Abstract
The glucagon receptor belongs to the B family of G-protein coupled receptors. Little structural information is available about this receptor and its association with glucagon. We used the substituted cysteine accessibility method and three-dimensional molecular modeling based on the gastrointestinal insulinotropic peptide and glucagon-like peptide 1 receptor structures to study the N-terminal domain of this receptor, a central element for ligand binding and specificity. Our results showed that Asp63, Arg116, and Lys98 are essential for the receptor structure and/or ligand binding because mutations of these three residues completely disrupted or markedly impaired the receptor function. In agreement with these data, our models revealed that Asp63 and Arg116 form a salt bridge, whereas Lys98 is engaged in cation-π interactions with the conserved tryptophans 68 and 106. The native receptor could not be labeled by hydrophilic cysteine biotinylation reagents, but treatment of intact cells with [2-(trimethylammonium)ethyl]methanethiosulfonate increased the glucagon binding site density. This result suggested that an unidentified protein with at least one free cysteine associated with the receptor prevented glucagon recognition and that [2-(trimethylammonium)ethyl]methanethiosulfonate treatment relieved this inhibition. The substituted cysteine accessibility method was also performed on 15 residues selected using the three-dimensional models. Several receptor mutants, despite a relatively high predicted cysteine accessibility, could not be labeled by specific reagents. The three-dimensional models show that these mutated residues are located on one face of the protein. This could be part of the interface between the receptor and the unidentified inhibitory protein, making these residues inaccessible to biotinylation compounds.
Keywords: Adenylate Cyclase (Adenylyl Cyclase), Cell Surface Receptor, G Protein-coupled Receptors (GPCRs), Protein Folding, Receptor Structure-Function
Introduction
The G-protein coupled receptors related to the secretin receptor (1), known as “family B” receptors, form a very small but very important subfamily of G protein-coupled receptors. This family includes, among others, the receptors for parathyroid hormone, glucagon, gastrointestinal insulinotropic peptide (GIP),4 glucagon-like peptide 1 (GLP-1), corticotrophin-related factor (CRF), or growth hormone-releasing hormone. Although the two receptor families probably evolved from a common ancestor, rhodopsin-related (family A) and family B receptors present few similarities (1). Except for two cysteine residues in the first and second extracellular loops, none of the signature amino acids defining family A G protein-coupled receptors are identified in family B receptors. It is therefore impossible to extrapolate the structural information gathered on rhodopsin activation (2, 3) to these receptors.
All family B receptors possess a large extracellular N-terminal domain of 100–150 amino acids, which contains highly conserved residues and three conserved disulfide bonds. Truncated and hybrid peptide recognition by chimeric receptors indicate that these domains anchor the cognate agonist peptides through their C-terminal region, whereas the N-terminal regions of the peptides interact with and activate the receptors via the heptahelical transmembrane domains (4, 5).
The three-dimensional structures of the N-terminal domain of the CRF2β, PAC1, GLP-1, and GIP receptors and of the CRF1 or parathyroid hormone receptor N-terminal region fused with the maltose-binding protein, in complex with their respective agonist or antagonist ligands, have been determined by NMR or x-ray diffraction studies (6–12). These structures share a common fold stabilized by three conserved disulfide bridges and by an ionic bridge involving a conserved aspartate.
The scanning cysteine accessibility method (SCAM), has been used to study the lining of the pore of channels and transport proteins (13, 14). It was then extended to the study of the binding site of Cys-loop acetylcholine receptors (15) and, more recently, to G protein-coupled receptors (16). It consists of the systematic individual substitution of the amino acids in the region of interest into cysteine, followed by analysis of the reactivity of the newly introduced side chain to cysteine reagents.
Before applying the SCAM to the glucagon receptor, we verified that the wild type receptor did not possess any accessible cysteine by testing whether the receptor could be extracted after biotinylation with a hydrophilic cysteine reagent. We also tested the effect of MTSET on glucagon binding and receptor activation. Our results suggested that 50% or more of the glucagon receptors transiently expressed on HEK cells are masked by an accompanying protein.
Based on the structures of the homologous receptors of GIP and of GLP-1, we constructed two models of the glucagon receptor N-terminal domain. Using these models to guide mutagenesis experiments and to help interpret the experimental results, we evaluated the accessibility of substituted cysteines predicted to lie on the protein surface. These data reveal a region inaccessible to biotinylation that is located in loops positioned on a common face, opposite to the junction between the N-terminal and transmembrane domains. We propose that this surface could associate with the accompanying protein already suggested by the MTSET experiments.
EXPERIMENTAL PROCEDURES
Modeling
The positions of the signal peptide (residues 1–26) and of the transmembrane helices, intracellular (IC) and extracellular (EC) regions were predicted using the SignalP 3.0 server and the Predict Protein server, respectively (17, 18). They correspond to the residue segment 27–144 for the N-terminal domain, 194–226 for EC1, 285–305 for EC2, and 369–384 for EC3 and 167–173 for IC1, 250–263 for IC2, 327–350 for IC3, and 405–477 for IC4.
A BLAST search (19) identified two proteins strongly homologous to the N-terminal extracellular domain of human glucagon receptor: the human GIP receptor (GIPR) N-terminal domain (Protein Data Bank code 2QKH) and the glucagon-like peptide-1 human receptor (GLP-1R) N-terminal domain (Protein Data Bank codes 3C59 and 3C5T). A multiple-sequence alignment of these sequences accessed at the Universal Protein Resource (UniProt) was performed using ClustalW2 (20) (see Fig. 1). This multiple-sequence alignment served as a basis to construct the models using MODELLER 9v1 (21). Modeler builds models by satisfying different types of spatial restraints, which include homology-derived restraints, stereochemical restraints obtained from the CHARMM22 force field, and statistical preferences for dihedral angles and non-bonded distances obtained from a representative set of protein structures. Two models were constructed using as templates the structure of GIPR (Protein Data Bank code 2QKH) and GLP-1R (Protein Data Bank code 3C5T).
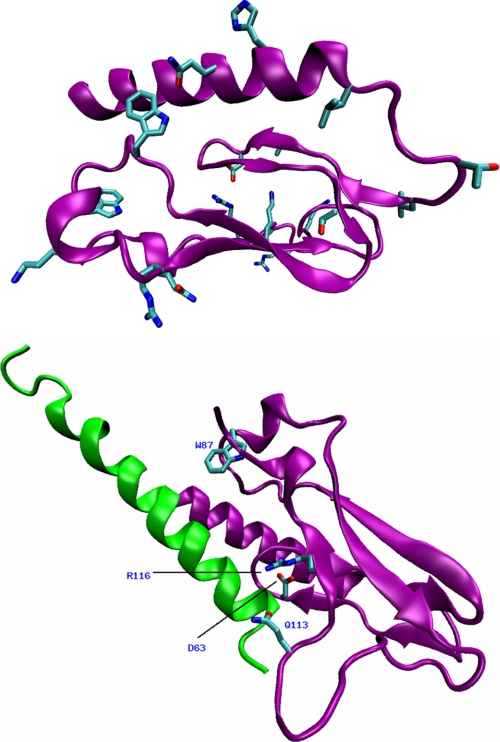
FIGURE 1.
Top, two views of the superposition of the three-dimensional models of the N-terminal domain of glucagon receptor constructed using the GIPR and GLP-1R crystal structures. The models built with GIPR and GLP-1R are represented by a purple and green ribbon, respectively. The three disulfide bridges, Asp63, Lys98, and Arg116 are depicted as sticks and colored according to the following color scheme (cyan, carbon; red, oxygen; blue, nitrogen; yellow, sulfur). A salt bridge is formed between Asp63 and Arg116. Similar interactions are observed in the respective x-ray structure templates. Bottom, multiple sequence alignment of the human N-terminal domains of the glucagon, GIP, and GLP-1 receptors used to generate the three-dimensional models of the glucagon receptor. Sequence identity is highlighted in gray.
Peptide Synthesis
Unlabeled glucagon was synthesized in our laboratory as described previously (22).
Receptor Expression
HEK 293-T cells were maintained in DMEM enriched with 10% fetal calf serum, penicillin (10 milliunits/ml), streptomycin (10 μg/ml) (PAA Laboratories, Linz, Austria). Generation of the truncated and point-mutated receptors was achieved using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as described previously (22). We transfected either 18 μg of plasmid per 10-cm Petri dish by the calcium phosphate precipitation method (23) to measure cAMP levels in response to glucagon or 1 μg of plasmid/well (6-well plate) by the PEI transfection method (24) for binding studies and to evaluate the glucagon receptor extraction by dot blot. The cells were used 48–72 h after transfection.
Cysteine Reagents
MTSET (Toronto Research Chemicals, Toronto, Canada) and maleimide-PEO2 biotin ((+)-biotinyl-3-maleimidopropionamidyl-3,6-dioxaoctanediamine, Pierce-Perbio Science (Aalst, Belgium)) were solubilized in ice-cold PBS immediately before use. N-Biotinylaminoethyl methanethiosulfonate (MTSEA)-biotin (Toronto Research Chemicals) was dissolved in DMSO and diluted in ice-cold PBS immediately before use.
Membrane Preparation
106 transfected HEK 293-T cells were harvested mechanically and rinsed in PBS. The cells were pelleted and lysed in 1 mm NaHCO3 solution followed by immediate freezing in liquid nitrogen; the lysate was unfrozen and centrifuged for 15 min at 20,000 × g, and the pellet was resuspended in PBS. When the membranes were prepared for Western blots or streptavidin extraction, the cell lysis and resuspension solutions were enriched with CompleteTM protein inhibitors (Roche Applied Science).
Binding Studies
Binding studies were performed as described previously (22). The density of the wild type receptors, evaluated by analysis of homologous 125I-glucagon/glucagon competition curves obtained in the presence of 10 μm GTP varied between 1.5 and 3.0 nmol/mg protein.
We also performed binding assays on control and KCl-treated membranes. Membranes prepared as above (22) were resuspended in 20 mm Tris-HC1 buffer (pH 7.5) containing 0.25 m sucrose and 5 mm MgC12. An equal volume of 20 mm Tris-HC1 buffer (pH 7.5), enriched with 0.25 m sucrose without (control) or with (treated) 2.5 m KC1, was added dropwise. This mixture was left for 1 h in an ice bath, under continuous stirring, and then centrifuged at 30,000 × g for 10 min. The resulting pellets were washed three times in 5 ml of 20 mm Tris-HC1 buffer (pH 7.5) containing 0.25 m sucrose.
Western Blots
Fifty μg of membrane proteins were solubilized at room temperature in Laemmli sample buffer, resolved by SDS-PAGE using a 10% gel, and electrotransferred on a PVDF membrane. This membrane was then blocked for 1 h in PBS enriched with 5% dry milk powder (Nestlé) and 1‰ Tween 20, incubated for 1 h in the presence of a rabbit antiserum raised against a peptide corresponding to the C-terminal glucagon receptor sequence (SAETPLAGGLPRLAESPF (1:25,000 in the blocking buffer)), and washed in PBS-Tween (1‰). The signal was detected by chemiluminescence using an HRP-coupled goat antirabbit antibody and the West-Pico Supersignal reagent (Pierce) according to the manufacturer's instructions.
Adenylate Cyclase Activity
For adenylate cyclase activation studies, transfected HEK 293-T cells were labeled in their culture medium for 3 h in the presence of 1 μCi/ml [3H]adenine, mechanically harvested, centrifuged at 500 g for 5 min, and rinsed in PBS. HEK cells were resuspended in 200 μl of PBS and treated for 10 min at 25 °C in the absence or presence of a fresh solution of MTSET (1 mm). The reaction was stopped by the addition of 3.5 ml of DMEM enriched with the phosphodiesterase inhibitors isobutylmethylxanthine (1 mm) and Ro 20–1724 (4-(3-butoxy-4-methoxybenzyl)-2-imidazolidinone) (0.1 μm) (Sigma). cAMP synthesis was evaluated after a 30-min incubation of 100,000 HEK cells in a total volume of 120 μl at 37 °C, in the absence or presence of 1 μm vasoactive intestinal peptide, 10 μm forskolin (used as positive controls), or 0.1 nm to 10 μm glucagon. The reaction was stopped by the addition of 1 ml of 1% SDS enriched with 0.5 mm ATP and 0.75 mm cAMP; [3H]cAMP was separated from the other nucleotides by chromatography (25).
Biotinylation and Biotin Pullout
Intact cells (or membranes) were treated for 10 min at 25 °C in 120 μl of PBS with MTSEA-biotin (1 mm, 1% DMSO) or maleimide-PEO2 biotin (0.4 mm) and then washed four times at 4 °C, resuspended in ice-cold 1 mm NaHCO3 enriched with CompleteTM, frozen in liquid nitrogen, and stored at −80 °C until use. The homogenate was unfrozen and centrifuged at 20,000 × g for 15 min. The membrane pellet was resuspended in radioimmune precipitation buffer (PBS enriched with 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 1 mm EDTA (pH 7.4)). Eight to ten μg of solubilized protein were incubated overnight at 4 °C under mild agitation in the absence or presence of immobilized streptavidin (50 μl of settled resin) (Pierce) in a total volume of 400 μl. We then allowed the resin to settle for at least 20 min at 4 °C and blotted 200 μl of the supernatant for glucagon receptor detection and 50 μl of the supernatant for biotin detection on PVDF membranes. The membrane was blocked and incubated with the antireceptor antibody as explained above (Western blot). The receptor signal was detected with an anti-rabbit antibody, and biotin was detected with streptavidin, both labeled with alkaline phosphatase, using the Western Blue reagent, according to the manufacturer's instructions (Promega, Leiden, Netherlands). The blots were immediately scanned, and the signal was quantified using the Scion Image program (National Institutes of Health). We verified by serial dilutions that both signals were proportional to the protein concentration. Membranes from non-transfected or non-biotinylated HEK 293T cells did not give any detectable signals in these assays.
Streptavidin-extracted MTSEA-biotinylated proteins can be cleaved off by reducing agents at high temperature or in the presence of 2 m guanidinium hydrochloride (26), but solubilized receptors treated under these conditions could not be immunodetected. We attempted to replace streptavidin by “soft link avidin” to facilitate the glucagon receptor release, but this induced a strong nonspecific signal in our glucagon receptor immunodetection system. Protein precipitation by ethanol (9 volumes, overnight at −20 °C) or TCA (10% at 4 °C) as well as solubilization in SDS resulted in cumulative decreases in the receptor immunodetection; it was not possible to detect the receptor in Western blot after extraction of small protein quantities (<10 μg) and larger protein concentrations saturated the streptavidin. We therefore resorted to dot blots of the streptavidin-extracted supernatant in view of the following arguments: 1) there was no background in either Western blot or dot blot after streptavidin extraction of non-transfected cells; 2) the wild type or mutant receptor degradation observed in Western blot was minimal; and 3) the biotinylated cysteine 42 (Q42C mutant) was not separated from the C-terminal epitope during the solubilization and extraction procedure.
Statistics
All competition curves and dose-effect curves were analyzed by non-linear regression (GraphPad Prism 3.0, GraphPad Software, San Diego, CA). Competition curves obtained in the absence of GTP were biphasic (not shown) but monophasic in the presence of GTP, making the estimate of the receptor concentration much easier. Statistical analyses were performed with the program “InStat” (GraphPad Software); the pEC50 values and effect of MTSET on mutant receptors were compared with wild type by one-way analysis of variance, using the Dunnett post-test. p < 0.05 was accepted as being significant.
RESULTS
Three-dimensional Models of the N-terminal Domain of the Glucagon Receptor
Two three-dimensional models of the human N-terminal domain of the glucagon receptor were constructed using as templates the three-dimensional structure of the GIPR in complex with the GIP hormone and of the GLP-1R in complex with exendin-4. The GIPR and GLP-1R sequences exhibit significant amino acid identity of 47 and 46%, respectively, to the glucagon N-terminal receptor region (see Fig. 1, bottom). Such percentages of identity make these templates good candidates to elaborate three-dimensional models of the glucagon receptor by comparative modeling. The stereochemistry of the models was assessed with Procheck (27). The Ramachandran plots indicate that a high percentage of the nonglycine and nonproline residues are located in the most favored allowed regions: 93.2 and 94.4% for the model built using the GIPR and GLP-1R receptors, respectively. The two models feature three disulfide bonds (Cys58–Cys100, Cys43–Cys67, and Cys81–Cys121) and one salt bridge, Asp63–Arg116, matching those in their respective template. Three hydrophobic clusters are pinpointed. The central one features intramolecular interactions involving conserved residues, Trp68 and Trp106, which sandwich Lys98 and Val96. The GIPR and GLP-1R three-dimensional models superpose rather well (root mean square deviations of the positions = 0.9 Å calculated for the Cα atoms excluding those of the loop, which is missing in GIP (see Fig. 1)). As shown in Fig. 1 (top), the main difference between the two models resides in the conformation of the 6-residue insertion 108–113, for which there is no equivalent in the GIPR template. In the GIPR-based model, this insertion was built ab initio.
Immunodetection of the Receptor
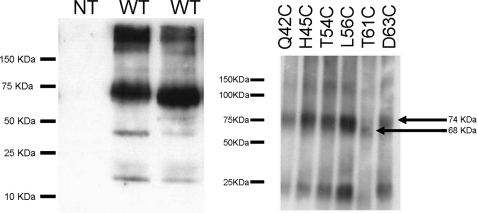
The glucagon receptor expression was analyzed by Western blot (Fig. 2). We observed major bands with apparent molecular mass of 74 and >100 kDa that may correspond to receptor dimer and aggregates and minor band(s) of <50 kDa. All of the mutant receptors gave the same profile in Western blot, with the exception of the T61C mutant that had a slightly lower apparent molecular mass (68 kDa), probably due to the disappearance of the Asn59 consensus glycosylation site (Fig. 2).
FIGURE 2.
Left, Western blot of membranes from non-transfected cells (NT; lane 1) and cells transiently expressing the wild type glucagon receptor (WT; lanes 2 and 3). Right, Western blot of the Q42C, H45C, T54C, L56C, T61C, and D63C mutant receptors (membranes from different transfections). The glucagon receptor was visualized using an antibody raised against a C-terminal peptide.
Effect of Mutations of Asp63, Lys98, and Arg116
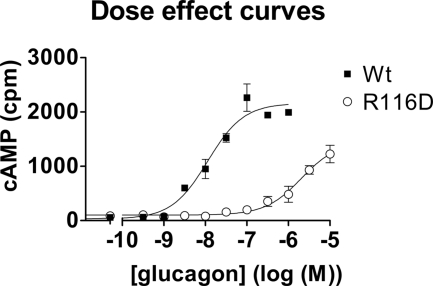
We studied the effect of mutation of the conserved Asp63 and Lys98 and also of Arg116 that was predicted by our three-dimensional models to form an ionic bond with Asp63. Adenylate cyclase stimulation through the D63K, D63R, or K98D receptors was barely detectable at 10 μm glucagon (not shown). Glucagon had also a markedly decreased affinity for the R116D receptor mutants (Fig. 3). The double mutants (charge switch) D63K/K98D and D63R/R116D did not restore the glucagon potency (data not shown). Specific 125I-glucagon binding to the mutant receptors was non-significant, but the expression of all mutants, verified by dot blot (not shown), was comparable with wild type.
FIGURE 3.
Comparison of glucagon dose-effect curves on HEK 293-T cells transfected with the “wild type” glucagon receptor (closed squares) or with the R116D mutant (open circles). The pEC50 values in this experiment were 7.89 ± 0.09 and 5.39 ± 0.05. Results shown are representative of three experiments. Error bars, S.E.
Effect of MTSET on the Wild Type Receptor
Before embarking on the glucagon receptor SCAM, we treated intact cells with the hydrophilic cysteine reagent, MTSET. We then examined whether this reagent impairs the receptor recognition. A positive result would indicate that the affected cysteine is close to the binding site. MTSET treatment of transiently transfected HEK cells did not inhibit but rather improved by 2–3-fold the glucagon potency for cAMP stimulation (Table 1 and Fig. 4, top).
TABLE 1.
Glucagon potency (pEC50) and effect of MTSET on the glucagon dose-effect curves
Experiments were performed in duplicate (n = 3).
| Receptor construct | pEC50 | MTSET-pEC50 shifta |
|---|---|---|
| Wild type | 8.5 ± 0.05 | 0.3 ± 0.1 |
| C171S | 7.0 ± 0.2b | 0.6 ± 0.3 |
| C224S | 6.6 ± 0.1b | 0.5 ± 0.2 |
| C240S | 8.3 ± 0.2 | 0.7 ± 0.2 |
| C287S | 8.5 ± 0.2 | 0.5 ± 0.2 |
| C294S | 9.1 ± 0.3 | 0.3 ± 0.3 |
| C401S | 7.9 ± 0.2 | 0.7 ± 0.3 |
a MTSET pretreatment significantly improved the glucagon potency (p < 0.01) at all of the receptors tested.
b Different from wild type (p < 0.05).
FIGURE 4.
Top, effect of MTSET on glucagon dose-effect curves. HEK 293-T cells transiently transfected with the glucagon receptor were treated in the absence (closed symbols) or presence (open symbols) of MTSET prior to cAMP measurement in the absence or presence of the indicated glucagon concentrations. The glucagon potency increased in this experiment from 7.7 ± 0.1 to 8.0 ± 0.08. Results shown are representative of 50 experiments. Bottom, 125I-glucagon competition curves obtained in the presence of 10 μm GTP, using membranes prepared from control (closed squares) or MTSET-treated cells (open triangles). The glucagon pIC50 in this experiment was 7.17 ± 0.15 in control membranes and 7.09 ± 0.09 in membranes from treated cells. Inset, Scatchard representation of the binding data. The receptor concentration, calculated under the assumption that glucagon and 125I-glucagon have the same affinity for the glucagon receptor, increased in this experiment from 1.13 pmol/mg protein (95% confidence interval; 1.08–1.75 pmol/mg protein) in membranes from control cells to 2.7 pmol/mg protein (95% confidence interval; 2.4–3.2 pmol/mg protein). Results shown are representative of five experiments. Error bars, S.E.
In order to test whether MTSET pretreatment increased glucagon receptor affinity, we analyzed 125I-glucagon saturation curves. The results suggested that the effect of MTSET on the glucagon dose effect curves was due to an increase of the 125I-glucagon receptor density (from 2 ± 0.5 to 6.5 ± 0.6 nmol/mg protein, n = 5 experiments), not of their affinity for glucagon (Fig. 4, bottom). The receptor concentration per se (evaluated in dot blot) did not change after MTSET treatment (not shown). This result suggested that, in transiently transfected HEK cells, at least half of the glucagon receptors were masked and became accessible to glucagon only after MTSET pretreatment.
Effect of Cysteine to Serine Mutations
If a single cysteine from the receptor was responsible for the effect of MTSET on glucagon recognition, its mutation to serine (an almost isosteric amino acid) should give rise to an MTSET-insensitive receptor. Our models suggested that all six N-terminal domain cysteines are involved in disulfide bridges. We mutated the other six cysteines (Cys171, Cys224, Cys240, Cys287, Cys294, and Cys401) to serine. MTSET had a similar effect on the glucagon potency of all of these mutant receptors (Table 1).
If mutation of cysteine 294 as well as cysteines 240, 287, and 401 to serine did not significantly affect the glucagon potency (Table 1), in contrast, mutation to serine of cysteine 224 (which may be close to the glucagon binding site) and of the intracellular cysteine 171 (which may belong to the G protein binding site) induced a significant decrease of the glucagon potency, despite almost normal receptor protein expression (Table 1). Binding to these two receptor mutants was not sufficient for meaningful competition curves analysis (Bmax and KD determination).
Biotinylation and Extraction of the Wild Type Receptor
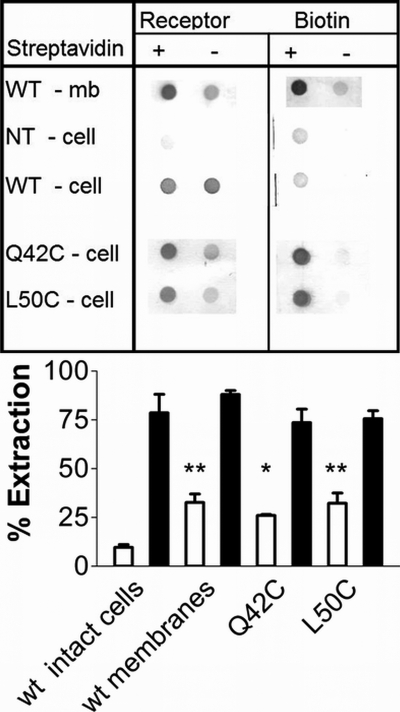
To further test the hypothesis that one or more of the receptor cysteines could react with hydrophilic reagents, we treated intact cells or membranes either with MTSEA-biotin (a MTSET-related reagent) or with maleimide-PEO2-biotin and then extracted the biotinylated proteins with streptavidin. The wild type receptor could be extracted only if membranes rather than intact cells were biotinylated (Fig. 5); neither PEO2-biotin nor MTSEA-biotin labeled extracellular cysteine(s).
FIGURE 5.
Top, membranes from cells expressing the wild type receptor (row 1); intact, non-transfected HEK cells (row 2); or cells expressing the transiently transfected wild type (row 3), Q42C (row 4), or L50C (row 5) glucagon receptors were treated with maleimide-PEO2 biotin before streptavidin extraction. Membrane samples were solubilized and extracted in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of immobilized streptavidin. The supernatants were blotted and tested for the presence of glucagon receptor (columns 1 and 2) or biotin (columns 3 and 4). Results shown are representative of 3–6 experiments. Bar graph, extraction of the glucagon receptor (gray bars) and of total biotin (used as control; white bars), expressed as a percentage of the signal observed in non-extracted samples. The average ± S.D. (error bars) of 3–6 experiments is shown. *, significantly different from WT (p < 0.05); **, significantly different from WT (p < 0.01).
Taken together, our results supported the hypothesis that the N-terminal cysteines 43–67, 58–100, and 81–121 indeed form disulfide bridges (as suggested in our two models) and that extracellular cysteines 224, 287, and 294 were buried in the protein interior. In contrast, at least one intracellular cysteine, possibly Cys171, was exposed and water-accessible. These data also suggested that MTSET did not affect directly the receptor but targeted another protein, non-covalently associated with the glucagon receptor.
Effect of High Salt Wash
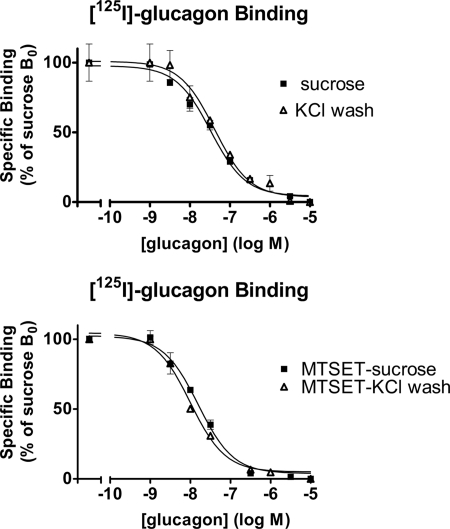
We washed membranes 1h in a high salt concentration to remove extrinsic proteins before measuring 125I-glucagon binding to control and MTSET-treated membranes. The high salt wash decreased the membrane protein yield but did not affect glucagon binding per mg of protein in the presence of GTP (Fig. 6), indicating that the receptor-associated protein is either transmembrane or anchored to the plasma membrane.
FIGURE 6.
125I-Glucagon competition curves obtained in the presence of GTP, on control (closed squares) and KCl-washed membranes (open triangles). The protein yield was reduced after a KCl wash of the membranes; 125I-glucagon binding was normalized with respect to the protein concentrations. The results are expressed as percentage of specific binding/mg of protein to control membranes in the absence of glucagon. Top, competition curves obtained on membranes from non-treated cells. The glucagon potencies (pEC50) were 7.48 ± 0.06 and 7.38 ± 0.1 on control and KCl-washed membranes, respectively. Bottom, competition curves obtained on membranes from MTSET-treated cells. The glucagon potencies (pEC50) were 7.80 ± 0.05 and 8.02 ± 0.09 on control and KCl-washed membranes, respectively. Results shown are representative of three experiments in duplicate. Error bars, S.E.
Effect of Cysteine Substitutions
D63C, K98C, and R116C receptor mutants could not be extracted after maleimide-biotin treatment (Table 2). This can be rationalized by our models which attest the low solvent surface accessibility (SASA) values of these residues (Table 2).
TABLE 2.
Glucagon potency (pEC50), receptor extraction, and cysteine accessibility
Experiments were performed in duplicate (n ≥ 3).
| Receptor construct | pEC50 | Extracted | SASAa GIPR-based model | SASAa GLP-1R-based model |
|---|---|---|---|---|
| Wild type | 8.5 ± 0.05 | No | ||
| D63C | 4.9 ± 0.1b | No | 0.1 | 0.2 |
| K98C | 4.7 ± 0.1b | No | 0.1 | 0.0 |
| R116C | 5.4 ± 0.3b | No | 0.4 | 0.2 |
| Q42C | 6.6 ± 0.1b | Yes | 0.7 | 0.5 |
| H45C | 6.5 ± 0.2b | Yes | 0.6 | 0.6 |
| L50C | 6.0 ± 0.2b | Yes | 0.3 | 0.4 |
| T54C | 6.7 ± 0.3b | No | 1.0 | 0.7 |
| L56C | 7.0 ± 0.3b | No | 0.8 | 0.5 |
| T61C | 5.9 ± 0.2b | No | 0.5 | 0.4 |
| T71C | 7.0 ± 0.3b | No | 0.1 | 0.0 |
| W83C | 5.9 ± 0.1b | No | 0.7 | 0.3 |
| W87C | 4.9 ± 0.1b | Yes | 0.2 | 0.3 |
| K90C | 6.0 ± 0.1b | Yes | 0.4 | 0.9 |
| R94C | 5.3 ± 0.1b | No | 0.4 | 0.0 |
| R108C | 6.5 ± 0.1b | No | 0.6 | 0.6 |
| R111C | 5.9 ± 0.1b | No | 1.0 | 1.0 |
| Q113C | 6.2 ± 0.1b | No | 0.9 | 0.8 |
| Q120C | 5.5 ± 0.3b | No | 0.4 | 0.5 |
a Fraction of solvent accessibility of the mutated residue, computed for the side chain in three-dimensional models featuring the Cys mutation, calculated by the Vadar program (35).
b Different from wild type (p < 0.05).
To identify the associated protein footprint on the glucagon receptor, 15 additional residues were selected using the three-dimensional models and replaced individually by a cysteine residue. Three are located in the long N-terminal helix, and six are distributed over the β-strands of the large and small β-sheets. The others belong to the loops or the small helices. These residues spanned almost all of the faces of the three-dimensional structures (see Fig. 7), and the majority of the introduced cysteines featured a non-negligible SASA value in our models (see Table 2). Despite a very low SASA value, Thr71 and Trp87 were also chosen for substitution because their thiol group in the modeled cysteine was oriented toward the protein surface and partially accessible.
FIGURE 7.
Top, representation of the 15 residues selected to be substituted individually by cysteine (Gln42, His45, Leu50, Thr54, Leu56, Thr61, Thr71, Trp83, Trp87, Lys90, Arg94, Arg108, Arg111, Gln113, and Gln120) in the three-dimensional model structure of glucagon receptor. Bottom, three-dimensional model of the glucagon receptor N-terminal domain complexed to the glucagon peptide. The purple and green ribbons depict the glucagon receptor and peptide, respectively. Asp63, Arg116, Trp87, and Gln113 are drawn as sticks and colored according to the following color scheme (cyan, carbon; red, oxygen; blue, nitrogen).
The receptor expression levels evaluated in dot blot were normal. The Q42C, H45C, L50C, W87C, and K90C mutants could be extracted by streptavidin after protein biotinylation (Fig. 5 and Table 2). All of the other mutants could not be labeled by maleimide-PEO2-biotin (Table 2). All of the N-terminal cysteine mutants had a reduced affinity for glucagon (Table 2).
DISCUSSION
As observed for other members of the secretin receptor-like receptor family (see Ref. 28 and references therein), mutation of the conserved aspartate (Asp63) impaired markedly the receptor function (Table 2). The importance of this residue is explained by our two three-dimensional models because it features a hydrogen bond with Trp68 and a salt bridge with Arg116. As for the CRF2β receptor (28), our attempts to reconstruct an ionic bridge by charge reversal (D63K/K98D and D63R/R116D glucagon receptor mutants) were unproductive, confirming that the conserved salt bridge is not the only major interaction involving Asp63. Mutation of Lys98 had a more profound inhibitory effect than mutation of Arg116, indicating a more important role for the lysine (Fig. 3 and Table 2). Both three-dimensional models suggest that formation of a salt bridge between Asp63 and Lys98 is possible only by means of a side chain rotation. Lys98 is, however, sandwiched between two tryptophans: Trp68, which also interacts with Asp63, and Trp106, with which it forms cation-π interactions. Substitution of Asp63 or Lys98 probably perturbed this cluster, which contributes to the structure scaffold.
Pretreatment of intact cells expressing wild type glucagon receptors with the hydrophilic reagent, MTSET, did not affect the glucagon affinity but increased the 125I-glucagon binding site density (Fig. 4). It is well known that if the receptor density is high, maximum effects can be achieved by an agonist concentration occupying only a small portion of the receptors, a phenomenon known as “spare receptors” or “receptor reserve” (29). When this occurs, the agonist potency (pEC50) is lower than expected from its affinity constant and further decreases with increasing receptor concentrations. The observation that MTSET increased the glucagon receptor accessibility therefore explains the increase of the apparent potency of glucagon in functional assays (Fig. 4 and Table 1).
It is often believed, by analogy with rhodopsin-related receptors, that the conserved cysteines 224 and 294 form a disulfide bridge, maintaining the first and second extracellular loops in close proximity to the glucagon binding site (30). If this hypothesis was correct, mutation of either or both cysteines should lead to a similar impairment of glucagon recognition (due to the loss of the same disulfide bridge) (31). Our results did not support this hypothesis because glucagon binding was different for each Cys → Ser mutant receptor.
Mutation of the transmembrane domain Cys171, Cys224, Cys240, Cys287, Cys294, and Cys401 did not hamper the effect of MTSET (Table 1). In addition, MTSEA-biotin and maleimide-PEO2-biotin did not label the glucagon receptor in intact cells, although they did label the Q42C mutant receptor as well as one or more intracellular cysteines, possibly Cys171, on the wild type receptor (Fig. 5). We therefore concluded that the glucagon receptor extracellular cysteines are either implicated in disulfide bridges (residues 43–67, 58–100, and 81–121 in our three-dimensional models) or buried inside the protein and that the extracellular cysteine(s) targeted by MTSET belong to another (receptor-associated) protein. Taken together, our results thus suggested that a significant proportion of the glucagon receptors expressed in HEK cells were masked by an unidentified protein (Fig. 4) that could not be washed away by high salt (Fig. 6) and that MTSET relieved this inhibition by targeting an accessible cysteine on this associated protein (Fig. 5).
At first sight, a potential candidate could be the transmembrane protein RAMP2, which is known to interact with the glucagon receptor (32). It belongs to a family of three receptor activity-modifying proteins (RAMP1, -2, and -3) that affect the glycosylation, membrane trafficking and ligand recognition properties of the calcitonin receptor-like receptor, a family B receptor distantly related to the glucagon receptor (33). However, similar to the homologous RAMP1 expressed in Escherichia coli (34), the four extracellular cysteines of RAMP2 are probably involved in two conserved disulfide bridges and therefore not available to cysteine reagents. RAMP2 is therefore unlikely to be responsible for the effect of MTSET on glucagon recognition in HEK cells.
To identify the associated protein footprint on the glucagon receptor, we evaluated the accessibility of 15 additional side chains, mutated into cysteine, by biotinylation and streptavidin extraction. Although our three-dimensional models led us to expect that the thiol group of most of our 15 mutated residues would be relatively well solvent-exposed, only five receptor mutants (Q42C, H45C, L50C, W87C, and K90C) could in fact be extracted after biotinylation. Reexamination of our three-dimensional models allowed us to propose an interpretation to some of these results, as described below.
Thr71 is one of the mutants that could not be extracted. Our models show that the T71C cysteine Sγ atom is only at about 4 Å from the sulfur atoms of the cystine 58–100, thus close enough to form non-native disulfide bonds with either Cys58 or Cys100. Similarly, the cysteine Sγ atoms of R94C and of Q120C are only 3.5–8 Å from the cystine 81–121 sulfur atoms, and the distances measured between the W83C Sγ atom and the cystine 43–67 sulfur atoms, although longer, were also in the range of potential disulfide bond formation (<9 Å). If the introduced cysteines were involved in non-native disulfide bonds, one of the native cysteines would become free but might remain buried in the protein interior. The glucagon potencies at three of the four mutant receptors (W83C, R94C, and Q120C) were low (<6.0), compatible with the idea that the mutant receptor structure had been altered. The T71C glucagon receptor probably retained a “native-like” structure, as suggested by its comparatively high glucagon potency; however, cysteine 71 featured a low fraction of SASA value (Table 1), which explains its lack of reactivity to the bulky maleimide molecule.
Surprisingly, one of the biotinylatable cysteines, W87C, displayed in our models a rather low SASA value. The models, however, showed that Trp87 is enclosed in a hydrophobic pocket shaped by Leu85, Pro86, and Met123 and hydrogen-bonds via its side chain NH group to Cys121, which is itself engaged in a disulfide bond. Mutation W87C is likely to perturb this cluster, leading to a disruption of the receptor structure, solvent exposure of the Cys87 residue, and receptor extraction after treatment by maleimide PEO2 biotin. This hypothesis is supported by the very low glucagon potency at this receptor mutant, comparable with the Asp63 or Lys98 receptor mutants (Table 2).
The T54C, L56C, T61C, R108C, R111C, and Q113C mutants display rather high cysteine SASA values in our models, with a thiol group pointing toward the protein exterior. Glucagon had a potency higher than 6.0 for four of these six mutants (see Table 2), suggesting that their three-dimensional structure was not notably altered. Thr54 and Leu56 are located in one of the receptor loops close to Thr61, whereas Arg108, Arg111, and Gln113 belong to an adjacent loop (see Fig. 7). They are positioned on a common face, opposite to the junction between the N-terminal and transmembrane domains. This face is a potential candidate to participate in the interface between the glucagon receptor and its associated protein.
We also modeled the structure of the N-terminal domain of the glucagon receptor complexed to glucagon. We resorted to our glucagon receptor model, the crystal structure of glucagon (Protein Data Bank code 1GCN), and the GIP-GIPR or GLP-1-GLP-1R x-ray structures (Fig. 7). In these modeled structures, the glucagon peptide is positioned at distances less than 6 Å from Asp63, Trp87, and Arg116 (three very important positions; see Table 2) and within 4–8 Å from Gln113. If the unidentified associated protein indeed interacts with (among other residues) Gln113 and neighboring residues, that would indeed prevent glucagon binding by steric hindrance. It will be important to identify this associated protein in order to evaluate its potential physiological importance in glucagon-responsive cells.
This work was supported by Fonds de la Recherche Scientifique Médicale Grant 3.4541.03 and by an “Action de Recherche Concertée” from the Communauté Française de Belgique.
- GIP
- gastrointestinal insulinotropic peptide
- GIPR
- GIP receptor
- GLP-1
- glucagon-like peptide 1
- GLP-1R
- GLP-1 receptor
- MTSET
- [2-(trimethylammonium)ethyl]methanethiosulfonate
- pEC50
- −log(EC50)
- pIC50
- −log(IC50)
- SCAM
- scanning cysteine accessibility method
- CRF
- corticotrophin-related factor
- MTSEA
- N-biotinylaminoethyl methanethiosulfonate.
REFERENCES
- 1.Fredriksson R., Lagerström M. C., Lundin L. G., Schiöth H. B. (2003) Mol. Pharmacol. 63, 1256–1272 [DOI] [PubMed] [Google Scholar]
- 2.Ahuja S., Smith S. O. (2009) Trends Pharmacol. Sci. 30, 494–502 [DOI] [PubMed] [Google Scholar]
- 3.Hofmann K. P., Scheerer P., Hildebrand P. W., Choe H. W., Park J. H., Heck M., Ernst O. P. (2009) Trends Biochem. Sci. 34, 540–552 [DOI] [PubMed] [Google Scholar]
- 4.Gourlet P., Vilardaga J. P., De Neef P., Waelbroeck M., Vandermeers A., Robberecht P. (1996) Peptides 17, 825–829 [DOI] [PubMed] [Google Scholar]
- 5.Runge S., Wulff B. S., Madsen K., Bräuner-Osborne H., Knudsen L. B. (2003) Br. J. Pharmacol. 138, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grace C. R., Perrin M. H., DiGruccio M. R., Miller C. L., Rivier J. E., Vale W. W., Riek R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12836–12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grace C. R., Perrin M. H., Gulyas J., Digruccio M. R., Cantle J. P., Rivier J. E., Vale W. W., Riek R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parthier C., Kleinschmidt M., Neumann P., Rudolph R., Manhart S., Schlenzig D., Fanghänel J., Rahfeld J. U., Demuth H. U., Stubbs M. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13942–13947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun C., Song D., Davis-Taber R. A., Barrett L. W., Scott V. E., Richardson P. L., Pereda-Lopez A., Uchic M. E., Solomon L. R., Lake M. R., Walter K. A., Hajduk P. J., Olejniczak E. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7875–7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pioszak A. A., Xu H. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pioszak A. A., Parker N. R., Suino-Powell K., Xu H. E. (2008) J. Biol. Chem. 283, 32900–32912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Runge S., Thøgersen H., Madsen K., Lau J., Rudolph R. (2008) J. Biol. Chem. 283, 11340–11347 [DOI] [PubMed] [Google Scholar]
- 13.Akabas M. H., Stauffer D. A., Xu M., Karlin A. (1992) Science 258, 307–310 [DOI] [PubMed] [Google Scholar]
- 14.Javitch J. A. (1998) Methods Enzymol. 296, 331–346 [DOI] [PubMed] [Google Scholar]
- 15.Stauffer D. A., Karlin A. (1994) Biochemistry 33, 6840–6849 [DOI] [PubMed] [Google Scholar]
- 16.Javitch J. A., Shi L., Liapakis G. (2002) Methods Enzymol. 343, 137–156 [DOI] [PubMed] [Google Scholar]
- 17.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 18.Rost B., Yachdav G., Liu J. (2004) Nucleic Acids Res. 32, W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) Curr. Protoc. Bioinformatics, Chapter 5, 5.6.1–5.6.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perret J., Van Craenenbroeck M., Langer I., Vertongen P., Gregoire F., Robberecht P., Waelbroeck M. (2002) Biochem. J. 362, 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan M., Schallhorn A., Wurm F. M. (1996) Nucleic Acids Res. 24, 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godbey W. T., Wu K. K., Mikos A. G. (1999) J. Control Release 60, 149–160 [DOI] [PubMed] [Google Scholar]
- 25.Salomon Y., Londos C., Rodbell M. (1974) Anal. Biochem. 58, 541–548 [DOI] [PubMed] [Google Scholar]
- 26.Savage M. D., Mattson G., Desai S., Nielander G. W., Morgensen S., Conklin E. G. (1992) Avidin-Biotin Chemistry: A Handbook, pp. 273–298, Pierce Chemical Co., Rockford, IL [Google Scholar]
- 27.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 28.Perrin M. H., Grace C. R., Digruccio M. R., Fischer W. H., Maji S. K., Cantle J. P., Smith S., Manning G., Vale W. W., Riek R. (2007) J. Biol. Chem. 282, 37529–37536 [DOI] [PubMed] [Google Scholar]
- 29.Stephenson R. P. (1956) Br. J. Pharmacol. Chemother. 11, 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Authier F., Desbuquois B. (2008) Cell. Mol. Life. Sci. 65, 1880–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vilardaga J. P., Di Paolo E., Bialek C., De Neef P., Waelbroeck M., Bollen A., Robberecht P. (1997) Eur. J. Biochem. 246, 173–180 [DOI] [PubMed] [Google Scholar]
- 32.Christopoulos A., Christopoulos G., Morfis M., Udawela M., Laburthe M., Couvineau A., Kuwasako K., Tilakaratne N., Sexton P. M. (2003) J. Biol. Chem. 278, 3293–3297 [DOI] [PubMed] [Google Scholar]
- 33.Morfis M., Christopoulos A., Sexton P. M. (2003) Trends Pharmacol. Sci. 24, 596–601 [DOI] [PubMed] [Google Scholar]
- 34.Kusano S., Kukimoto-Niino M., Akasaka R., Toyama M., Terada T., Shirouzu M., Shindo T., Yokoyama S. (2008) Protein Sci. 17, 1907–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willard L., Ranjan A., Zhang H., Monzavi H., Boyko R. F., Sykes B. D., Wishart D. S. (2003) Nucleic Acids Res. 31, 3316–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]