Abstract
Mrp systems are widely distributed and structurally complex cation/proton antiporters. Antiport activity requires hetero-oligomeric complexes of all six or seven hydrophobic Mrp proteins (MrpA–MrpG). Here, a panel of site-directed mutants in conserved or proposed motif residues was made in the Mrp Na+(Li+)/H+ antiporter from an alkaliphilic Bacillus. The mutant operons were expressed in antiporter-deficient Escherichia coli KNabc and assessed for antiport properties, support of sodium resistance, membrane levels of each Mrp protein, and presence of monomeric and dimeric Mrp complexes. Antiport did not depend on a VFF motif or a conserved tyrosine pair, but a role for a conserved histidine in a potential quinone binding site of MrpA was supported. The importance of several acidic residues for antiport was confirmed, and the importance of additional residues was demonstrated (e.g. three lysine residues conserved across MrpA, MrpD, and membrane-bound respiratory Complex I subunits (NuoL/M/N)). The results extended indications that MrpE is required for normal membrane levels of other Mrp proteins and for complex formation. Moreover, mutations in several other Mrp proteins lead to greatly reduced membrane levels of MrpE. Thus, changes in either of the two Mrp modules, MrpA–MrpD and MrpE–MrpG, influence the other. Two mutants, MrpB-P37G and MrpC-Q70A, showed a normal phenotype but lacked the MrpA–MrpG monomeric complex while retaining the dimeric hetero-oligomeric complex. Finally, MrpG-P81A and MrpG-P81G mutants exhibited no antiport activity but supported sodium resistance and a low [Na+]in. Such mutants could be used to screen hypothesized but uncharacterized sodium efflux functions of Mrp apart from Na+ (Li+)/H+ antiport.
Keywords: Bacteria, Enzyme Mutation, Enzyme Structure, Proton Pumps, Sodium Proton Exchange, Bacillus pseudofirmus, Na+/H+ Antiporter, Alkaliphile, pH Homeostasis
Introduction
Na+/H+ antiporters have an essential role in supporting cytoplasmic pH homeostasis in alkaliphilic bacteria (1–3). These Na+/H+ antiporters catalyze active efflux of Na+ in electrogenic exchange for H+, with the number of H+ entering exceeding the number of Na+ exiting during each turnover. This property of the antiport enables Mrp to achieve accumulation of cytoplasmic H+ against its concentration gradient (4, 5). The coupled Na+ efflux activity of the antiporters supports Na+ resistance and produces an inwardly directed gradient of Na+ that drives solute transport and flagellar rotation in alkaliphilic bacteria (6–9). Although extremely alkaliphilic Bacillus species contain multiple cation/proton antiporters (1, 10), the Mrp-type antiporter has a dominant and essential role in alkaliphily (11, 12). Mrp antiporters are also broadly distributed among non-alkaliphilic Gram-positive and Gram-negative bacteria and play roles in diverse physiological processes that may depend upon pH and Na+ homeostasis (2). In addition to Na+/H+ antiport, several of these Mrp antiporters support resistance to anions, such as arsenite and bile salts (13–15). We hypothesize that Mrp complexes are consortiums of more than one transporter type (2).
The most intensively studied Mrp systems have seven (MrpA–MrpG) or six (a fused MrpA/B–MrpG) hydrophobic proteins that are encoded in operons (2, 10, 16, 17), in contrast to most other bacterial Na+/H+ antiporters (e.g. NhaA from Escherichia coli) that are hydrophobic products of a single gene (18). Kajiyama et al. (19) first showed evidence for a physical complex of Mrp proteins in B. subtilis that contained all seven Mrp proteins and accounts for earlier findings that all of the Mrp proteins are required for Na+/H+ antiport activity (20, 21). Our group subsequently expressed the mrp operon from alkaliphilic Bacillus pseudofirmus OF4 in antiporter-deficient E. coli strain KNabc after engineering the operon so that each of the seven Mrp proteins could be detected using antibody probes. We identified hetero-oligomeric Mrp complexes that corresponded to sizes expected for monomeric and dimeric complexes containing all seven Mrp proteins. The analyses also revealed a MrpA–MrpD subcomplex that could form in the absence of MrpE–MrpG but has no antiport activity (22). Although MrpA and MrpD proteins do not exhibit antiport activity in this subcomplex or when they are expressed individually (10, 23), one or both of these proteins may have antiport potential. A MrpA/MrpD homologue encoded by a “stand alone” gene from polyextremophilic Natranaerobius thermophilus was recently reported to exhibit Na+/H+ and K+/H+ antiport activity in anaerobically grown E. coli KNabc (24). MrpA and MrpD are the two largest Mrp subunits. They both have 14 transmembrane segments (TMSs)2 that share homology with each other and with the H+-translocating NuoL, -M, and -N subunits of NADH:quinone oxidoreductases from E. coli or mitochondria (i.e. respiratory chain Complex I) (2, 11, 25, 26). MrpA and its closest Complex I homologue, NuoL, both have a C-terminal extension beyond the core 14 TMSs (26). The MrpA and NuoL extensions are not homologous to each other (supplemental Fig. S1). Except for MrpC, which exhibits some homology to NuoK, none of the other Mrp proteins resembles respiratory chain subunits (17).
Site-directed mutagenesis studies of Bacillus subtilis Mrp by Kosono and co-workers (19, 27) have identified six acidic residues of MrpA and one acidic residue in MrpB and MrpD that are required for normal antiport but are apparently not required for Mrp complex formation. The phenotypic effects of mutations observed in E. coli KNabc were consistent with those observed in the native B. subtilis except for one mrpF mutant. However, characterization of B. subtilis Mrp antiport activity by the standard fluorometric assay in E. coli KNabc vesicles (28) is limited by the low activity of B. subtilis Mrp (i.e. there is <20% dequenching of the acridine orange fluorescence, which is the metric used for antiport assessment) (19, 27, 29). Here, we took advantage of the greater activity of the B. pseudofirmus OF4 Mrp system in E. coli KNabc vesicles (>40% dequenching), which facilitates better assessment of antiport deficits of mutant forms and determinations of apparent Km values for Na+. The engineered antiporter construct also made it possible to measure levels of each Mrp protein and visualize the complexes and subcomplexes formed by mutant antiporters. We probed the effects of mutations in 28 different sites throughout the Mrp subunits. The goals were to develop an extensive mutant profile to support or negate the importance of particular motifs in antiport activity, to identify additional residues across Mrp that are required for antiport, and to identify residues that affect membrane levels of other Mrp proteins or formation of one or both of the hetero-oligomeric complexes.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in supplemental Table S1. The media for routine growth of E. coli were LB medium (pH 7.0) (30) for strain DH5αMCR and LBK medium plus 50 mm NaCl (pH 7.5) (28) for strain E. coli KNabc (31). Cells were grown with shaking, at 200 rpm, at 37 °C. For growth experiments comparing E. coli transformants expressing wild-type and mutant forms of Mrp for Na+ sensitivity, the strains were grown in LBK, pH 7.5, under the same conditions except that test concentrations between 100 and 700 mm NaCl were added as indicated in connection with specific experiments. Ampicillin at 100 μg/ml and kanamycin at 25 μg/ml were added to media for plasmid-bearing E. coli KNabc cells and selecting transformants. Transformation of E. coli strains and all recombinant DNA manipulations were carried out by standard methods (30).
Introduction of the Site-directed Mutations in the mrp Operon
pGEMmrpTFCHS7, in which different epitope tags are linked to the C terminus of MrpA (T7 tag), MrpB (FLAG tag), MrpC (c-myc tag), MrpD (His7 tag), and MrpG (S tag), respectively, was constructed previously (22). In this study, a series of amino acid-substituted mutation plasmids was constructed using pGEMmrpTFCHS7. For introducing site-directed mutagenesis in vitro, The Gene TailorTM site-directed mutagenesis system (Invitrogen) was used, following the protocols from the instruction manual supplied by Invitrogen. Details of mutant construction are provided in the supplemental material.
Preparation of Membrane Vesicles
E. coli KNabc transformants were grown in LBK medium plus 50 mm NaCl at 37 °C. Everted membrane vesicles were prepared as described previously (22). The membrane vesicles were resuspended in TCDG buffer (10 mm Tris-HC1, 140 mm choline chloride, 0.5 mm dithiothreitol, and 10% (v/v) glycerol, pH 7.5) for antiporter assays or in ACA buffer (750 mm ϵ-aminocaproic acid, 50 mm Bis-tris propane, 20% (w/v) glycerol, pH 7.0) for blue native PAGE analyses. Vesicles were stored at −80 °C.
Antiport Assays
Fluorescence-based assays of the Na+/H+ antiport activities were conducted on everted membrane vesicles suspended in TCDG buffer. Protein content was determined using the Lowry method with bovine serum albumin as the protein standard (32). Assays were conducted by the method described by others (9) at pH values indicated for particular experiments. Briefly, each vesicle preparation was assayed in 2 ml containing 50 mm BTP-chloride buffer, 140 mm choline chloride, 5 mm MgCl2, 1 μm acridine orange, and 66 μg of vesicle protein. Measurements were conducted using a Hitachi High-Technologies model F-4500 fluorescence spectrophotometer with excitation at 420 nm (10-mm slit) and emission at 500 nm (10-mm slit). Respiration was initiated by the addition of Tris-succinate to a final concentration of 2.5 mm. After steady-state fluorescence quenching was reached, NaCl was added to a final concentration of 10 mm. The Na+-dependent dequenching of fluorescence is the measure of Na+/H+ antiport activity. Assays were conducted in duplicate on two independent membrane preparations. The concentration of Na+ yielding the half-maximal dequenching has been validated as a good estimate of the apparent Km of Na+/H+ antiporters (29). The apparent Km was determined using antiport activity assays at several concentrations of Na+ for selected mutants that had phenotypic changes as compared with the wild-type and had sufficient antiport activity (>10% of the wild-type level) to facilitate an assessment of Km.
Immunoblot Analyses of each Mrp Protein in Transformant Membrane Fractions
3 μl of a membrane suspension (10 μg of membrane protein/μl) suspended in TCDG buffer was used for SDS-PAGE analyses on membrane samples. The same volume of SDS loading buffer was added for each sample, after which the proteins were separated on 15% polyacrylamide SDS gels (33). The gels were then transferred to nitrocellulose filters (Bio-Rad) electrophoretically in Tris-glycine-methanol buffer (25 mm Tris, 192 mm glycine, 20% (v/v) methanol, pH 8.3). MrpA, MrpB, MrpC, MrpD, MrpE, MrpF, and MrpG proteins were detected by anti-T7 antibody (Abcam), anti-DDDDK (anti-FLAG tag) antibody (Abcam), anti-c-myc antibody (Abcam), anti-His antibody (Qiagen), anti-MrpE antibody, anti-MrpF antibody, and anti-S tag antibody (Abcam), respectively. Goat anti-rabbit HRP (Bio-Rad) was also used as the second antibody for detection of anti-MrpE and anti-MrpF antibodies. ECL solution (Amersham Biosciences) was the detection reagent. A quantitative imaging system, Pluor-S MAX (Bio-Rad), was used for detection and analysis of chemiluminescence images.
Blue Native PAGE (BN-PAGE)
Protein content of the membrane fraction suspended in ACA buffer was determined using the Micro-BCA assay (Pierce). Protein complexes (10 mg/ml) were solubilized at 4 °C for 20 min, in ACA buffer containing dodecyl maltoside at concentrations of 1.0% (w/v) dodecyl maltoside. Following solubilization, samples were cleared by centrifugation at 40,000 rpm for 90 min in a Beckman Ti70 rotor at 4 °C. The supernatant was divided into 20-μl aliquots, which were stored at −80 °C if not used immediately. Protein content was determined using the micro-BCA assay (Pierce). 30 μg of solubilized protein was loaded for BN-PAGE. BN-PAGE was performed in an XCell SureLockTM minicell using a Native PAGETM 4–16% gel, 15-well apparatus (Invitrogen). The details of the BN-PAGE protocols were those provided in the instruction manual from Invitrogen. High molecular mass markers, the Native MarkTM unstained protein standard, were obtained from Invitrogen.
RESULTS AND DISCUSSION
Computer Analysis of Mrp Protein Topologies and Selection of Residues for Mutation
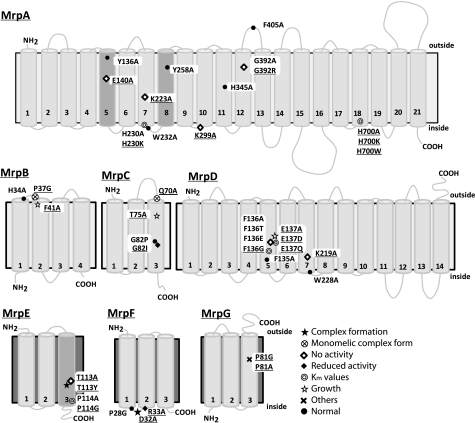
The secondary structure of each Mrp protein was predicted by ConPred II, HMMTOP, and TMHMM (all available on the World Wide Web). All three programs predicted the number of TMSs of MrpB, MrpC, MrpD, MrpF, and MrpG to be 4, 3, 14, 3, and 3, respectively, as depicted in Fig. 1. HMMTOP and TMHMM predicted the number of TMSs of MrpA and MrpE as 21 and 2, respectively, whereas ConPred II predicted 19 and 3 TMSs, respectively, for MrpA and MrpE. Those helices for which there was not full agreement are indicated by dark gray columns in Fig. 1. The core 14 TMSs of MrpA and MrpD differ somewhat from earlier topological predictions by others (31, 35) but are in agreement with the secondary structure revealed by the recent three-dimensional crystallographic data of Sazanov and co-workers (26) for the E. coli homologues NuoL, -M, and -N. A piston-like involvement of the MrpA C-terminal extension was hypothesized to energize Mrp antiport in analogy to the proposal for the role of the NuoL C-terminal extension observed in the structure (26). However, the Mrp and Nuo systems may have diverged before development of the piston mechanism because the C-terminal extensions of MrpA and NuoL are distinct, and MrpA is lacking in functional Mrp systems from some groups of bacteria (e.g. cyanobacterial Mrp systems) (2, 10).
FIGURE 1.
Transmembrane topology of Mrp proteins and positions of mutations induced in this study. Transmembrane segments were predicted by ConPred II, HMMTOP, and TMHMM (available on the World Wide Web) were used in the analyses of the secondary structure predictions for each Mrp subunit. Light gray transmembrane segments were predicted by all algorithms. Dark gray shading in MrpA indicates transmembrane segments that were predicted by only two of the algorithms (HMMTOP and TMHMM). Dark gray shading in MrpE indicates transmembrane segments that were predicted by only ConPred II. Underlined mutations indicate positions at which a mutation produced a phenotype that differed from wild-type Mrp. ●, positions at which the mutations complemented an antiporter-deficient E. coli KNabc transformant and exhibited normal Na+/H+ antiport activity; *, mutations that affected the level of Mrp proteins in the membrane; ♢, mutations that led to loss of the Na+/H+ antiport activity and loss of Na+ tolerance; ♦, mutations that decreased the Na+/H+ antiport activity, without an effect on growth of the E. coli transformant;  , mutations that affected the Km values of Mrp-dependent antiport activity;
, mutations that affected the Km values of Mrp-dependent antiport activity;  , mutations that affected transformant cell growth; ⊗, mutations that affected Mrp complex formation; ×, the two mutants in MrpG-P81 that had a unique phenotype.
, mutations that affected transformant cell growth; ⊗, mutations that affected Mrp complex formation; ×, the two mutants in MrpG-P81 that had a unique phenotype.
Multiple alignments of each Mrp subunit from alkaliphile, neutralophile, and pathogens were analyzed by ClustalW software (available on the World Wide Web). They are shown in supplemental Fig. S1. The sequence of NuoL, NuoM, and NuoN, subunits of NADH dehydrogenases from E. coli and Rhodobacter capsulatus, were also aligned with sequences of MrpA and MrpD. 21 highly conserved amino acid residues of the Mrp complex from alkaliphilic B. pseudofirmus OF4 were chosen for inclusion in the mutant panel (Table 1). Two of these residues, MrpD-Phe135 and -Phe136, were in a VFF sequence that we identified in MrpD. Kuroda et al. (34) reported that a VFF sequence is present in many Na+-coupled transport proteins. They proposed that VFF might be a motif involved in Na+ binding. When the phenylalanines of the VFF motif were replaced, there was decreased binding of amiloride, a sodium site probe, to the NhaA antiporter of Vibrio parahemeolyticus, whereas a mutation of valine to leucine had no effect. Including the two MrpD phenylalanines, the full group of highly conserved Mrp residues that were mutated as part of a larger panel was as follows: MrpA-Glu140, -Lys223, -Lys299, -Trp232, and -Phe405; MrpB-Phe41, -Pro37, and -His34; MrpC-Gln70 and -Thr75; MrpD-Phe135, -Phe136 (the two VFF phenylalanines), -Glu137, -Lys219, and -Trp228; MrpE-Thr113 and -Pro114 (Table 1 contains data of previously reported mutations in each of these MrpE positions (22)); MrpF-Asp32, -Arg33, and -Pro28; and MrpG-Pro81. Each of these conserved residues was replaced with alanine, except for MrpB-Pro37 and MrpF-Pro28, which were replaced by glycine, whose flexibility is closer than that of alanine to proline. More than one replacement was made in several instances. The additional replacements were glycine, threonine, and glutamic acid for MrpD-Phe136, aspartic acid and glutamine for MrpD-Glu137, tyrosine for MrpE-Thr113, and glycine for MrpG-Pro81. The following additional seven residues were chosen for inclusion in the mutant panel for reasons other than conservation alone. MrpA-Gly392 and MrpC-Gly82 were included because they correspond to the Mrp residues of alkaliphilic Bacillus halodurans C-125 whose mutations (BH-MrpA-G393R and BH-MrpC-G82E) led to Mrp discovery when they caused loss of alkaliphily (11, 12). In this study, MrpA-G392 from alkaliphilic B. pseudofirmus OF4 was replaced by alanine as well as arginine, whereas MrpC-Gly82 was replaced by proline and isoleucine. Two histidine residues of 341AAIFHLIN348 and 695LAFYHLPEL704 in MrpA were included in the panel as potential quinone binding sites based on criteria for such sites from Fisher and Rich (aliphatic-X3HX2/3(L/T/S) (35) and Yang et al. (AX3HX2S) (36). These histidine residues were both replaced by alanine, and MrpA-H700 was additionally replaced by lysine and tryptophan. A third histidine residue, MrpA-His230, was also considered a potential quinone binding site because of a report that a histidine residue in the corresponding position of NuoN, NuoN-His224, from E. coli interacts with quinones (37). MrpA-His230 was replaced by both alanine and lysine. Finally, two tyrosine residues in MrpA, MrpA-Tyr136 and MrpA-Tyr258, were included in the panel and replaced with alanine because of a suggestion that these tyrosines play a role in Na+ movement in homologous respiratory complex subunits (38). Each of the mutations is listed in Table 1, and their locations are schematically shown on the topological models in Fig. 1.
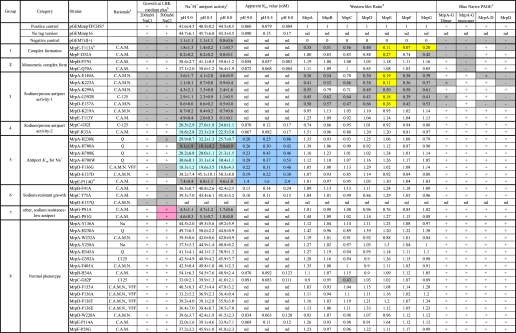
TABLE 1.
Summary of growth, antiport activity, membrane expression level, and complex formation of each mutation and positive and negative controls
a C.A.M., conserved across Mrps; C.A.M.N., conserved across MrpA, MrpD, NuoL, NuoM, and NuoN; Q, hypothesized quinone-binding site, partial LXXXH motif (35–37); VFF, Na+-binding motif (34); Na+, proposed Na+-binding residue (38).
b Data from Morino et al. (22).
c Details of the methods for preparation and assays of membrane vesicles are described under “Experimental Procedures.” The values presented for the percentage dequenching are average values from duplicate assays from three independent experiments. These values for percentage dequenching are shown with the S.D. values.
d A quantitative imaging system, Pluor-S MAX (Bio-Rad), was used for detection and analysis of a chemiluminescence image. Values in the table indicate the ratio of expression level of each Mrp protein calculated by comparing with that of pGEMmrp THCHS7.
e The clear presence of multiple Mrp proteins at hetero-oligomer positions is scored as a “+” in the MrpA–MrpG dimer or monomer column even if one or more other Mrp proteins is not well resolved. Dark gray highlights indicate mutant phenotypes that differ markedly from the wild-type transformant (e.g. inability to grow in the presence of 200 or 500 mm added NaCl, major defects in Na+/H+ antiporter activity, decreases in the membrane levels of Mrp proteins of ≥70%, or absence of one or more Mrp complexes in BN-PAGE analyses. Mutants in which the Na+/H+ antiporter activity is at least 25% lower than wild type are highlighted in light blue. Mutants in which the apparent Km is at least 2.5 higher than that of wild-type Mrp are highlighted in dark blue. Mutants in which the expression level of Mrp proteins in membrane vesicles is ≤30% of the positive control are highlighted in yellow. nd, not determined (see “Experimental Procedures” for the criteria for which assays were omitted).
Classification of the Mutant Phenotypes into Groups
We classified the mutant phenotypes into eight mutant groups. The mutants of Group 8 had mutations that produced no discernable effects on Mrp or had an effect that did not meet criteria for significance, as indicated below. The following assays were conducted on each mutant and generated the data on which the categorization was based: (i) growth of antiporter-deficient E. coli KNabc transformants at high sodium concentrations; (ii) Na+/H+ antiporter activity in membrane vesicles and, for selected samples, its Km value for Na+ as a function of pH; (iii) level of each Mrp protein in the E. coli KNabc transformant membranes, as assessed by SDS-PAGE and immunoblot detection; (iv) Mrp complex formation assessed in the membrane by BN-PAGE and immunoblot detection (Table 1). The data for the growth experiments, the Na+/H+ antiport assays, immunoblot analyses of SDS-PAGE-fractionated membranes, and immunoblot analyses of BN-PAGE-fractionated membranes are shown in supplemental Figs. S2–S6, respectively.
Group 1 contains two Mrp mutants with changes in a residue of two different “small Mrp subunits,” MrpE-T113A, and MrpF-D32A. The defining feature of these two mutants is the low level of membrane-associated MrpE accompanied by the absence of both MrpA–MrpG monomer and dimer forms of the hetero-oligomeric Mrp complexes (Table 1 and supplemental Fig. S5). Also part of the Group 1 profile was the absence of growth in the presence of 200 mm sodium and almost no detectable Na+/H+ antiport activity in vesicle assays (supplemental Figs. S2 and S3). The MrpA–MrpD subcomplex was detected in both mutants (supplemental Fig. S5). The lack of antiport activity is consistent with the earlier conclusion that full Mrp complexes are active but the stable MrpA–MrpD subcomplex has no antiport activity (22). The MrpE-T113A mutant profile was reported earlier (22), including reduced levels of all the Mrp proteins in the membrane. By contrast the MrpF-D32A mutant exhibited almost normal levels of Mrp proteins except for MrpE and MrpG, which were present at 27 and 42% of wild-type levels, respectively. MrpD dimer was present in the antiport-inactive MrpF-D32A mutant, indicating that, like the MrpA–MrpD subcomplex, the MrpD dimer lacks antiport activity.
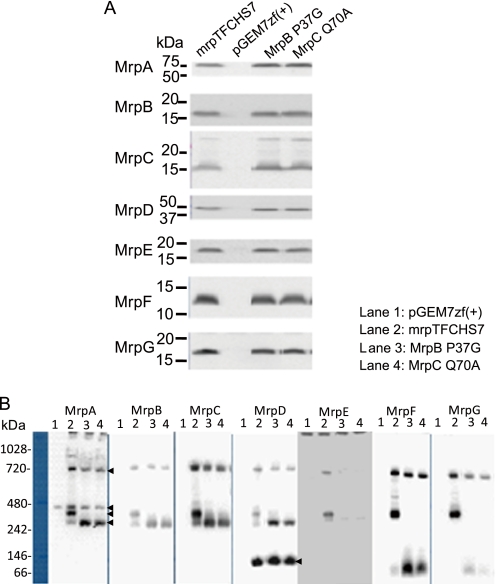
Group 2 contains two mutants with mutations in MrpB and MrpC, MrpB-P37G and MrpC-Q70A. The defining feature of this pair of mutants is their effect on only one form of the MrpA–MrpG complex. The two Group 2 mutants reproducibly exhibited absence of the monomeric MrpA–MrpG complex, whereas the dimeric complex was detected (Table 1 and Fig. 2). As shown in Fig. 2, both Group 2 mutants had elevated levels of the MrpA–MrpD subcomplex relative to the wild type. They also had elevated amounts of a putative subcomplex composed of MrpE, MrpF, and MrpG (Fig. 2 and supplemental Fig. S6). Because of limitations of the MrpE antibody, MrpE protein was detected only by two-dimensional blue native SDS-PAGE followed by immunoblotting (supplemental Fig. S6), so its identification as part of a subcomplex is more tenuous than earlier observations of a MrpE–MrpG subcomplex (22). Both MrpA–MrpD and MrpE–MrpG subcomplexes were observed previously in wild-type preparations and are hypothesized to be assembly intermediates and/or breakdown products of larger hetero-oligomeric forms (22). The two Group 2 mutants also have modestly higher levels of a MrpD dimer that is another presumed breakdown product/assembly intermediate that was seen before in BN-PAGE analyses of Mrp samples (22). The Na+/H+ antiport levels of the two mutants were comparable with the wild-type controls although the monomeric complex was absent (Table 1 and supplemental Fig. S3). Possibly, the monomeric form of the full Mrp hetero-oligomer is present in the mutant membranes, but this mutant complex is more unstable upon extraction with detergents than the dimeric hetero-oligomer. Alternatively, the dimeric hetero-oligomeric Mrp complex may account for the vast majority of antiport activity.
FIGURE 2.
Levels of Mrp proteins and Mrp complexes in membranes in which MrpB-P37G and MrpC-Q70A mutant Mrp systems are expressed. A, detection of the Mrp proteins in membranes from E. coli KNabc transformants expressing full MrpTFCHS7, a vector control, or MrpB-P37G and MrpC-Q70A mutants by SDS-PAGE. B, detection of the Mrp complexes from E. coli KNabc transformants with empty vector pGEM7zf(+) (lane 1, negative control), expressing full MrpTFCHS7 (lane 2, positive control) or MrpB-P37G (lane 3) and MrpC-Q70A (lane 4) mutants by BN-PAGE. The arrows on the right of the MrpA panel indicate MrpA–MrpG dimer (top), nonspecific band (second), MrpA–MrpG monomer (third), and MrpA–MrpD subcomplex (bottom). The arrow on the right of the MrpD panel indicates the MrpD dimer (MrpDD). The methodological details are described under “Experimental Procedures.”
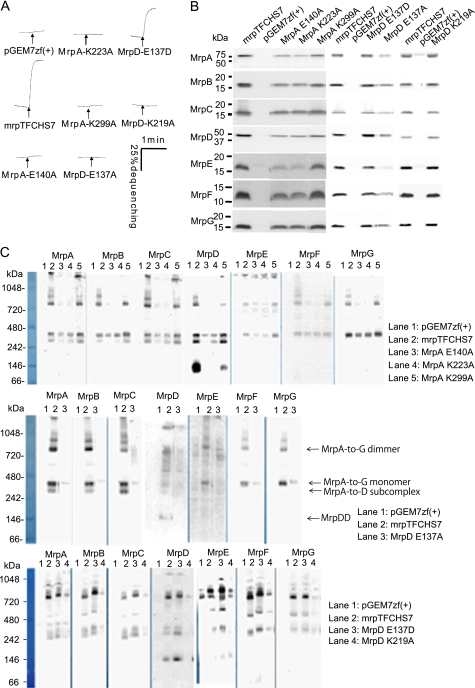
Group 3 mutants all had very low antiport activity levels. These Mrp systems were unable to support Na+ resistance (Fig. 3 and Table 1). These phenotypes may be secondary to reduced membrane levels of all the Mrp proteins, especially MrpE (Table 1 and supplemental Fig. S4). The mutant also had reduced levels of the dimeric Mrp complex and lacked the MrpD dimer, whose presence tends to correlate with robust presence of large oligomeric complexes of Mrp (supplemental Fig. S5) (22). However, complex formation of MrpA-K299A on BN-PAGE was normal as compared with that of wild type. Some members of this group had mutations in charged residues of either MrpA or MrpD. The mutations were MrpA-E140A, MrpA-K223A, MrpA-K299A, MrpD-E137A, and MrpD-K219A (Fig. 3). The MrpA-E140A and MrpD-E137A mutants in homologous positions had been identified as important for antiport of B. subtilis Mrp by Kosono's group (27, 39). This study suggests, for the first time, roles in antiport for three conserved lysine residues, MrpA-Lys233, homologous MrpD-Lys219, and MrpA-Lys299. Alanine replacements at all three locations resulted in greatly reduced antiport activity although both hetero-oligomeric Mrp complexes were observed. Like MrpA-Glu140 and MrpD-Glu137, the three lysines that are here implicated in antiport roles are conserved among the NuoL, -M, and -N proteins of Complex I as well as across Mrp systems. Mutations in these positions of the respiratory protein homologues have been shown to reduce activity of the proton pumping complex (37, 40, 41). MrpA-G392R and MrpE-T113Y were also included in Group 3 because of their very low antiport level. They have patterns of Mrp protein and complex contents that are similar to MrpA-K223A and MrpD-K219A, respectively.
FIGURE 3.
Effects of mutations of charged amino acids of MrpA and MrpD, MrpA-Glu140, MrpA-Lys223, MrpA-Lys299, MrpD-Glu137, and MrpD-Lys219, that are conserved across MrpA, MrpD, and NADH-dehydrogenase-1 subunits NuoL/M/N. A, antiport activity is shown as the percentage of dequenching of acridine orange fluorescence when 10 mm Na+ was added (at the arrow) to energized everted vesicles that had achieved a steady-state pH, acid in, by respiratory proton pumping. Na+/H+ antiport activity was assayed at pH 8.5 in vesicles from transformants with the empty vector pGEM7zf(+) or the vector expressing full Mrp (pGEMmrpTFCHS7) or the mutant Mrp systems with the following point mutations: MrpA-E140A, -K223A, or -E137A or MrpD-E137A, -E137D, or -K219A. B, Western analyses of SDS-PAGE-fractionated membranes from the wild-type and the mutant transformants using antibodies against each Mrp subunit. The plasmid present in the membranes is indicated above each lane. C, detection of Mrp complexes by immunoblotting analysis of BN-PAGE. 30 μg of protein extracted from membrane was loaded onto gels for BN-PAGE fractionation and then transferred to PVDF membrane for immunoblotting. Mrp subunits were detected with antibodies against unique epitope tags linked to the C terminus of MrpA–MrpD and MrpG. MrpE and MrpF proteins were detected with polyclonal antibodies directed toward these proteins. Methodological details are described under “Experimental Procedures.”
Group 4 contains two mutants, MrpC-G82I and MrpF-R33A. These mutant Mrp systems were able to support Na+ tolerance although they had reduced Na+/H+ antiport activity. The apparent Km for Na+ was not greatly elevated, and the membrane levels of Mrp proteins and the complex contents of these mutants were comparable with the wild type pattern.
MrpC-G82I and MrpA-G392R (Group 3) were the sites of the identified original mutations, which led to the non-alkaliphilic phenotype in B. halodurans C-125. The MrpC-G82I mutation was in the position corresponding to that of the original B. halodurans C-125 Mrp mutation. This mutation of MrpC-Gly82 to isoleucine was considered more temperate than the original MrpC-G82E mutation in B. halodurans C-125 (12). The replacement of MrpC-Gly82 with isoleucine led to an approximately 50% loss of antiport activity with retention of the capacity to confer Na+ resistance. There was little reduction in Mrp protein levels and no change in the pattern of complexes and subcomplexes observed in BN-PAGE analyses (Table 1). Neither of the two glycines that were mutated in the original B. halodurans C-125 mutagenesis is crucial for antiport inasmuch as the MrpA-G392A and MrpC-G82P mutants of B. pseudofirmus OF4 yielded mutant Mrp systems with no functional phenotype of significance and only a partial reduction in the membrane levels of MrpC itself for the MrpC-G82P mutant (see Group 8).
Group 5 contains seven mutants whose common feature is a more than 2.5-fold increase in the Km for Na+ of the antiport activity (highlighted in blue in Table 1). The mutants vary in overall antiport activity level. Unlike the MrpD-E137A mutant in Group 3 that lacked antiport capacity, the conservative replacement in the MrpD-E137D supported almost normal antiport activity levels (Fig. 3) but with a significant change in Km (Table 1). Four of the mutants in Group 5 have replacements in two histidine residues that are part of potential quinone binding sites in MrpA, with three mutations in MrpA-His700 (MrpA-H700A, -H700K, and -H700W) and one in MrpA-His230 (MrpA-H230K). Among the MrpA-His700 mutations, the MrpA-H700A had less than 22% of the wild-type antiport activity as well as an elevated Km. The activity level was less severely reduced in the three other histidine mutants of this subset, MrpA-H700K and -H700W and MrpA-H230K. The MrpA-H230K mutant had >50% wild-type activity with an elevated Km for Na+. Hypothetically, sensitivity of the electrogenic Mrp antiporter to the redox state of the quinone pool could modulate antiport activity in a manner that is responsive to the energy state. However, histidine is not required for normal function at this position because the MrpA-H230A mutant did not have a significant phenotype and is in Group 8 (Table 1). Included in Group 5 is the MrpE-P114G mutant. This mutant had earlier been shown to have a significantly reduced level of antiport activity, an elevated Km for Na+, and reduced capacity for Na+ resistance but has normal membrane levels of the Mrp proteins and a normal BN-PAGE profile. The new MrpE-P114A mutant, however, had no phenotype (see Group 8, Table 1). Finally, Group 5 includes a mutant with an MrpD-F136G mutation that led to reduced Na+ resistance (supplemental Fig. S2), reduced antiport levels, and elevation of the Km for Na+, without an effect on the levels of Mrp proteins or the profile of complex formation (Table 1). Other mutations at MrpF-136 and the MrpD-F135A mutation in the putative VFF motif did not affect levels of antiport or any other antiporter properties; these mutants are included in Group 8 (Table 1).
Group 6 contains two mutants with single mutations in MrpB and MrpC, MrpB-F41A and MrpC-T75A. The defining feature of this group is a deficiency in growth in the presence of added NaCl although there is no observable deficit in antiport levels or properties that accounts for the phenotype (Table 1). One more mutant, MrpD-E137Q, is listed in Group 6 in Table 1 because of its Na+ sensitivity. This mutant is distinct in that the transformant of E. coli KNabc expressing this mutant Mrp grew poorly on LBK medium, even in the absence of added NaCl, and could not be assayed for the parameters shown in Fig. 1. Perhaps all three of these Group 6 mutants produce a Mrp system that is toxic to the host in some way that increases its sensitivity to Na+, with MrpD-E137Q being an extreme example.
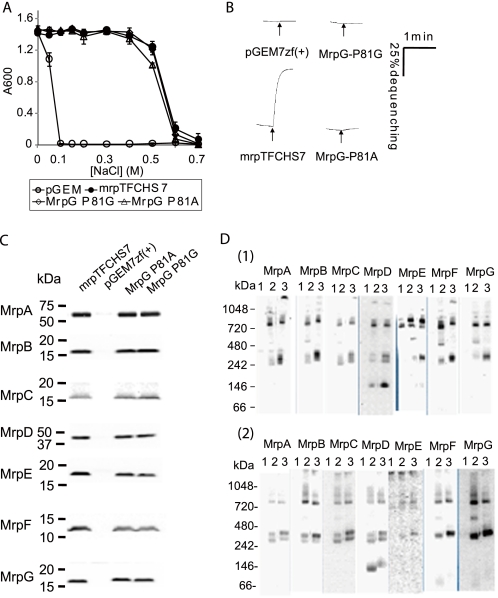
Group 7 contains two mutants in a single residue of MrpG, MrpG-Pro81, which was replaced with either alanine or glycine. These mutants, like the Group 3 mutants and the Group 5 MrpE-P114G mutant, had normal membrane levels of Mrp proteins and normal profiles in BN-PAGE analyses of Mrp complexes and subcomplexes but had very low levels of antiport activity. But unlike those Group 3 mutants, both the MrpG-P81A and MrpG-P81G mutant Mrp systems exhibited an unusual phenotypic feature. The MrpG-P81A and -P81G mutants retained a wild type Mrp capacity to confer NaCl resistance to cells of Na+/H+ antiporter-deficient E. coli KNabc despite their very low antiport activity. We also measured intracellular Na+ concentrations of the transformants with MrpG-P81G, pGEM7zf(+) vector (negative control), and pGEMmrpTFCHS7 (engineered wild-type Mrp, positive control) in LBK medium with either 100 or 300 mm NaCl. The values of the MrpG-P81G transformant were maintained at 17.4 ± 0.7 and 41.5 ± 3.8 mm Na+ in the medium with 100 and 300 mm NaCl, respectively. These values were close to the positive control levels of 16.8 ± 2.0 and 39.2 ± 5.7, whereas the levels in the negative control transformant were 107.1 ± 7.1 and 196.5 ± 8.8. The phenotype of these mutants raises the possibility that they retain a Na+ efflux activity of Mrp that is distinct from Na+/H+ antiport. The existence of such additional transport activities of Mrp systems has been hypothesized, but none of the activities has yet been biochemically characterized. Alternatively, the mutations at MrpG-Pro81 may change the catalytic properties of the Mrp complex, conferring a new property that accounts for the capacity to support Na+.
Group 8 contains the remaining mutations: MrpA-Y136A, MrpA-H230A, MrpA-W232A, MrpA-Y258A, MrpA-H345A, MrpA-G392A, MrpA-F405A, MrpB-H34A, MrpC-G82P, MrpD-F135A, MrpD-F136A, MrpD-F136E, MrpD-F136T, MrpD-W228A, MrpE-P114A, and MrpF-P28G. The phenotype of the mutants in this group was almost identical to that of wild type, except for differences that were assessed as minor in the context of normal levels of antiport activity and capacity to confer Na+ resistance. For example, the amount of MrpC protein observed in membranes of the MrpC-G82P mutant was 43% of the wild-type MrpC subunit level, but this mutant was included in the “normal” group. Residues mutated in Group 8 mutants are presumed not to be essential. Among such residues is a pair of tryptophan residues in homologous positions of MrpA and MrpD, MrpA-Trp232 and MrpD-Trp228. They are conserved in NuoL, NuoM, and NuoN subunits as well as Mrp systems and had been proposed to have a subunit linking function in the respiratory complex setting (42). Their mutation had no effect on Mrp.
Interactions between MrpA–MrpD and MrpE–MrpG Subcomplexes
This study produced evidence for roles of MrpE, -F, and -G in antiport activity and in formation of Mrp complexes that contain all seven proteins of the alkaliphile Mrp. The MrpA, -B, -C, and -D proteins (MrpA–MrpD) and MrpE, -F, and -G proteins (MrpE–MrpG) are thought to comprise two modules that together constitute an antiport-active Mrp system. Both modules are found as subcomplexes in BN-fractionated membranes of cells expressing the full alkaliphile mrp operon (22). The MrpA–MrpD module, which is thought to be the catalytic module for antiport, contains the MrpA, -C, and -D proteins that share homology with modules of respiratory complexes and also contains an MrpB protein that is homologous to a MrpB-like domain present in MrpA (2, 16–17, 27). The MrpE–MrpG module is unique to Mrp systems and is usually encoded by contiguous genes mrpEFG (2). We hypothesized the MrpE–MrpG module is required to induce an antiport-active conformation of the MrpA–MrpD module. We also hypothesized that MrpE is required for maintenance of normal membrane levels of other Mrp proteins and to stabilize hetero-oligomeric Mrp complexes. Here, we confirmed the earlier finding that a MrpE-T113A mutant lacked all Mrp complexes except for the MrpA–MrpD subcomplex, which had no antiport activity. We further showed that all of the Mrp proteins were present in significantly reduced levels in this mutant (Table 1 and supplemental Fig. S4). By contrast, a new mutant with a different replacement at this position, MrpE-T113Y, had normal membrane levels of all Mrp complexes and subcomplexes as well as normal levels of each individual Mrp protein. Nonetheless, this mutation led to an antiport activity level <5% of wild-type, which was insufficient to support Na+ resistance. We hypothesize that the threonine → tyrosine change at the 113 position of MrpE prevents the MrpE–MrpG module from producing the conformational activation of the MrpA–MrpD subcomplex. A similar defect is hypothesized for the MrpE-P114G mutant that exhibited greatly reduced antiport activity together with an increased Km although levels of Mrp proteins and all Mrp complexes were normal; a mutant with an alanine substitution at the same position had no phenotype, underscoring the distinct properties of alanine and glycine in putative TMS regions (Group 8) (Table 1 and supplemental Fig. S3). Among the other MrpE–MrpG proteins, the MrpG-P81A and -P81G mutants and MrpF-R33A mutation also led to reductions in antiport activity that were particularly severe in the MrpG mutants (Table 1 and Fig. 4 and supplemental Fig. S3). These reductions in antiport, as in the MrpE-T113Y and MrpE-P114G mutants, were not correlated with reduced Mrp protein or complex levels (Table 1 and supplemental Fig. S3). Taken together, the results strongly support a role for the MrpE–MrpG module proteins in conferring antiport activity upon Mrp hetero-oligomers.
FIGURE 4.
Effect of mutations at MrpG-Pro81. A, tests of complementation capacity of MrpG-P81A and MrpG-P81G mutant forms of Mrp on the Na+ sensitivity of E. coli KNabc transformants. Each of the transformants was grown in 2 ml of LBK medium containing the concentration of added NaCl indicated in the figure. After incubation, with shaking at 200 rpm, for 16 h at 37 °C, the A600 value of each culture was measured. The error bars indicate S.D. for duplicate cultures in three independent experiments. B, Na+/H+ antiport activity was assayed at pH 8.0 in everted vesicles from transformants with the empty vector pGEM7zf(+) or the vector expressing the full tagged wild-type Mrp (pGEMmrpTFCHS7) or mutant forms with MrpG-P81A and -P81G point mutations. C, immunoblot analyses of SDS-PAGE-fractionated membranes from the wild type and the mutant transformants using antibodies against each Mrp subunit. The plasmid present in the membranes is indicated above each lane. D, 1, detection of Mrp complexes from the membrane fraction of E. coli KNabc transformed with the empty vector (lane 1), pGEMmrpTFCHS7 (lane 2), and pTFCHS7MrpG-P81G (lane 3). 2, detection of Mrp complexes from the membrane fraction of E. coli KNabc transformed with the empty vector (lane 1), pGEMmrpTFCHS7 (lane 2), and pTFCHS7MrpG-P81A (lane 3). Details of the procedures are described under “Experimental Procedures.”
The mutant panel also revealed that interactions between the MrpA–MrpD and MrpE–MrpG modules are reciprocal. Among the Group 3 mutants with greatly reduced antiport activity were those with mutations MrpA-E140A, MrpA-K223A, and MrpD-137A of the MrpA–MrpD module that led to profound reductions in the levels of MrpE as well as reductions in levels of additional Mrp proteins (Table 1 and Fig. 3). Kajiyama et al. (27) had similarly noted reductions in MrpE levels in mutants with site-directed changes in two other Mrp proteins of B. subtilis Mrp. The profiles of the MrpE-deficient Group 1 and Group 3 mutants support the conclusion that levels of MrpE affect membrane levels of other Mrp proteins and hence of Mrp complexes. This complicates the assignment of direct roles to particular Mrp proteins in ion binding or translocation involved in Mrp-dependent Na+/H+ antiport. Nonetheless, this study produced evidence for the involvement of several newly examined Mrp residues in antiport activity.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM28454 (to T. A. K.). This work was also supported by a special grant-in-aid from Toyo University, a grant from a High-Tech Research Center program of the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M. I.), and a grant from the Japan Society for the Promotion of Science Research Fellowships for Young Scientists (to M. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S6.
- TMS
- transmembrane segment
- Bis-tris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- BN-PAGE
- blue native PAGE.
REFERENCES
- 1.Padan E., Bibi E., Ito M., Krulwich T. A. (2005) Biochim. Biophys. Acta 1717, 67–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swartz T. H., Ikewada S., Ishikawa O., Ito M., Krulwich T. A. (2005) Extremophiles 9, 345–354 [DOI] [PubMed] [Google Scholar]
- 3.Slonczewski J. L., Fujisawa M., Dopson M., Krulwich T. A. (2009) Adv. Microb. Physiol. 55, 1–79, 317 [DOI] [PubMed] [Google Scholar]
- 4.Booth I. R. (1985) Microbiol. Rev. 49, 359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macnab R. M., Castle A. M. (1987) Biophys. J. 52, 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krulwich T. A., Federbush J. G., Guffanti A. A. (1985) J. Biol. Chem. 260, 4055–4058 [PubMed] [Google Scholar]
- 7.Hirota N., Kitada M., Imae Y. (1981) FEBS Lett. 132, 278–280 [Google Scholar]
- 8.Ito M., Hicks D. B., Henkin T. M., Guffanti A. A., Powers B. D., Zvi L., Uematsu K., Krulwich T. A. (2004) Mol. Microbiol. 53, 1035–1049 [DOI] [PubMed] [Google Scholar]
- 9.Ito M., Xu H., Guffanti A. A., Wei Y., Zvi L., Clapham D. E., Krulwich T. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10566–10571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krulwich T. A., Hicks D. B., Ito M. (2009) Mol. Microbiol. 74, 257–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamamoto T., Hashimoto M., Hino M., Kitada M., Seto Y., Kudo T., Horikoshi K. (1994) Mol. Microbiol. 14, 939–946 [DOI] [PubMed] [Google Scholar]
- 12.Seto Y., Hashimoto M., Usami R., Hamamoto T., Kudo T., Horikoshi K. (1995) Biosci. Biotechnol. Biochem. 59, 1364–1366 [DOI] [PubMed] [Google Scholar]
- 13.Ito M., Guffanti A. A., Oudega B., Krulwich T. A. (1999) J. Bacteriol. 181, 2394–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashyap D. R., Botero L. M., Lehr C., Hassett D. J., McDermott T. R. (2006) J. Bacteriol. 188, 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzioba-Winogrodzki J., Winogrodzki O., Krulwich T. A., Boin M. A., Häse C. C., Dibrov P. (2009) J. Mol. Microbiol. Biotechnol. 16, 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathiesen C., Hägerhäll C. (2002) Biochim. Biophys. Acta 1556, 121–132 [DOI] [PubMed] [Google Scholar]
- 17.Mathiesen C., Hagerhall C. (2003) FEBS Lett. 249, 7–13 [DOI] [PubMed] [Google Scholar]
- 18.Padan E. (2008) Trends Biochem. Sci. 33, 435–443 [DOI] [PubMed] [Google Scholar]
- 19.Kajiyama Y., Otagiri M., Sekiguchi J., Kosono S., Kudo T. (2007) J. Bacteriol. 189, 7511–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiramatsu T., Kodama K., Kuroda T., Mizushima T., Tsuchiya T. (1998) J. Bacteriol. 180, 6642–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito M., Guffanti A. A., Wang W., Krulwich T. A. (2000) J. Bacteriol. 182, 5663–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morino M., Natsui S., Swartz T. H., Krulwich T. A., Ito M. (2008) J. Bacteriol. 190, 4162–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krulwich T. A., Hicks D. B., Swartz T. H., Ito M. (2007) in Physiology and Biochemistry of Extremophiles (Gerday C., Glansdorff N. eds) pp. 311–329, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 24.Mesbah N. M., Cook G. M., Wiegel J. (2009) Mol. Microbiol. 74, 270–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich T., Weiss H. (1997) J. Theor. Biol. 187, 529–540 [DOI] [PubMed] [Google Scholar]
- 26.Efremov R. G., Baradaran R., Sazanov L. A. (2010) Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 27.Kajiyama Y., Otagiri M., Sekiguchi J., Kudo T., Kosono S. (2009) Microbiology 155, 2137–2147 [DOI] [PubMed] [Google Scholar]
- 28.Goldberg E. B., Arbel T., Chen J., Karpel R., Mackie G. A., Schuldiner S., Padan E. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2615–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swartz T. H., Ito M., Ohira T., Natsui S., Hicks D. B., Krulwich T. A. (2007) J. Bacteriol. 189, 3081–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J., Frisch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 31.Nozaki K., Kuroda T., Mizushima T., Tsuchiya T. (1998) Biochim. Biophys. Acta 1369, 213–220 [DOI] [PubMed] [Google Scholar]
- 32.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 33.Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 34.Kuroda T., Shimamoto T., Mizushima T., Tsuchiya T. (1997) J. Bacteriol. 179, 7600–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher N., Rich P. R. (2000) J. Mol. Biol. 296, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 36.Yang X., Yu L., He D., Yu C. A. (1998) J. Biol. Chem. 273, 31916–31923 [DOI] [PubMed] [Google Scholar]
- 37.Amarneh B., Vik S. B. (2003) Biochemistry 42, 4800–4808 [DOI] [PubMed] [Google Scholar]
- 38.Steuber J. (2001) J. Bioenerg. Biomembr. 33, 179–186 [DOI] [PubMed] [Google Scholar]
- 39.Kosono S., Kajiyama Y., Kawasaki S., Yoshinaka T., Haga K., Kudo T. (2006) Biochim. Biophys. Acta 1758, 627–635 [DOI] [PubMed] [Google Scholar]
- 40.Torres-Bacete J., Sinha P. K., Castro-Guerrero N., Matsuno-Yagi A., Yagi T. (2009) J. Biol. Chem. 284, 33062–33069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Bacete J., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2007) J. Biol. Chem. 282, 36914–36922 [DOI] [PubMed] [Google Scholar]
- 42.Euro L., Belevich G., Verkhovsky M. I., Wikström M., Verkhovskaya M. (2008) Biochim. Biophys. Acta 1777, 1166–1172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.