-containing Streptococcal Protein
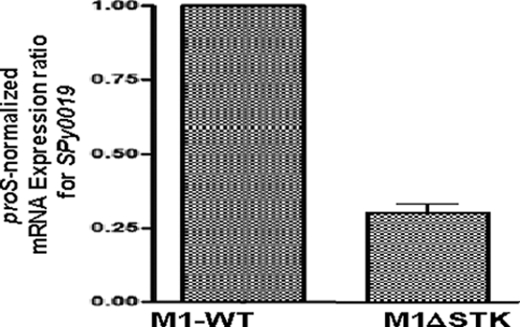
Based on our earlier reports, GAS lacking a eukaryotic-type serine threonine kinase (M1ΔSTK) is defective in cell division (13). Phenotypic characteristics of this mutant were found to mimic growth- and cell division-defective S. agalactiae (group B Streptococcus) and S. pneumoniae mutants lacking PcsB protein (15, 36), S. mutans lacking GbpB protein (19, 37), and E. faecalis/faecium lacking SagA/SalA (22, 21). PcsB, GbpB, and Saga/SalA are orthologs and are reported to possess similar structural characteristics, including the CHAP domain at their C-terminal ends (24, 38, 39). Based on this previous study, we hypothesized that the cell division defects observed in the M1ΔSTK mutant are probably due to the down-regulation of a PcsB ortholog SPy0019/CdhA. To confirm this, first, we complemented M1ΔSTK GAS mutant with the wild-type spy0019 and observed by light microscopy that the cell division defects of M1ΔSTK were restored to the wild-type chain-forming strain. The complemented strain displaying normal chain forming ability indicated that SP-STK directly or indirectly regulates the expression of CdhA. In the subsequent experiments, by employing real-time quantitative RT-PCR, we observed a significant decrease in the cdhA transcript abundance in the M1ΔSTK mutant strain (Fig. 1) and thus established a direct link between SP-STK and CdhA. Earlier, CdhA/SPy0019 was identified as SibA because of its ability to bind to human immunoglobulin (18). However, apart from this function, the physiologically relevant role of this protein in GAS has not been described.
FIGURE 1.
Physiological relationship between SP-STK and SPy0019/CdhA. mRNA expression levels of spy0019/cdhA in the M1ΔSTK mutant strain in comparison with that in the wild-type M1SF370 strain. Shown is the mean ± S.E. (error bars) of three independent experiments, each in triplicate wells.
CdhA Regulates GAS Growth
Based on the structural similarity of CdhA with six other Gram-positive orthologs (15, 19, 21, 22, 36, 37), we predicted that the N-terminal coiled-coil portion may have a role in cell division/separation/aggregation, and the C-terminal region containing CHAP domain may have a cell wall hydrolysis/synthesis function. CdhA possesses a typical motif of the CHAP-domain at its C-terminal end (SSX7CX13GX30GX10GX5YGH*VXVV, where X represents a non-conserved amino acid and H* is a putative catalytic histidine residue) (24, 38, 39). Because the precise functional roles of these regions in Gram-positive pathogens in general and GAS physiology or pathogenesis in particular are not known, we created two mutants using pFW5 suicide vector: one with complete deletion of the cdhA gene (M1ΔCdhA) and the second with deletion of the region corresponding to the C-terminal CHAP domain (M1CdhAΔC55). The latter expressed truncated CdhA (amino acids 1–343) lacking the last 55 amino acids constituting the conserved and putative catalytic histidine residue (His354) responsible for conferring the cell wall hydrolase activity (24).
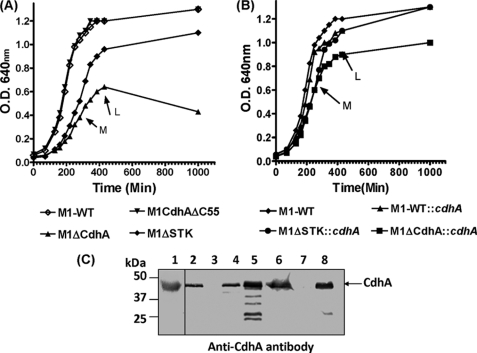
Growth characteristics of the mutants (M1ΔCdhA, M1CdhAΔC55, and M1ΔSTK) and the cdhA-complemented strains (M1ΔCdh::cdhA, M1-WT::cdhA, M1ΔSTK::cdhA) determined over a period of ∼17 h revealed unique patterns (Fig. 2, A and B). Among all mutants, M1ΔCdhA exhibited retarded growth, attaining a maximum A600 of 0.62 in ∼8 h. This mutant was unable to grow further and in fact showed a decrease in the A value (0.42) after an overnight incubation (∼17 h), indicating a progressive lysis of the mutant. The corresponding complemented strain (M1ΔCdhA::cdhA) grew faster and displayed a maximum A value of 1.0 (versus wild-type A600 of ∼1.3), indicating no degradation in the overnight grown culture and hence the restoration of CdhA function. The growth patterns of mutants expressing the truncated CdhA lacking the CHAP domain containing C-terminal 55-residue (M1CdhAΔC55) and that of the wild-type strain were comparable. M1CdhAΔC55, despite being devoid of the CHAP domain with putative cell wall synthesis function, in fact grew faster than the CdhA-overexpressing complemented strain, M1-WT::cdhA. This indicated that the growth defect observed in M1ΔCdhA is primarily due to the absence of the N-terminal region of CdhA. The growth pattern of M1ΔSTK, which displayed slow growth as reported previously (13), when complemented with cdhA, grew as fast as the wild-type strain, indicating that the slow growth observed of the M1ΔSTK mutant (13) could be due to the down-regulation of cdhA.
FIGURE 2.
Growth curves and integrity of CdhA-specific mutant and complemented strains. Wild-type (M1-SF370 or M1-WT) and various isogenic mutant strains and corresponding complemented strains were created with different strategies and were grown in THY broth as described under “Experimental Procedures.” A, growth curves of the wild-type (M1-WT) and isogenic mutant strains (M1ΔCdhA mutant lacking CdhA, M1CdhAΔC55 mutant expressing truncated-CdhA, and M1ΔSTK mutant lacking SP-STK (13)); B, corresponding complemented strains (M1ΔCdhA::cdhA and M1ΔSTK::cdhA (SP-STK mutant complemented with cdhA) and CdhA-overexpressing strain (M1-WT::cdhA) were monitored spectrophotometrically by measuring A600 (O.D.600 nm) over a period of 18 h. Each data point represents an average of three independent readings. For clarity, error bars have been omitted. Arrows M and L indicate middle and late log phase points for each growth curve. C, Western blot analysis showing the presence and absence of CdhA expression in the whole lysates of mutant, wild-type, and complemented strains using anti-CdhA IgG (lane 1), recombinant CdhA protein, and whole cell lysates of wild type M1-SF370 (lane 2), M1ΔCdhA (lane 3), M1CdhAΔC55 (lane 4), M1ΔCdhA::cdhA (lane 5), M1-WT::cdhA (lane 6), M1ΔSTK (lane 7), and M1ΔSTK::cdhA (lane 8).
Western blot analysis of the recombinant CdhA protein and the proteins present in the whole cell lysates of the strains described above (Fig. 2, A and B) using affinity-purified monospecific anti-CdhA IgG showed the presence of a 43-kDa CdhA-specific reactive protein band in the wild-type and the cdhA-complemented strains and its absence in the M1ΔCdhA and M1ΔSTK mutant strains, thus confirming the integrity of these strains also at the protein levels (Fig. 2C). The truncated CdhA found in the M1CdhADC55 mutant despite the loss of the C-terminal 55 residues migrates similarly to wild-type CdhA (Fig. 2C, lane 4). The recombinant truncated CdhA (amino acids 1–270) also migrates similarly to the wild type (data not shown), indicating that the CHAP domain of CdhA does not contribute to the migration of the whole protein in the SDS-PAGE.
CdhA Regulates GAS Cell Division and Belongs to a Class of Proteins That Participates in the Formation of Cell Septum and Determines Bacterial Chain Length
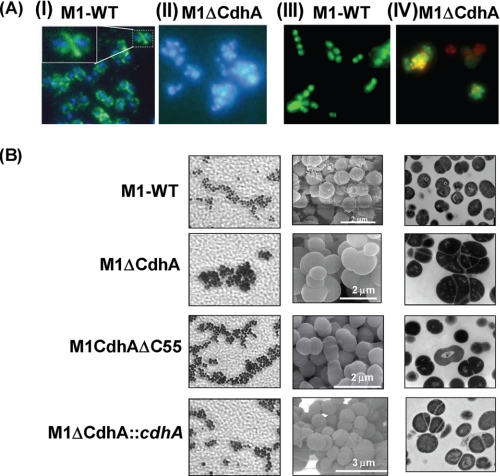
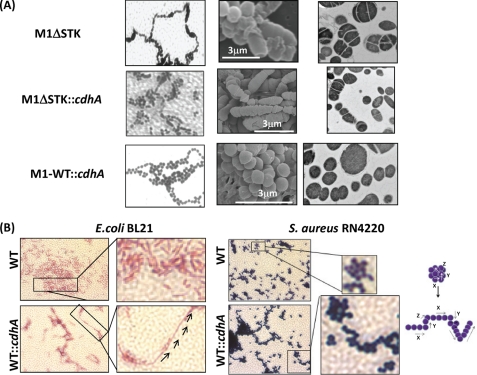
Indirect immunofluorescence microscopy of the wild-type and M1ΔCdhA mutant strains using FITC-labeled affinity-purified CdhA-specific IgG (Fab′ fragment) revealed localization of CdhA protein primarily in the septum (Fig. 3A, I, inset). Further, the absence of CdhA resulted in aggregation of the bacterial cells, as shown in Fig. 3A (DAPI stain in II). Electron microscopy of M1ΔCdhA clearly revealed defective cell division, resulting in the formation of incomplete and unorganized septa and in turn bacterial aggregates (Fig. 3B). This random septa formation resulted in the generation of ghost cells devoid of the genetic content. These cells were confirmed as dead by fluorescence microscopy using Live-Dead bacterial stain (Fig. 3A, IV). As shown in Fig. 3B, M1CdhAΔC55 mutant strain expressing truncated CdhA did not display defect in cell division and the cdhA-complemented mutant strain (M1ΔCdhA::cdhA) reverted back to the wild-type phenotype, supporting our hypothesis that the cell division function is restricted to the N-terminal region. Further, as was also reported for the PscB protein of S. pneumoniae (15, 16), CdhA-overexpressing M1-WT::cdhA strain displayed increased chain length, confirming that CdhA indeed plays an important role in the chain formation in GAS (Fig. 4A). However, the SEM- and TEM-based analyses of M1ΔSTK::cdhA revealed striking differences in the cell division restoration pattern. M1ΔSTK::cdhA strain primarily showed increase in only chain length; however, the organized cell septation was not completely restored, indicating that the primary function of SPy0019/CdhA is more likely in the recognition of the division plane but not the whole complex phenomenon of cell division governed by a battery of proteins (40–42) and possibly governed directly or indirectly by SP-STK (13). To confirm this in heterologous bacterial species, we examined the CdhA-overexpressing E. coli BL21 strain before and after IPTG induction. It was evident that upon CdhA expression, the cococbacillary form of uninduced E. coli was indeed transformed into the long chain-forming bacilli (Fig. 4B). Further, the cdhA-complemented S. aureus RN4220 (which is known to divide in x, y, and z planes and form clusters) using an S. aureus-compatible complementation vector, pCN40tetR (26), also formed chains with intermittent kinks (Fig. 4C). Together, light microscopy, SEM, and TEM of the wild-type, mutant, and complemented strains confirmed unequivocally that N-terminal domain of CdhA is responsible for the cell division plane recognition and chain formation.
FIGURE 3.
Light, scanning, and transmission electron microscopy of M1-SF370 wild-type and M1ΔCdhA, M1CdhAΔC55, and M1ΔCdhA::cdhA mutant GAS strains. A, indirect immunofluorescence microscopy of intact wild-type SF370 and the mutant M1ΔCdhA strain using anti-CdhA antibody, FITC-labeled conjugate antibody, and DAPI (blue nuclear stain). CdhA is primarily located at the septum (green fluorescence in I) of M1-SF370. The inset in the first panel shows the magnification of a region localized with CdhA in one of the dividing bacteria. II, the M1ΔCdhA strain forming cluster and the absence of green fluorescence (only nuclear staining with DAPI). III and IV were stained with Live-Dead bacteria stain. The presence of red stain in some portion of the clusters of M1ΔCdhA (IV) indicates the presence of dead bacteria. B, images acquired for the M1-WT, M1ΔCdhA, M1CdhAΔC55, and M1ΔCdhA::cdhA strains employing light microscopy, SEM, and TEM are shown in left, middle, and right panels, respectively. The indicated scale mark applies to both SEM and TEM for each strain. Light microscopy was carried using Gram stain.
FIGURE 4.
CdhA is a bacterial cell division plane-recognizing and chain-forming factor. A, light and electron microscopy (SEM and TEM) of M1ΔSTK and M1ΔSTK::cdhA and M1-WT::cdhA mutant strains. B, light microscopy of uninduced E. coli BL21 and IPTG-induced pET14B-His-CdhA-transformed (His-CdhA-overexpressing) BL21::cdhA strains. C, S. aureus RN4220 wild-type and pCN40tetR-cdhA-transformed (CdhA-overexpressing) RN4220::cdhA strains.
CdhA Does Not Contribute Significantly to the Overall GAS Ability to Bind hIgGs
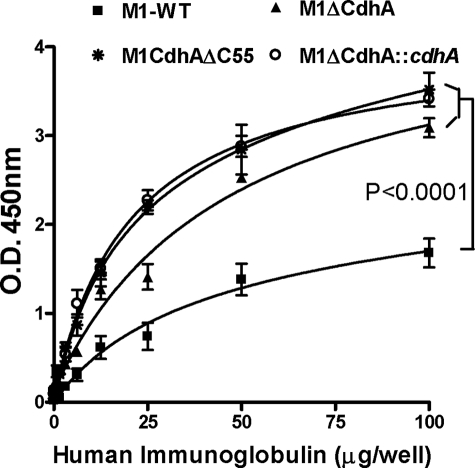
SPy0019 is a secretory protein and was previously termed as SibA based on its ability to bind hIgG (hence named SibA) (18). Fluorescence microscopy with anti-SPy0019 antibody indicated that SPy0019/SibA is located both at the septum and on the surface. We therefore hypothesized that the hIgG-binding ability of GAS may be attributed to SPy0019, and the corresponding mutant lacks hIgG-binding ability. In fact, to our surprise, the bacterial ligand binding assays revealed significantly increased hIgG binding activity in the mutant strains as compared to that in wild type (p < 0.0001), indicating that the absence of CdhA on the surface somehow exposes the native GAS hIgG binding activity (Fig. 5). This hIgG binding activity of the mutant was not reduced by complementation with the wild-type cdhA gene, possibly due to homologous functions possessed by CdhA and other bacterial immunoglobulin-binding proteins. Together, these results indicate that although CdhA is indeed an hIgG-binding protein, it does not primarily contribute to the overall GAS hIgG binding activity.
FIGURE 5.
Comparison of human immunoglobulin binding property of CdhA mutants with that of the wild-type GAS strain. Ligand-binding assays were performed in 96-well formats, as described under “Experimental Procedures.” Each data point represents an average ± S.E. (error bars) of three individual experiments performed in triplicates. Final readings represent values obtained after subtracting the average value obtained from 12 control wells.
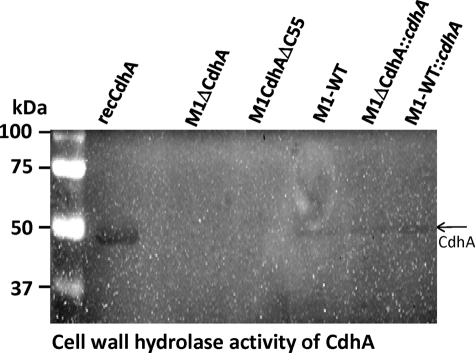
CdhA Hydrolyzes GAS Cell Walls
The CHAP domain-specific cell wall hydrolase/amino peptidase activity for CdhA orthologs (15, 18, 19, 21, 22) in Gram-positive organisms has not been reported so far. To determine the putative peptidoglycan hydrolase activity of CdhA, the recombinant CdhA protein and ammonium sulfate precipitates of culture supernatants were subjected to renaturing zymogram analysis using purified M1-SF370 peptidoglycan because our initial attempts using acetone powder of Micrococcus luteus and S. pygenes M1-SF370 did not yield detectable activity (data not shown). As shown in Fig. 6, the recombinant CdhA protein and the ammonium sulfate precipitates of culture supernatants of the M1-WT, M1ΔCdhA::cdhA, and M1-WT::cdhA showed 43-kDa protein-associated hydrolysis of the GAS peptidoglycan. The absence of similar activities in M1ΔCdhA and M1CdhAΔC55 supernatants clearly indicated that the CHAP domain of CdhA indeed possesses cell wall hydrolase activity.
FIGURE 6.
In vitro cell wall hydrolyzing property of native CdhA expressed in GAS and recombinant CdhA. Zymographic analysis of GAS cell wall hydrolase activity of purified CdhA and ammonium sulfate precipitates of culture supernatants of the wild-type, CdhA-specific mutant, and cdhA-complemented GAS strains as indicated.
Absence of CdhA Adversely Affects GAS Adherence to Human Pharyngeal Detroit 562 Cells
Electron microscopy of M1ΔCdhA and M1CdhAΔC55 revealed significant reduction in the size of surface electron dense fuzzy layer (Fig. 3B). The presence of the latter is essentially attributed to many surface proteins (43, 44) that play a crucial role in GAS adherence to host cells (45). We therefore speculated that the reduced expression of crucial surface proteins may significantly affect the bacterial adherence ability to host cells. Direct bacterial adherence assays, using Detroit 562 pharyngeal carcinoma cell lines, detected significantly reduced (cfu/well, p < 0.001) adherence property of M1ΔCdhA mutant strain in comparison with the wild-type strain (Fig. 7). The mutants expressing truncated CdhA (M1CdhAΔC55), which although adhered better (cfu/well) than M1ΔCdhA to pharyngeal cells and displayed their surface structure nearly normally, also adhered significantly less efficiently when compared with the wild-type strain. This indicated that CdhA directly or indirectly controls expression of certain surface proteins. The number of cfu in control wells containing minimal essential medium without pharyngeal cells remained the same at the end of each bacterial adherence assay, indicating that bacterial cell death or multiplication did not contribute to these changes.
FIGURE 7.
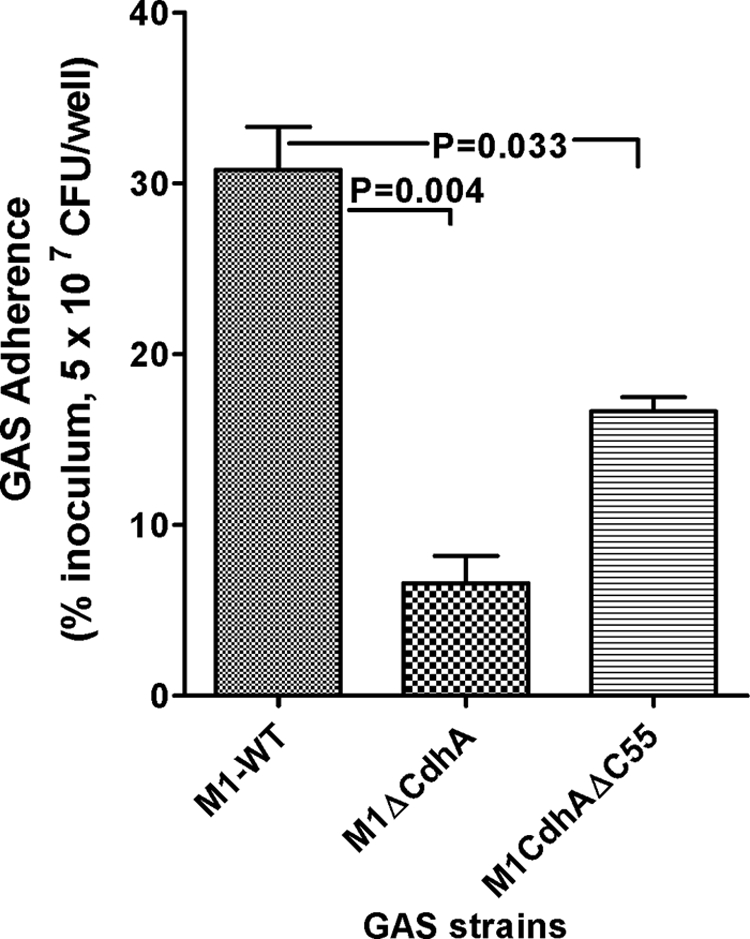
Lack of CdhA adversely affects bacterial adherence to human pharyngeal cells. The adherence index was determined using Detroit 562 human pharyngeal carcinoma cell lines and as a percentage of initial inoculum (MOI 50:1 (bacteria/cell); i.e. 5 × 107 cfu/well). Shown is the mean ± S.E. (error bars) of three independent experiments, each performed in at least triplicate wells.
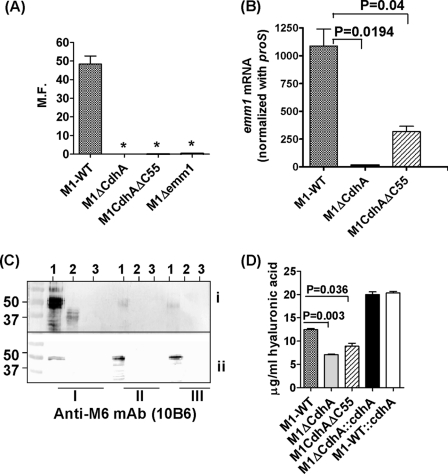
Lack of CdhA Abrogates Antiphagocytic Function
Because the M protein is a major virulence factor and an important antiphagocytic molecule that contributes significantly to the external electron dense layer of GAS (44), we predicted that the loss of electron-dense surface layer in the M1ΔCdhA mutant may in part be attributed to the loss of the M or M-like proteins. Alternatively, we also predicted that the aggregated forms of the mutant might not be efficiently phagocytosed because of their size limitation. To determine whether the predicted loss of the M protein in the M1ΔCdhA mutant in fact affects bacterial ability to resist phagocytosis, whole blood phagocytosis assays were performed to determine the ability of GAS to survive and multiply. Both M1ΔCdhA and M1CdhAΔC55 mutant strains, in comparison with the wild-type strain, were readily phagocytosed like the Δemm1 mutant GAS strain (Fig. 8A), although the M1ΔCdhA mutants showed a tendency to form aggregates, and M1CdhAΔC55 did not show any growth defect. The loss of the ability to resist phagocytosis in these two mutants was therefore attributed to the loss of the M protein as determined by Western blot analysis and not simply due to some defects in the cell growth observed only for M1ΔCdhA mutant (Fig. 8C). These results corroborated with the observed 4-fold (M1CdhAΔC55) to 50-fold (M1ΔCdhA) decrease in the emm1-specific mRNA level as determined by quantitative real-time PCR analyses (Fig. 8B). Further, both mutants displayed significantly reduced (p < 0.05) capsular formation as determined by hyaluronic acid contents (Fig. 8D). Almost 2-fold increased capsular contents in the cdhA-complemented mutant and the wild-type strains (M1ΔCdhA::cdhA and M1-WT::CdhA) suggested that CdhA somehow regulates the expression of the M protein and the capsule formation.
FIGURE 8.
Lack of CdhA abrogates GAS antiphagocytic function by down-regulating the M protein expression and capsule formation. A, phagocytic index of M1-WT, M1ΔCdhA, M1CdhAΔC55, and M1ΔEmm. The phagocytic index was measured as a multiplication factor (M.F.), indicating the ability of the GAS strain to multiply in the whole human defibrinated blood as described under “Experimental Procedures.” M1ΔEmm lacks the antiphagocytic M protein encoded by the emm1 gene and was used as positive control (13). B, quantitative real-time PCR to determine emm1-specific mRNA in the late log phase culture (growth level III) of M1-WT, M1ΔCdhA, and M1CdhAΔC55 GAS strains in comparison with the internal control gene (proS)-specific mRNA. C, Western blot analysis to determine the expression of the M-protein in the (i) culture supernatants and (ii) cell wall extracts of early (I), middle (II), and late (III) log phase-grown cultures of M1-WT (lane 1), M1ΔCdhA (lane 2), and M1CdhAΔC55 (lane 3) strains using the Emm1-reactive 10B6 monoclonal antibody (33). D, comparison of hyaluronic acid contents of the mutant M1ΔCdhA, M1CdhAΔC55, M1ΔCdhA::cdhA, and M1-WT::cdhA strains with the wild-type M1SF370 (M1-WT) GAS strains. Vertical bars represent mean ± S.E. (error bars) of three independent experiments. *, p < 0.001.
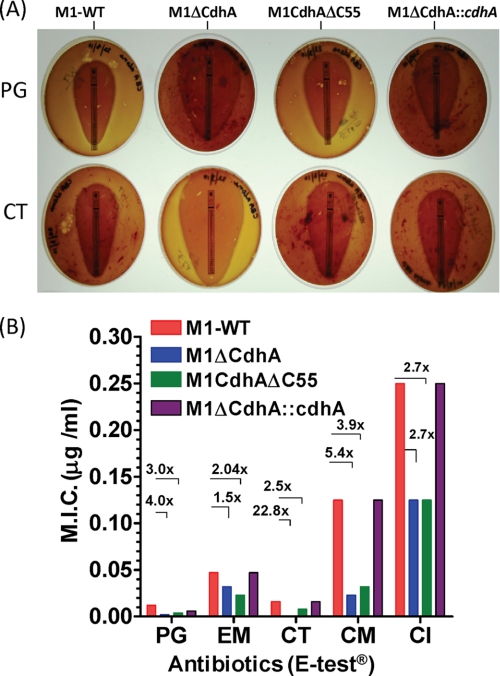
Absence of CdhA Increases Bacterial Susceptibility to Antibiotics
On the basis of the functional properties of CdhA to regulate cell division and cell wall hydrolase activity, we hypothesized that the lack of multifunctional CdhA (cell division and CHAP domain) may affect susceptibility to certain antibiotics that target peptidoglycan biosynthesis. To determine this, we measured minimum inhibitory concentrations of various antibiotics for the wild-type and M1ΔCdhA and M1CdhAΔC55 strains. We included two bacterial cell wall synthesis-targeting (PG and CT), two protein synthesis-targeting (clindamycin and erythromycin), and one DNA gyrase/topoisomerase IV-targeting (ciprofloxacin) antibiotics using an AB-BIODISK E-test. Significant reduction in MICs of PG (4-fold for M1ΔCdhA, 3.0-fold for M1CdhAΔC55), CT (22.8-fold for M1ΔCdhA and 2.5-fold for M1CdhAΔC55), and clindamycin (5.4-fold for M1ΔCdhA, 3.9-fold for M1CdhAΔC55) and only marginal reduction (1.5–3-fold) in MICs of erythromycin and ciprofloxacin for both mutants indicated that presence of CdhA protein plays an important role in the maintenance of the cell wall integrity; however, the hydrolase activity CdhA may have a limited contribution in the perceived role of controlling cell wall composition and modeling (Fig. 9, A and B).
FIGURE 9.
Lack of CdhA affects susceptibility of GAS to various antibiotics. Comparison of antibiotic susceptibility profiles (changes in MICs) of M1ΔCdhA, M1CdhAΔC55, and M1ΔCdhA::cdhA with respect to those of M1-WT GAS strain. MIC values were determined by E-test AB biodisk-recommended procedures. A, representative antibiotic susceptibility profiles of PG and CT for the wild-type, mutant, and cdhA-complemented strains. B, each bar represents an average reading obtained with three E-strips. The -fold changes (numbers followed by ×) in MIC values are calculated as the ratio of MIC for the wild type/MIC for the mutant. EM, erythromycin; CL, clindamycin; CI, ciprofloxacin.
SPy0019 Mutants Are Attenuated for Virulence in Mice
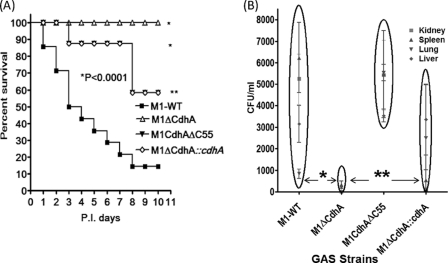
Collectively, the growth pattern, in vitro adherence, and bactericidal/phagocytosis assays strongly suggest that CdhA could serve as a potential virulence factor and thus may have a direct or indirect role in GAS pathogenicity. To determine this, we employed the mouse intraperitoneal infection model. As shown in Fig. 10, animals infected with the wild-type strain showed 50% mortality on day 4, whereas the M1ΔCdhA GAS mutant was completely attenuated for virulence because no morbidity or mortality was observed in infected mice during the entire observation period. In contrast, M1CdhAΔC55, despite its growth pattern similar to that of the wild-type strain, displayed significantly reduced virulence because only 20% mortality was observed for the infected mice by day 10 postinfection. The virulence property of the complemented strain, M1ΔCdhA::cdhA, which was able to regain the normal wild-type growth function (Fig. 10A), was significantly restored (40% mortality, p < 0.05), although it was partial. Further significantly reduced bacterial load in the spleen, kidney, and other visceral organs of the M1ΔCdhA-infected mice and increased bacterial load in the complemented strain indicated that the CHAP domain contribute significantly to the virulence-regulating properties of CdhA (Fig. 10, A and B). Although truncated CdhA expressing M1CdhAΔC55 mutant was readily phagocytosed by human neutrophils, total bacterial load in the lungs of mice infected with M1CdhAΔC55 was found to be significantly higher than those in the wild-type GAS (M1SF370)-infected mice. The resulting partial loss of GAS virulence indicated that in the absence of the CHAP domain, the CdhAΔC55 protein somehow helps to modify the host response (more specifically associated with mouse lung or other organ macrophages) in a manner favorable for GAS (Fig. 10B). Thus, the CHAP domain of CdhA seems to play a critical role in GAS virulence.
FIGURE 10.
Absence of CdhA or its catalytic CHAP domain attenuates GAS for virulence in an experimental mouse intraperitoneal infection model. A, effects due to the deletion of the entire cdhA gene or its CHAP domain-specific region (C-terminal 55 residues) on GAS virulence in mice were observed for M1-WT, M1ΔCdhA, M1CdhAΔC55, and M1ΔCdhA::cdhA GAS strains using the mouse intraperitoneal infection model. Survival/mortality curves for all strains were monitored for 10 days and statistically evaluated and plotted by the log rank test using GraphPad Prism 4 software. The SHAM-infected mice survived throughout the observation period (not shown). B, bacterial load determined in different organs (lung, liver, spleen, and kidney) from three infected animals from each group (10 mice each) sacrificed on day 4. *, p < 0.001; **, p < 0.05.
CHAP Domain of CdhA Directly or Indirectly Regulates GAS Virulence
The results described above indicated that CdhA is a multifunctional protein, the cell division plane recognition/chain-forming domain of which is located in the N-terminal two-thirds of the protein, and virulence is probably regulated by the CHAP domain. To determine the mechanism underlying the CHAP domain-mediated regulation of virulence, we analyzed microarray-based differential global gene expression profiles for M1ΔCdhA and M1CdhAΔC55 mutants (supplemental Tables S4–S6), validated with quantitative RT-PCR. Comparative analyses revealed several similarities in terms of genes belonging to specific functional categories that showed significant difference in their expression levels (≥2-fold, p < 0.05). Of 67 and 106 significantly differentiated genes in M1ΔCdhA and M1CdhAΔC55, respectively, the 50 differentiated genes were identical in both strains (Table 1 and supplemental Tables S4 and S5). The most affected and notable categories of genes in this common group belonged to virulence (7 of 50), carbohydrate transport/metabolism (11 of 50), amino acid metabolism and transport (7 of 50), and energy production (putative V-type Na+ ATPase pump; 10 of 50). Among the 11 major virulence-related genes affected in M1ΔCdhA, 10 genes (SPy0165-nga, SPy0167-slo, SPy0738-sagA, SPy1302-amyA, SPy1983-scl, SPy2016-sic, SPy2018-emm1, SPy2039-speB, SPy2043-mf, and SPy2200-hasA) were severely down-regulated (5–128-fold), and three genes (SPy1357-grab, SPy712-mf2, and SPy0711-speC) were up-regulated (3–4-fold) (Tables 1 and 2 and supplemental Tables S4– S6). In M1CdhAΔC55 expressing truncated CdhA lacking the last 55 residues, six (scl, hasA, amyA, emm1, sagA, and mf) and two (mf2 and speC) of the above mentioned virulence-related genes were down- and up-regulated, respectively. These results concur with electron microscopy and in vitro (adherence, antiphagocytic studies) and in vivo (experimental infection model) virulence studies.
TABLE 1.
Microarray-based global gene expression profiles for M1ΔCdhA and M1CdhAΔC55 and distribution of the number of differentially expressed genes into major functional categories, including those related to GAS virulence
| Functional categories | M1ΔCdhA |
M1ΔCdhA and M1CdhAΔC55 |
M1CdhAΔC55 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Upa (n = 7) | Downa (n = 60) | Total (n = 67) | Up (n = 3) | Down (n = 47) | Total (n = 50) | Up (n = 6) | Down (n = 100) | Total (n = 106) | |
| Amino acid transport | 1 | 6 | 7 | 0 | 5 | 5 | 0 | 7 | 7 |
| Carbohydrate transport | 1 | 11 | 12 | 0 | 11 | 11 | 0 | 28 | 28 |
| Cell envelope biogenesis, outer membrane | 0 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 |
| Cell motility and secretion | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| Co-enzyme metabolism | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 |
| DNA replication and recombination | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 |
| Energy production and conversion | 0 | 5 | 5 | 0 | 5 | 5 | 0 | 10 | 10 |
| Unknown function including CdhA | 1 | 19 | 20 | 1 | 13 | 14 | 2 | 21 | 23 |
| General functions only | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 8 | 9 |
| Lipid metabolism | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 3 |
| Post-translational modification/chaperones | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 3 | 3 |
| Secondary metabolism | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Signal transduction | 0 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 |
| Transcription and translation | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 4 | 4 |
| Virulence | 3 | 9 | 12 | 2 | 5 | 7 | 3 | 6 | 9 |
a Ratios of mRNA expression levels (mutant versus wild type) >2-fold (up or down) with p < 0.05 denote significantly differentiated gene (see also supplemental Tables S4 and S5 for more details).
TABLE 2.
Microarray-based differentially expressed virulence-related genes and their distribution in M1ΔCdhA and M1CdhAΔC55 mutant strains
| M1ΔCdhA |
M1ΔCdhA and M1CdhAΔC55 |
M1CdhAΔC55 |
||||
|---|---|---|---|---|---|---|
| Upa (n = 3) | Downa (n = 9) | Up (n = 2) | Down (n = 5) | Up (n = 3) | Down (n = 6) | |
| NAD-glycohydrolase | SPy0165(×25)b | |||||
| Streptolysin-O precursor | SPy0167(×13.4) | |||||
| Exotoxin C/SpeC | SPy0711(×3.4) | SPy0711 | SPy0711(×3.1) | |||
| DNase/mitogenic factor | SPy0712(×3.2) | SPy0712 | SPy0712(×3.1) | |||
| Streptolysin-S/SagA | SPy0738(×2.3) | SPy0738(×5.7) | ||||
| Cyclomaltodextrin glucanotransferase/AmyA | SPy1302(×6.5) | SPy1302 | SPy1302(×12.3) | |||
| GRAB | SPy1357(×3.6) | |||||
| DNase-mitogenic factor-3/Mf3 | SPy1436(×4.4) | |||||
| Collagen-like surface protein/Scl | SPy1983(×42.2) | SPy1983 | SPy1983(×21.1) | |||
| M-protein/Emm1 | SPy2018(×17.1) | SPy2018 | SPy2018(×7.5) | |||
| Exotoxin-B/SpeB | SPy2039(×12.1) | |||||
| Mitogenic factor | SPy2043(×4.7) | SPy2043 | SPy2043(×3.2) | |||
| Hyaluronate synthetase/HasA | SPy2200(×147) | SPy2200 | SPy2200(×24.5) | |||
a Expression profile (up or down), followed by the number of virulence genes in parentheses.
b Values in parentheses following multiplication symbols denote “x-fold” changes (up or down) in the transcript levels of the corresponding gene.
DISCUSSION
In the present study, we describe novel domain-specific functions of group A streptococcal secreted protein SPy0019/CdhA, which was described earlier as “SibA” based on the hIgG-binding ability of its recombinant form (18). Based on our earlier observations for the GAS mutants lacking SPy1625 or SP-STK (M1ΔSTK) showing cell division-defective phenotypes (13), the present study was carried out to address two important questions. (i) Are SP-STK-regulated cell division defects controlled by CdhA, as Gram-positive pathogens lacking CdhA orthologs display similar cell division defects? (ii) What is the contribution of CdhA in GAS pathogenesis? The answer to the first question was readily addressable by complementing and restoring the cell division defects of the GAS mutant lacking SP-STK with the wild-type cdhA. We believe that, unlike S. pneumonia PscB (15, 36) and S. mutans SagA/GtfB (46), the dispensable nature of CdhA allowed us to further study its domain-specific biochemical and biological functions in GAS pathogenesis using domain-specific knock-out mutants. Additionally, by complementing heterologus wild-type strains, E. coli and S. aureus, and the mutant strain, M1ΔSTK, with the wild-type cdhA, we also delineated the specific role of CdhA as a cell division plane recognition protein. The latter property seemed to be utilized as one of the earliest events in bacterial chain formation attributed previously to streptococcal PcsB (16, 17). Recent reports showing the autoregulatory loop between the Ser/Thr kinase and a CdhA homolog, YocH, found in Bacillus subtilis (47, 48) and the down-regulated pcsB gene in S. pneumoniae ΔSTK mutant (49) support our hypothesis. The designation assigned to SPy0019 as CdhA in this study was thus based on its biochemical and biological functions.
By light microscopy and complementation of the M1ΔSTK mutant with the wild-type cdhA, we provided an initial proof that CdhA is regulated by SP-STK because, by increasing the expression of CdhA in M1ΔSTK mutant, it was possible to recover the wild-type chain-forming characteristics, growth pattern and partial restoration of cell division defects. These phenotypes looked similar to the M1ΔCdhA mutant complemented with wild-type cdhA. Although SEM and TEM of the cdhA-complemented strain, M1ΔCdhA::cdhA, indeed confirmed the restoration of cell division defects, a similar study for M1ΔSTK::cdhA revealed the restoration of chain formation but not the defective cell division. These results indicated that CdhA plays an important role in certain aspects of the complex cell division process (40–42). In particular, the ability of CdhA to induce chain formation was confirmed when the CdhA-overexpressing wild-type M1-SF370 strain (M1-WT::cdhA) exhibited an extremely long chain. The ability of CdhA to confer similar phenotypic effects in heterologous systems like E. coli (BL21::cdhA) and S. aureus (RN4220::cdhA), which do not normally form chains, unequivocally indicated that CdhA is a chain-forming protein. This is in contrast to PcsB from S. mutans and S. agalactiae, which do not replace pneumococcal PcsB function (16). In fact, the underexpression and not the overexpression of PcsB in pneumococci leads to formation of long chains of cells and excessive septal wall synthesis (16). These contrasting results indicate that despite the structural similarities, their biochemical and biological activities are unique. Because S. pyogenes and E. coli divide in one plane, the long chain formation in these organisms was very obvious as an outcome of CdhA overexpression. In S. aureus, however, complementation with wild-type cdhA resulted in loosely formed clusters with short chains. Characteristically, in S. aureus, unlike the other two strains, these chains contained frequent kinks. We believe that the kink formation in S. aureus is due to its characteristic division pattern in three specific planes, x, y, and z (Fig. 4C), and a long chain in one plane is due to the S. aureus obligation to divide under the influence of CdhA until the onset of y- and z-specific second and third signals that will divide S. aureus in three different planes. Accordingly, it is likely that S. aureus may possess more than one plane-recognizing protein including an ortholog of CdhA. Although SA2093 (SSA) of S. aureus is a likely ortholog (50), its function has not been revealed so far. Although CdhA is a secreted protein, its role in cell division is underscored by its location in the septum, as seen by fluorescence microscopy (Fig. 3A). It is not known how CdhA is functionally integrated with the temporally regulated orchestration of several proteins, such as FtsZ, FtsA, and/or DivA, that are involved in the complex cell division process to achieve specific shape and forms of bacteria (41, 51, 52). Although this relation is presently under investigation, the results showing unregulated cell division with multiple septa in different planes in M1ΔCdhA and restoration of these defects in M1ΔCdhA::cdhA clearly indicated that the cdhA is a cell division plane recognition protein playing a crucial role in early events of cell division and ultimately chain formation in S. pyogene. Several decades ago, the induction of long chains in GAS was attributed to the presence of type M-specific antibody (53). However, the molecular and genetic basis of bacterial chain formation in GAS that is described in this study is distinct from the trypsin-sensitive, and type-specific antibody-mediated physical associations of more than one bacterial chain (53).
Although the mechanisms behind the direct relationship of the absence and overexpression of CdhA with up- and down-regulation of capsule formation are presently not known, a similar corroborating report for pneumococcal PcsB (54) suggests that a certain capsule regulator(s) may potentially interact with CdhA and modulate their regulatory functions.
CdhA belongs to a class of proteins that contains the CHAP domain (24, 38). The CHAP domain-containing proteins are in general known to degrade the cell wall cross-links or, as an amidase, to disrupt acetylmuramyl bonds (24, 38). Interestingly, none of the CdhA orthologs in Gram-positive pathogens have been conclusively shown to possess either in vitro or in vivo hydrolase activity or specific biologically relevant functions. The ability of CdhA to reveal in vitro zymogram-based cell wall hydrolase activity only in the presence of purified GAS cell wall peptidoglycan (Fig. 6) is probably due either to uniform distribution of peptidoglycan in the SDS gel or to specificity of CdhA-CHAP for M1-SF370 peptidoglycan because earlier attempts failed when acetone powder of M. luteus and S. pyogenes were employed. Because cell wall synthesis and cell division are physiologically closely linked (40, 52), we initially attributed the cell division regulatory function to the CHAP domain of CdhA, which, unlike other orthologs from Gram-positive pathogens (19, 16, 22), is not essential for GAS survival. Hence, it was possible to derive GAS mutants lacking CdhA and that expressing truncated CdhA lacking the C-terminal 55 residues encompassing the catalytic site of the CHAP domain and to investigate their physiologically relevant functions based on phenotypic characteristics of the derived mutants. The M1CdhAΔC55 mutant, although it did not display any in vitro hydrolase activity, to our surprise grew like the wild-type strain and did not show any defects in cell division, indicating that the CHAP domain is not involved in cell division or the cell division plane recognition process as described above (Fig. 3B). To reveal the precise biological role of the CHAP domain that is directly relevant to GAS pathogenesis, we initially performed several in vitro virulence-determining assays. Significant reduction in the abilities of M1ΔCdhA as well as M1CdhAΔC55 mutants to adhere to human pharyngeal cells and resist phagocytosis, most likely due to significant down-regulation of the M-protein in the corresponding cell wall extracts and a significantly reduced amount of hyaluronic acid, clearly indicated that CdhA, and more specifically the CHAP domain, directly or indirectly regulate GAS virulence. The complete attenuation of M1ΔCdhA as determined by a mouse virulence assays and a partial yet significant reduction in mouse mortality by intraperitoneal injection of M1CdhAΔC55 substantiate results obtained by employing in vitro virulence factor-determining assays. These results indicate that the CHAP domain of CdhA, although does not participate in cell the division process and the mutant lacking this domain grows like the wild-type strain, it plays a significant role in the GAS virulence. In other words, the growth defects observed in M1ΔCdhA (which grows maximally at A of 0.6) contribute minimally for the significantly decreased multiplication factor values in in vitro phagocytosis assays and in vitro virulence assays.
Because bacterial adherence and subsequent colonization play a primary role in bacterial virulence, the decreased virulence of M1ΔCdhA and M1CdhAΔC55 GAS mutants is also due to their significantly decreased ability to adhere to human pharyngeal cells in addition to loss of antiphagocytic activity. The mechanism underlying the ability of CdhA to control the expression of other surface proteins, which may play an important role in GAS adherence, is not apparent at present. We initially speculated that the incomplete septum formation might cause missorting of many surface proteins and virulence factors and even their spilling in the supernatant instead of linking covalently to the cell wall (43, 55, 56). The microarray analysis of the CdhA-specific mutants, however, offered a more plausible explanation because, in addition to Emm1, at least three other well established surface proteins, ScpA (57), Sic (58), and Scl (59), were found to be significantly down-regulated. This may result in cell walls without adhesins.
A functionally active CHAP domain indicated that cell wall hydrolase activity of CdhA may play an important role in cell wall synthesis and modulation. However, the absence of the CHAP domain of CdhA does not influence growth and cell division. Further, the increase in susceptibility to cell wall-acting antibiotics (penicillin and cefotaxime), especially in the absence of the CHAP domain, was only limited to 2–3-fold. The antibiotic susceptibility pattern thus invokes a question whether the CHAP domain of CdhA cell plays a role in cell wall synthesis or modulation (24, 38). Thus, defective cell walls of CdhA knock-out mutants, as revealed by high susceptibility to the cell wall-acting antibiotics, may be an indirect outcome of improper cell division. The recently reported mass spectrometric analysis for a pneumococcal mutant lacking the PcsB protein (CdhA homolog) revealing apparently defective wall without any change in the relative amount or composition of the peptidoglycan peptides lend support to our hypothesis (16, 54). The function of the CHAP domain could therefore be only to release muropeptides (57) during multiplication and until stationary phase and not the modulation of the cell wall.
The precise role of the CHAP domain of the bacterial peptidoglycan hydrolases in controlling bacterial virulence is presently not known. In this regard, in the present study, the results obtained from the animal experiments showing significantly increased streptococcal colony-forming units in the lungs of M1CdhAΔC55-infected mice as compared with those found in the lungs of the wild-type strain-infected mice draw special attention with regard to the specific virulence-related role of the CHAP domain in CdhA. The presence of a large number of cfu of M1CdhAΔC55 in lungs despite it being depleted of the M protein and capsule indicate that lung macrophages of the infected mouse could not phagocytose GAS expressing truncated CdhA, or possibly they were phagocytosed but not killed. The CHAP domain-containing bacterial peptidoglycan hydrolases have been shown to modulate muropeptide release and host innate immune responses (60). Certain members of the Nod-like receptor family, which are generally expressed constitutively by antigen-presenting cells (e.g. dendritic cells and macrophages) and epithelia, are capable of detecting specific muropeptides. Signaling through the Nod-like receptor family results in the coordinated activation of cytoplasmic, multimolecular protein complexes (inflammasomes), which in turn orchestrates massive inflammation and tissue injury (61). Thus, the mutant strain expressing the truncated CdhA lacking the catalytic site of its CHAP domain may not be able to release the required muropeptides from the GAS that has metastatically localized in lungs after experimental intraperitoneal infection (Fig. 10B). This may not result in activation of Nod-like receptor-associated inflammasomes in lung macrophages and killing of the M1CdhAΔC55 in lungs. Although this could be a valid reason for the significantly increased number of M1CdhAΔC55 in lungs and partial attenuation for mouse virulence (Fig. 10A), the proposed hypothesis needs experimental verification.
In addition to apparent changes in the transcription profiles of several virulence-related genes, we also observed drastic changes in the carbohydrate and amino acid metabolism-related genes. This is in contrast to recently published results for S. pneumoniae mutants depleted of PcsB displaying significant defects in cell shape and morphology showed changes in only a few genes belonging to the VicRK regulon (54). In the present study, the most notable differentially down-regulated carbohydrate genes (2–4-fold) in both M1ΔCdhA and M1CdhAΔC55 were the malM operon, lacA.1, lacA.2, lacB.1, lacD.2, and lacE. The down-regulation of these genes in GAS has recently been reported to inversely affect GAS virulence (62–66). Thus, the attenuation of virulence in mutants as a result of a complete or partial deletion of the cdhA genes can also be attributed to the down-regulation of carbohydrate metabolism genes. Both mutants, unlike the wild-type GAS strain, were unable to grow efficiently in the chemically defined medium supplemented with maltose/maltodextrin instead of glucose (data not shown).
Transcription of PscB of S. pneumoniae R6 and GbpB (SagA) of S. mutans is positively regulated by their respective VicR/K TCRSs, indicating that CdhA and its orthologs in other Gram-positive bacteria could be a common target of VicR/K (15, 16, 46, 67). In the present study, we showed that SP-STK also somehow regulates the transcription of cdhA. Although the function of VicR is regulated by its cognate histidine kinase, VicK, the streptococcal VicR has been shown to function independently of its cognate VicK (15, 67). It is therefore conceivable that VicR-mediated regulation of CdhA expression is fine tuned by SP-STK by phosphorylating VicR at its threonine residues. The mechanism of this regulation is presently under investigation.
In summary, this is the first report showing that CdhA is an SPy1625(SP-STK)-dependent cell wall-associated as well as secreted GAS protein, displaying a multifunctional nature with a cell division function located in its N-terminal two-thirds portion and the cell wall hydrolase function and major virulence functions located in the remaining C-terminal region. Because CdhA does not significantly contribute to the human immunoglobulin binding property of GAS, we have annotated SPy0019 as CdhA. The mechanisms by which the CHAP domain of CdhA controls GAS virulence and the N-terminal domain recognizes the division plane and orchestrates cell division are presently unknown. Although host innate immune responses may be a target for this protein for the observed virulence, the increased antibiotic susceptibility of GAS in the absence of SPy0019/SibA/CdhA and the virulence attenuation especially in the absence of its C-terminal region favor this protein being an important candidate/target for the development of newer immunotherapeutic as well as chemotherapeutic agents.
Supplementary Material
Acknowledgments
We thank Amanda Beltramini for the S. aureus complement construct and Shivani Agarwal, Shivangi Agarwal, and Haritha Adhikarla for critical reading of the manuscript and helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant AI64912 (to V. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S6.
- GAS
- group A Streptococcus
- CHAP
- cysteine and histidine-dependent aminohydrolases/peptidases
- SP-STK
- S. pyogenes Ser/Thr kinase
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- hIgG
- human IgG
- MIC
- minimal inhibitory concentration
- PG
- benzylpenicillin
- CT
- cefotaxime
- TEM
- transmission electron microscopy.
REFERENCES
- 1.Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. (2005) Lancet Infect. Dis. 5, 685–694 [DOI] [PubMed] [Google Scholar]
- 2.Cone L. A., Woodard D. R., Schlievert P. M., Tomory G. S. (1987) N. Engl. J. Med. 317, 146–149 [DOI] [PubMed] [Google Scholar]
- 3.Stevens D. L. (1995) Emerg. Infect. Dis. 1, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson D. R., Stevens D. L., Kaplan E. L. (1992) J. Infect. Dis. 166, 374–382 [DOI] [PubMed] [Google Scholar]
- 5.Steer A. C., Carapetis J. R., Nolan T. M., Shann F. (2002) J. Paediatr. Child Health 38, 229–234 [DOI] [PubMed] [Google Scholar]
- 6.McDonald M., Currie B. J., Carapetis J. R. (2004) Lancet Infect. Dis. 4, 240–245 [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Iturbe B., Musser J. M. (2008) J. Am. Soc. Nephrol. 19, 1855–1864 [DOI] [PubMed] [Google Scholar]
- 8.Carapetis J. R. (2008) Circulation 118, 2748–2753 [DOI] [PubMed] [Google Scholar]
- 9.Musser J. M., Krause R. M. (1998) in Emerging Infections (Krause R. M. ed) pp. 185–218, Academic Press, Inc., New York [Google Scholar]
- 10.Krause R. M. (2002) Indian J. Med. Res. 115, 215–241 [PubMed] [Google Scholar]
- 11.Cunningham M. W. (2000) Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stock A. M., Robinson V. L., Goudreau P. N. (2000) Annu. Rev. Biochem. 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 13.Jin H., Pancholi V. (2006) J. Mol. Biol. 357, 1351–1372 [DOI] [PubMed] [Google Scholar]
- 14.Churchward G. (2007) Mol. Microbiol. 64, 34–41 [DOI] [PubMed] [Google Scholar]
- 15.Ng W. L., Robertson G. T., Kazmierczak K. M., Zhao J., Gilmour R., Winkler M. E. (2003) Mol. Microbiol. 50, 1647–1663 [DOI] [PubMed] [Google Scholar]
- 16.Ng W. L., Kazmierczak K. M., Winkler M. E. (2004) Mol. Microbiol. 53, 1161–1175 [DOI] [PubMed] [Google Scholar]
- 17.Reinscheid D. J., Gottschalk B., Schubert A., Eikmanns B. J., Chhatwal G. S. (2001) J. Bacteriol. 183, 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagan P. K., Reinscheid D., Gottschalk B., Chhatwal G. S. (2001) Infect. Immun. 69, 4851–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattos-Graner R. O., Jin S., King W. F., Chen T., Smith D. J., Duncan M. J. (2001) Infect. Immun. 69, 6931–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borges F., Layec S., Thibessard A., Fernandez A., Gintz B., Hols P., Decaris B., Leblond-Bourget N. (2005) J. Bacteriol. 187, 2737–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breton Y. L., Mazé A., Hartke A., Lemarinier S., Auffray Y., Rincé A. (2002) Curr. Microbiol. 45, 434–439 [DOI] [PubMed] [Google Scholar]
- 22.Teng F., Kawalec M., Weinstock G. M., Hryniewicz W., Murray B. E. (2003) Infect. Immun. 71, 5033–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubert K., Bichlmaier A. M., Mager E., Wolff K., Ruhland G., Fiedler F. (2000) Arch. Microbiol. 173, 21–28 [DOI] [PubMed] [Google Scholar]
- 24.Bateman A., Rawlings N. D. (2003) Trends Biochem. Sci. 28, 234–237 [DOI] [PubMed] [Google Scholar]
- 25.Ferretti J. J., McShan W. M., Ajdic D., Savic D. J., Savic G., Lyon K., Primeaux C., Sezate S., Suvorov A. N., Kenton S., Lai H. S., Lin S. P., Qian Y., Jia H. G., Najar F. Z., Ren Q., Zhu H., Song L., White J., Yuan X., Clifton S. W., Roe B. A., McLaughlin R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beltramini A. M., Mukhopadhyay C. D., Pancholi V. (2009) Infect. Immun. 77, 1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podbielski A., Spellerberg B., Woischnik M., Pohl B., Lütticken R. (1996) Gene 177, 137–147 [DOI] [PubMed] [Google Scholar]
- 28.Boël G., Jin H., Pancholi V. (2005) Infect. Immun. 73, 6237–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaffin D. O., Rubens C. E. (1998) Gene 219, 91–99 [DOI] [PubMed] [Google Scholar]
- 30.Jin H., Song Y. P., Boël G., Kochar J., Pancholi V. (2005) J. Mol. Biol. 350, 27–41 [DOI] [PubMed] [Google Scholar]
- 31.de Jonge B. L., Chang Y. S., Gage D., Tomasz A. (1992) J. Biol. Chem. 267, 11248–11254 [PubMed] [Google Scholar]
- 32.Lepeuple A. S., Van Gemert E., Chapot-Chartier M. P. (1998) Appl. Environ. Microbiol. 64, 4142–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones K. F., Khan S. A., Erickson B. W., Hollingshead S. K., Scott J. R., Fischetti V. A. (1986) J. Exp. Med. 164, 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rainer J., Sanchez-Cabo F., Stocker G., Sturn A., Trajanoski Z. (2006) Nucleic Acids Res. 34, W498–W503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaffl M. W. (2001) Nucleic Acids Res. 29, 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinscheid D. J., Ehlert K., Chhatwal G. S., Eikmanns B. J. (2003) FEMS Microbiol. Lett. 221, 73–79 [DOI] [PubMed] [Google Scholar]
- 37.Smith D. J., King W. F., Barnes L. A., Peacock Z., Taubman M. A. (2003) Infect. Immun. 71, 1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigden D. J., Jedrzejas M. J., Galperin M. Y. (2003) Trends Biochem. Sci. 28, 230–234 [DOI] [PubMed] [Google Scholar]
- 39.Anantharaman V., Aravind L. (2003) Genome Biol. 4, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabeen M. T., Jacobs-Wagner C. (2005) Nat. Rev. Microbiol. 3, 601–610 [DOI] [PubMed] [Google Scholar]
- 41.Rothfield L., Taghbalout A., Shih Y. L. (2005) Nat. Rev. Microbiol. 3, 959–968 [DOI] [PubMed] [Google Scholar]
- 42.Lock R. L., Harry E. J. (2008) Nat. Rev. Drug Discov. 7, 324–338 [DOI] [PubMed] [Google Scholar]
- 43.Navarre W. W., Schneewind O. (1999) Microbiol. Mol. Biol. Rev. 63, 174–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischetti V. A. (2006) in Gram-Positive Pathogens, 2nd Ed (Fischetti V. A. ed) pp. 12–25, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 45.Pancholi V. (2006) in Gram-Positive Pathogens 2nd Ed. (Fischetti V. A. ed) pp. 100–112, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 46.Senadheera M. D., Guggenheim B., Spatafora G. A., Huang Y. C., Choi J., Hung D. C., Treglown J. S., Goodman S. D., Ellen R. P., Cvitkovitch D. G. (2005) J. Bacteriol. 187, 4064–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah I. M., Laaberki M. H., Popham D. L., Dworkin J. (2008) Cell 135, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah I. M., Dworkin J. (2010) Mol. Microbiol. 75, 1232–1243 [DOI] [PubMed] [Google Scholar]
- 49.Sasková L., Nováková L., Basler M., Branny P. (2007) J. Bacteriol. 189, 4168–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dubrac S., Msadek T. (2004) J. Bacteriol. 186, 1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Margolin W. (2005) Nat. Rev. Mol. Cell Biol. 6, 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheffers D. J., Pinho M. G. (2005) Microbiol. Mol. Biol. Rev. 69, 585–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stollerman G. H., Ekstedt R. (1957) J. Exp. Med. 106, 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barendt S. M., Land A. D., Sham L. T., Ng W. L., Tsui H. C., Arnold R. J., Winkler M. E. (2009) J. Bacteriol. 191, 3024–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlsson F., Stålhammar-Carlemalm M., Flärdh K., Sandin C., Carlemalm E., Lindahl G. (2006) Nature 442, 943–946 [DOI] [PubMed] [Google Scholar]
- 56.Scott J. R., Barnett T. C. (2006) Annu. Rev. Microbiol. 60, 397–423 [DOI] [PubMed] [Google Scholar]
- 57.Ji Y., McLandsborough L., Kondagunta A., Cleary P. P. (1996) Infect. Immun. 64, 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoe N. P., Ireland R. M., DeLeo F. R., Gowen B. B., Dorward D. W., Voyich J. M., Liu M., Burns E. H., Jr., Culnan D. M., Bretscher A., Musser J. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7646–7651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lukomski S., Nakashima K., Abdi I., Cipriano V. J., Shelvin B. J., Graviss E. A., Musser J. M. (2001) Infect. Immun. 69, 1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Humann J., Lenz L. L. (2009) J. Innate Immun. 1, 88–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stutz A., Golenbock D. T., Latz E. (2009) J. Clin. Invest. 119, 3502–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shelburne S. A., 3rd, Keith D., Horstmann N., Sumby P., Davenport M. T., Graviss E. A., Brennan R. G., Musser J. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1698–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shelburne S. A., Davenport M. T., Keith D. B., Musser J. M. (2008) Trends Microbiol. 16, 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loughman J. A., Caparon M. G. (2007) Mol. Microbiol. 64, 269–280 [DOI] [PubMed] [Google Scholar]
- 65.Loughman J. A., Caparon M. G. (2006) EMBO J. 25, 5414–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosch J. W., Tuomanen E. (2007) Mol. Microbiol. 64, 257–259 [DOI] [PubMed] [Google Scholar]
- 67.Liu M., Hanks T. S., Zhang J., McClure M. J., Siemsen D. W., Elser J. L., Quinn M. T., Lei B. (2006) Microbiology 152, 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.