Abstract
The present study demonstrated that murine protein serine/threonine kinase 38 (MPK38) coimmunoprecipitates with Smad proteins (Smad2, -3, -4, and -7) and that this association is mediated by the catalytic kinase domain of MPK38. The association between MPK38 and Smad2, -3, and -4 was significantly increased by TGF-β or ASK1 signals, whereas these signals decreased association of MPK38 with Smad7. MPK38 stimulated TGF-β-induced transcription required for TGF-β-mediated biological functions, such as apoptosis and cell growth arrest, in a kinase-dependent manner. Knockdown of endogenous MPK38 showed an opposite effect, inhibiting TGF-β signaling. MPK38-mediated phosphorylation of Smad proteins (Ser245 of Smad2, Ser204 of Smad3, Ser343 of Smad4, and Thr96 of Smad7) was also found to be crucial to the positive regulation of TGF-β signaling induced by MPK38. In addition, MPK38 enhanced nuclear translocation of Smad3, as well as redistribution of Smad7 from the nucleus to the cytoplasm, in response to TGF-β. Together, these results indicate that MPK38 functions as a stimulator of TGF-β signaling through direct interaction with and phosphorylation of Smad proteins.
Keywords: Protein Kinases, Protein Phosphorylation, Protein-Protein Interactions, SMAD Transcription Factor, Transforming Growth Factor β (TGFβ), Murine Protein Serine/Threonine Kinase 38 (MPK38), Maternal Embryonic Leucine Zipper Kinase (Melk)
Introduction
Type I transforming growth factor-β (TGF-β) receptor kinases are serine/threonine kinases with roles in a wide array of cellular processes. Phosphorylation of the C-terminal SXS motif in receptor-regulated Smads (R-Smads) by these kinases is a crucial step in TGF-β family signaling (1–3). R-Smad phosphorylation results in the formation of heterodimeric complexes with the common Smad, Smad4. In addition, the phosphorylation of serine or threonine residues in the N-terminal or linker region has been observed in endogenous R-Smads (4). Several lines of evidence have demonstrated the existence of kinases responsible for N-terminal or linker region phosphorylation events, including mitogen-activated protein kinases (MAPKs), Ca2+ and calmodulin-dependent kinase II, cyclin-dependent kinase (CDK),3 protein kinase C (PKC), and G protein-coupled receptor kinase 2 (5). Common Smad, Smad4, is constitutively phosphorylated in cells, although not all of the phosphorylation sites are known. Extracellular signal-regulated kinase (ERK) has been shown to phosphorylate Smad4 at Thr277, resulting in the prevention of nuclear translocation of Smad4 (6). The inhibitory Smads (I-Smads), Smad6 and Smad7, are phosphorylated by uncharacterized kinases (7, 8). This provides a mechanism for integration of the Smad pathway with other signaling pathways, in which kinases act as essential mediators, which modulate Smad-mediated signaling. Thus, the identification and characterization of additional cellular kinases responsible for Smad phosphorylation could lead to a better understanding of the regulatory role of Smad phosphorylations, especially in the N-terminal and linker region, in TGF-β signaling.
Murine protein serine/threonine kinase 38 (MPK38), also known as pEg3 kinase and maternal embryonic leucine zipper kinase (Melk), is a member of the AMP-activated protein kinase family of serine-threonine kinases (9, 10). The expression patterns of MPK38 during maturation of oocytes and pre-implantation development were found to be typical of that described for maternally expressed genes, including β-actin and E-cadherin (11, 12). This suggested that MPK38 may play a role in transducing signals in embryonic cells during pre-implantation, although its exact roles in development were not revealed. Additionally, MPK38 is also expressed in several other tissues, including embryonic stem cells (13), adult germ cells (14), hematopoietic stem cells (15), adult hematopoietic cells (9), and neural stem cells (13). At the functional level, MPK38 has been implicated to be involved in several cellular functions, including cell cycle, spliceosome assembly, gene expression, cell proliferation, carcinogenesis, and apoptosis (15–19). However, the exact biological function of MPK38 remains unclear.
In this study, we show that there are direct physical and functional interactions between MPK38 and Smad proteins (Smad2, -3, -4, and -7), and that these interactions may play an important role in the regulation of Smad activities involved in TGF-β signaling.
MATERIALS AND METHODS
Cell Culture, Transfection, and in Vitro Interaction
HEK293, HepG2, Hep3B, and HaCaT cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Invitrogen). Transfection using WelFect-ExTM Plus has been previously described (17, 20). To determine the in vitro interaction between MPK38 and Smad proteins, purified recombinant MPK38 was autophosphorylated as described previously (17). Autophosphorylated MPK38 (∼3 μg) was incubated with unlabeled recombinant GST or GST-Smad proteins (each ∼5 μg) at room temperature for 1 h, and then analyzed using native PAGE.
Plasmids, Antibodies, and the Inducible MPK38 shRNA Cell Line
The wild-type and kinase-dead (K40R) MPK38 plasmids, p3TP-Lux and p21-Luc reporter plasmids, FLAG-tagged Smad2, -3, -4, and -7, and four Smad3 deletion constructs (MH1(L), MH1, MH2(L), and MH2) have been described previously (17, 20). Anti-Smad2, anti-Smad3, anti-Smad4, anti-Smad7, anti-FLAG (M2), anti-hemagglutinin (HA), anti-histone H2B, anti-CDK4, anti-Cyclin D1, anti-PAI-1, anti-p21, anti-β-actin, and anti-phospho-Ser/Thr antibodies have been described previously (17, 20, 22). Anti-MPK38 and anti-GST antibodies were described previously (23, 24). Alexa Fluor-594 anti-mouse and Alexa Fluor-488 anti-rabbit secondary antibodies were purchased from Molecular Probes Inc. (Eugene, OR). An inducible MPK38 shRNA NIH 3T3 cell line was generated using the following oligonucleotides: forward primer, 5′-TCGAGGCAGGCAGACAATGGAGGATTTCAAGAGAATCCTCCATTGTCTGCCTGCTTTTTTA-3′, containing an MPK38 sequence (underlined); and reverse primer, 5′-AGCTTAAAAAAGCAGGCAGACAATGGAGGATTCTCTTGAAATCCTCCATTGTCTGCCTGCC-3′, containing an MPK38 sequence (underlined), as described previously (25). Inducible MPK38 shRNA stable clones were screened in the presence of 450 μg/ml G418 for 14 days until all control parental NIH 3T3 cells completely died.
Construction of Smad Mutants
Smad mutants used for the in vitro kinase assays were generated by PCR under reaction parameters described previously (25). The following primers were used: for Smad2: forward (5′-GCGAATTCATGTCGTCCATCTTGCCATTC-3′) and reverse (5′-GCCTCGAGTTATGACATGCTTGAGCAACG-3′); for Smad3: forward (5′-GCGAATTCATGTCGTCATCCTGCCTTTC-3′) and reverse (5′-GCCTCGAGCTAAGACACACTGGAACAGCG-3′); for Smad4: forward (5′-GCGAATTCATGGACAATATGTCTATTACG-3′) and reverse (5′-GCGCCTCGAGTCAGTCTAAAGGTTGTGGGTC-3′); for Smad7: forward (5′-GCGAATTCATGTTCAGGACCAAACGATCT-3′) and reverse (5′-GCCTCGAGCTACCGGCTGTTGAAGATGAC-3′). These primers were used in conjunction with one of the following mutant primers: for Smad2(S245A): forward (5′-ATGGACACAGGCGCTCCAGCAGAACTA-3′) and reverse (5′-TAGTTCTGCTGGAGCGCCTGTGTCCAT-3′); for Smad2(S465A): forward (5′-TCAGTGCGTTGCTCAGCCATGTCATAACTCGAGGC-3′) and reverse (5′-GCCTCGAGTTATGACATGGCTGAGCAACGCACTGA-3′); for Smad2(T8A): forward (5′-GCGAATTCATGTCGTCCATCTTGCCATTCGCGCCG-3′) and reverse (5′-CGGCGCGAATGGCAAGATGGACGACATGAATTCGC-3′); for Smad3(S204A): forward (5′-ATGGACGCAGGTGCACCAAACCTATCC-3′) and reverse (5′-GGATAGGTTTGGTGCACCTGCGTCCAT-3′); for Smad3 (S423A): forward (5′-AGCATCCGCTGTTCCGCTGTGTCTTAGCTCGAGGC-3′) and reverse (5′-GCCTCGAGCTAAGACACAGCGGAACAGCGGATGCT-3′); for Smad3(T8A): forward (5′-GCGAATTCATGTCGTCCATCCTGCCTTTCGCTCCC-3′) and reverse (5′-GGGAGCGAAAGGCAGGATGGACGACATGAATTCGC-3′); for Smad4(S343A): forward (5′-TTTAAGGTTCCTGCAAGCTGCCCTATT-3′) and reverse (5′-AATAGGGCAGCTTGCAGGAACCTTAAA-3′); for Smad4 (S344A): forward (5′-AAGGTTCCTTCAGCCTGCCCTATTGTT-3′) and reverse (5′-AACAATAGGGCAGGCTGAAGGAACCTT-3′); for Smad4 (S403A): forward (5′-GTCAGGTGCCTTGCTGACCACGCGGTC-3′) and reverse (5′-GACCGCGTGGTCAGCAAGGCACCTGAC-3′); for Smad7(T96A): forward (5′-CTGAAGGCGCTCGCGCACTCGGTGCTC-3′) and reverse (5′-GAGCACCGAGTGCGCGAGCGCCTTCAG-3′); for Smad7(S249A): forward (5′-GGGCTTTCAGATGCCCAACTTCTTCTG-3′) and reverse (5′-CAGAAGAAGTTGGGCATCTGAAAGCCC-3′); and for Smad7(S365A): forward (5′-TTCCCCGGTTTCGCCATCAAGGCTTTC-3′) and reverse (5′-GAAAGCCTTGATGGCGAAACCGGGGAA-3′). The amplified PCR products were digested with EcoRI and XhoI, and ligated into pGEX4T-1 (Amersham Biosciences).
Preparation of Recombinant Proteins and MPK38 Kinase Assay
Recombinant glutathione S-transferase (GST) fusion vectors (pGEX4T-1) containing wild-type and mutant forms of Smads (Smad2, -3, -4, and -7) and wild-type MPK38, as well as recombinant His-tagged wild-type MPK38, which were expressed in Escherichia coli, were purified by affinity chromatography on glutathione-Sepharose 4B or His columns (Amersham Biosciences). MPK38 proteins purified from HEK293 cells transfected with GST-MPK38 by glutathione-Sepharose beads or the recombinant MPK38 proteins were incubated at 37 °C for 15 min with the recombinant GST-tagged Smads (each ∼5 μg) or ZPR9 (∼500 ng) substrates in kinase buffer (50 mm HEPES, pH 7.4, 1 mm DTT, and 10 mm MgCl2) and 5 μCi of [γ-32P]ATP as described previously (17). The reaction mixtures were separated by SDS-PAGE and analyzed by autoradiography. Protein concentration was determined by the Bradford assay.
RNA Interference
MPK38-specific siRNA (5′-CAGGCAGACAAUGGAGGAUTT-3′) targeting a coding region (amino acids 297–303) of MPK38 (17) and a control scrambled siRNA (5′-GCGCGGGGCACGUUGGUGUTT-3′) (26) were transfected into HEK293, HepG2, or HaCaT cells using the WelFect-ExTM Plus method according to the manufacturer's instructions.
Luciferase Reporter Assay
HepG2 cells were transfected with the p3TP-Lux or p21-Luc reporter plasmids, along with each expression vector as indicated, using WelFect-ExTM Plus. The cells were lysed, and luciferase activity was detected using the dual luciferase assay kit (Promega, Madison, WI) (17). The data were normalized to the expression levels of a cotransfected β-galactosidase reporter control.
Apoptosis Assay
The number of HaCaT cells undergoing apoptosis after treatment with TGF-β1 (2 ng/ml for 20 h) was quantified using green fluorescent protein (GFP) (17, 27). The percentage of apoptotic cells was calculated as the number of GFP-positive cells with apoptotic nuclei divided by the total number of GFP-positive cells.
FACS Analysis
Assays were performed using HaCaT cells (2 × 105/60-mm dish) transfected with the indicated combinations of plasmid vectors (empty vector and Smad3) and siRNA duplexes (MPK38 and control scrambled siRNAs) as described previously (21). The fraction of cells in each stage of the cell cycle was analyzed after 10% serum treatment for 24 h in the presence or absence of TGF-β1 (2 ng/ml). Flow cytometry analysis was performed using the FACSCalibur-S system (BD Biosciences).
Confocal Microscopy
Assays were performed using Hep3B cells transfected with FLAG-Smads (Smad3 and -7) and/or GST-tagged wild-type and kinase-dead (K40R) MPK38 on sterile coverslips as described previously (20). In brief, the cells were incubated with mouse anti-FLAG (M2), diluted 1:1000 in PBS, or rabbit anti-MPK38, diluted 1:200 in PBS, for 2 h at 37 °C. The cells were then incubated with Alexa Fluor-594 anti-mouse or Alexa Fluor-488 anti-rabbit secondary antibodies, diluted 1:1000 in PBS, at 37 °C for 1 h. Proteins were visualized using a Leica Dmire2 confocal microscope (Germany).
Statistical Analysis
All experiments were repeated at least three times, and results are expressed as the mean ± S.D. A p value relative to control of p < 0.05, as calculated by Student's t test, was considered statistically significant.
RESULTS
MPK38 Interacts with Smad Proteins
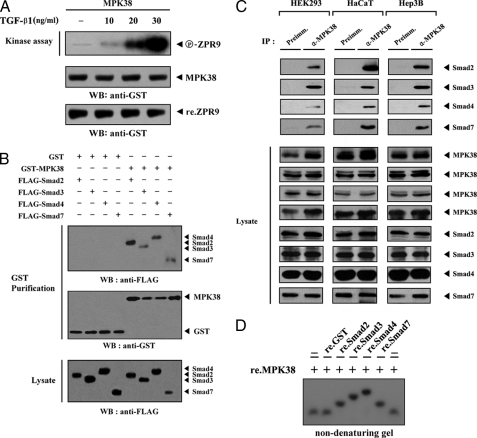
We have found previously that MPK38 physically interacts with ASK1 and stimulates H2O2-mediated apoptosis (17), suggesting the involvement of MPK38 in other death signaling in cells. In addition, our previous results showed that MPK38 can interact with serine-threonine kinase receptor-associated protein (28), which binds to Smad proteins such as Smad2, Smad3, and Smad7 (data not shown). Based on these findings, we hypothesized that MPK38 might regulate TGF-β signaling through the modulation of Smad proteins. To test this, we first analyzed MPK38 activity using an in vitro kinase assay in TGF-β-stimulated Hep3B cells. As shown in Fig. 1A, MPK38 precipitate from cell lysates treated with TGF-β1 significantly induced MPK38 kinase activity in a dose-dependent manner compared with untreated MPK38 precipitate. These results indicated that a functional link between MPK38 and TGF-β signaling pathways may exist in cells. Next, to examine whether MPK38 physically binds to Smad proteins, we performed binding assays in HEK293 cells. These results showed that FLAG-tagged Smad proteins (Smad2, -3, -4, and -7) interact with GST-MPK38 (Fig. 1B). The kinase-dead (K40R) MPK38 also retained the ability to associate with Smad proteins (data not shown), suggesting that the kinase activity of MPK38 is not required for Smads binding. To confirm the interaction of MPK38 with Smad proteins in cells, we also performed coimmunoprecipitation experiments with endogenous MPK38 and Smad proteins using three different cell lines, including HEK293, HaCaT, and Hep3B. As shown in Fig. 1C, MPK38 endogenously interacted with Smad proteins (Smad2, -3, -4, and -7). This interaction was also confirmed by reciprocal coimmunoprecipitation experiments in which anti-Smad antibodies, instead of anti-MPK38 antibody, were used for immunoprecipitation (supplemental Fig. S1). To assess direct interaction between MPK38 and Smad proteins, we also carried out a non-denaturing PAGE analysis using purified recombinant MPK38 and Smad proteins. Autophosphorylated recombinant MPK38 was incubated with unlabeled, recombinant Smad proteins, or with GST alone as a nonspecific control. As shown in Fig. 1D, a shift in the mobility of 32P-labeled MPK38 was clearly evident upon incubation in the presence of Smad proteins, but was undetectable when 32P-labeled MPK38 was incubated with GST alone (in the absence of Smad proteins), providing clear evidence of a physical association between MPK38 and Smad proteins.
FIGURE 1.
Interaction between MPK38 and Smad proteins. A, TGF-β1 stimulates MPK38 kinase activity. Hep3B cells transfected with wild-type GST-MPK38 were incubated with increasing amounts of TGF-β1 for 20 h, and MPK38 was purified on glutathione-Sepharose beads. The GST precipitates were subjected to an in vitro kinase assay using ZPR9 as a substrate (17), followed by SDS-PAGE and autoradiography. The circled P-ZPR9 indicates the phosphorylated ZPR9. B, FLAG-tagged Smads (Smad2, -3, -4, and -7) were cotransfected with GST-MPK38 or vector alone (GST) into HEK293 cells. GST fusion proteins were purified on glutathione-Sepharose beads (GST Purification), and complex formation between MPK38 and Smad proteins (top panel) was determined by immunoblot analysis using an anti-FLAG antibody. C, association of MPK38 with Smad proteins in cells. Cell lysates from HEK293, HaCaT, or Hep3B cells were subjected to immunoprecipitation using either rabbit preimmune serum (Preimm.) or anti-MPK38 antibody (α-MPK38), followed by immunoblot analyses using anti-Smad2, -3, -4, and -7 antibodies, to determine the complex formation between endogenous MPK38 and Smad proteins (Smad2, -3, -4, and -7). As a control, the expression levels of MPK38 and Smads in the total cell lysate were analyzed by immunoblot using anti-MPK38 and anti-Smad2, -3, -4, and -7 antibodies, respectively (Lysate). D, in vitro association of MPK38 with Smad proteins. For native PAGE (8%) of the MPK38-Smads complex, autophosphorylated MPK38 was incubated with unlabeled recombinant GST alone or GST-Smads as described under “Materials and Methods.” IP, immunoprecipitation; re., recombinant; WB, Western blot.
Identification of the Interaction Domains between MPK38 and Smad Proteins
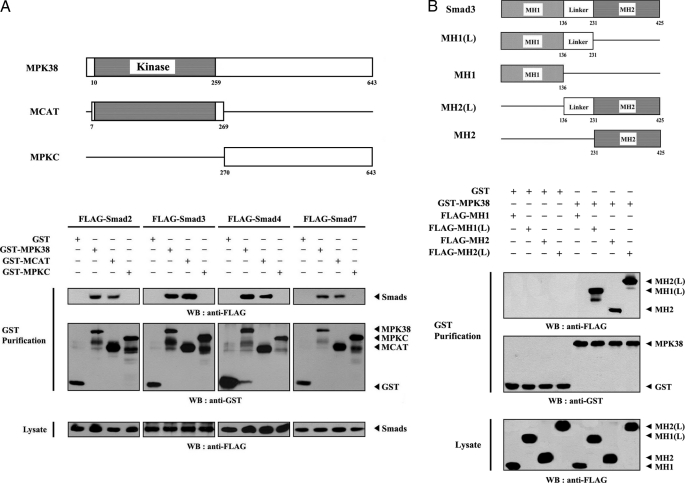
To examine the binding domain of MPK38 involved in association with Smad proteins, we performed binding assays in cells using two MPK38 deletion constructs: GST-MCAT, comprising the N-terminal kinase domain (amino acids 7–269), and GST-MPKC, harboring the C-terminal regulatory domain (amino acids 270–643), as described previously (17). Wild-type MPK38 and MCAT were shown to interact with Smad2, -3, -4, and -7, whereas MPKC (the construct with the N-terminal kinase domain deletion) was unable to interact with these Smad proteins (Fig. 2A), indicating that the interaction with Smad proteins is mediated via the N-terminal kinase domain of MPK38. Next, to determine the critical domains in Smad3 for MPK38 binding, four FLAG-tagged Smad3 deletion constructs (20), FLAG-MH1 (amino acids 1–136), FLAG-MH1(L) (amino acids 1–231), FLAG-MH2 (amino acids 231–425), and FLAG-MH2(L) (amino acids 136–425), were cotransfected with GSA-MPK38 or GST alone into HEK293 cells. The interaction between MPK38 and Smad3 deletion constructs was then examined via Western blot with an anti-FLAG antibody using GST precipitates. As shown in Fig. 2B, MPK38 interacted with three Smad3 deletion constructs (MH1(L), MH2, and MH2(L)), but not with the MH1 construct. Together, these results imply that the association of MPK38 and Smad proteins in cells is mediated via the N-terminal kinase domain of MPK38 and the Linker/MH2 region of Smad3.
FIGURE 2.
Mapping of the binding site involved in MPK38-Smads complex formation. The schematic structures of wild-type and deletion constructs of MPK38 (A) and Smad3 (B) are indicated. Numbers indicate the amino acid residues corresponding to the domain boundaries. A, mapping of MPK38 domains involved in Smads binding. HEK293 cells were cotransfected with GST alone or GST-MPK38 constructs (wild-type, MCAT, and MPKC), together with FLAG-Smads (Smad2, -3, -4, and -7), and purified with glutathione-Sepharose beads (GST purification). The amount of Smad proteins bound to MPK38 constructs was determined by Western analysis using an anti-FLAG antibody (top panel). The same stripped blot was re-probed with an anti-GST antibody to determine the expression of GST fusion proteins in the coprecipitates (middle panel). B, mapping of Smad3 domains involved in MPK38 binding. HEK293 cells were transfected with vector alone (GST) or GST-MPK38 in combination with the indicated FLAG-Smad3 deletion constructs (MH1, MH1(L), MH2, and MH2(L)), and cell lysates were purified using glutathione-Sepharose beads (GST purification). Complex formation between MPK38 and Smad3 deletion constructs was determined by immunoblotting with anti-FLAG antibody (top panel).
MPK38 Phosphorylates Smad Proteins
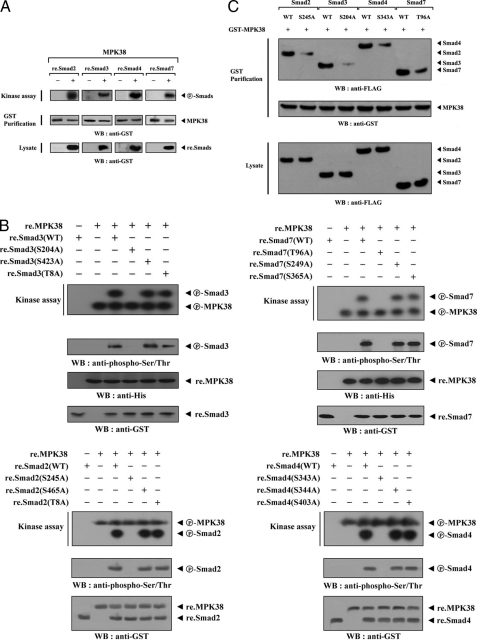
To determine whether Smad proteins (Smad2, -3, -4, and -7) can act as MPK38 substrates, recombinant wild-type Smad proteins were expressed in E. coli, purified, and used for the MPK38 kinase assay. Extracts from HEK293 cells expressing GST-MPK38 were purified with glutathione-Sepharose beads and incubated with [γ-32P]ATP to allow phosphorylation of the recombinant Smad proteins. The phosphorylation of all recombinant Smad proteins in the presence of MPK38 was observed, indicating that Smad2, -3, -4, and -7 can act as substrates for MPK38 (Fig. 3A). Next, to better characterize the MPK38 phosphorylation sites of Smad proteins, we performed an alignment analysis using the AMP-activated protein kinase consensus sequence (29) Hyd-(X,Basic)XX(S/T)XXX-Hyd (where Hyd = M, L, I, F, or V and Basic = R > K > H), because MPK38 is a member of the AMP-activated protein kinase family and three potential MPK38 phosphorylation sites on each Smad protein were selected. Through in vitro kinase assays using recombinant MPK38 and Smad substitution mutants, we found that the Smad3(S204A) mutant completely abolished MPK38-mediated Smad3 phosphorylation (Fig. 3B, upper left, 3rd lane versus 4th lane). However, MPK38-mediated phosphorylation was clearly observed in the presence of other Smad3 substitution mutants (S423A and T8A). These results indicate that MPK38 phosphorylation might occur at Ser204 of Smad3. Similarly, the mutation of Smad7 Thr96 to Ala96 completely abolished MPK38-mediated Smad7 phosphorylation. Other Smad7 mutants (S249A and S365A), on the other hand, permitted phosphorylation, indicating that the Thr96 of Smad7 represents a potential phosphorylation site for MPK38 (Fig. 3B, upper right, 3rd lane versus 4th lane). We also identified Ser245 of Smad2 and Ser343 of Smad4 as potential phosphorylation sites for MPK38 (Fig. 3B, lower panels). These results suggest that MPK38 directly phosphorylates Smad proteins through physical interaction. To investigate whether MPK38-mediated phosphorylation of Smad proteins can influence MPK38-Smads interaction, we compared the association of MPK38 and either wild-type or mutant Smad proteins. Four Smad substitution mutants found to be defective in MPK38-mediated phosphorylation, as described above, were used for this comparison (S245A of Smad2, S204A of Smad3, S343A of Smad4, and T96A of Smad7) (see Fig. 3B). The association between MPK38 and Smad proteins was considerably decreased for all four Smad mutants when compared with the wild-type controls (Fig. 3C), indicating an important role for MPK38 phosphorylation at Ser245 of Smad2, Ser204 of Smad3, Ser343 of Smad4, and Thr96 of Smad7 in the regulation of MPK38-Smads interaction.
FIGURE 3.
Phosphorylation of Smad proteins by MPK38. A, MPK38-mediated phosphorylation of Smad proteins. Approximately 3–4 μg of recombinant Smad proteins (Smad2, -3, -4, and -7) were mixed with MPK38 proteins purified with glutathione-Sepharose beads (GST Purification) from HEK293 cell lysates containing GST-MPK38, and in vitro kinase assays were then performed as described under “Materials and Methods.” B, identification of MPK38 phosphorylation sites on Smad proteins. For an in vitro kinase assay, 5 μg of recombinant wild-type Smad proteins (Smad2, -3, -4, and -7) or one of their substitution mutants were mixed with 10 μm ATP, 5 μCi of [γ-32P]ATP, and 10 mm MgCl2 in 20 μl of kinase buffer and incubated with the recombinant wild-type MPK38 (10 μg) for 15 min at 37 °C (top panels). The phosphorylation of Smads was also determined by immunoblotting with an anti-phospho-Ser/Thr antibody (middle panels). The circled P-MPK38 and circled P-Smads indicate the autophosphorylated MPK38 and phosphorylated Smads, respectively. C, effect of MPK38-mediated phosphorylation of Smad proteins on MPK38-Smad complex formation. FLAG-tagged, wild-type Smad proteins (Smad2, -3, -4, and -7) or their substitution mutants (S245A of Smad2, S204A of Smad3, S343A of Smad4, and T96A of Smad7), defective in MPK38-mediated phosphorylation, were cotransfected with GST-MPK38 into HEK293 cells. GST fusion proteins were purified on glutathione-Sepharose beads (GST Purification), and complex formation between MPK38 and Smad proteins (top panel) was determined by immunoblot analysis using an anti-FLAG antibody. WT, wild-type.
TGF-β and ASK1 Signals Differentially Regulate MPK38-Smads Complex Formation
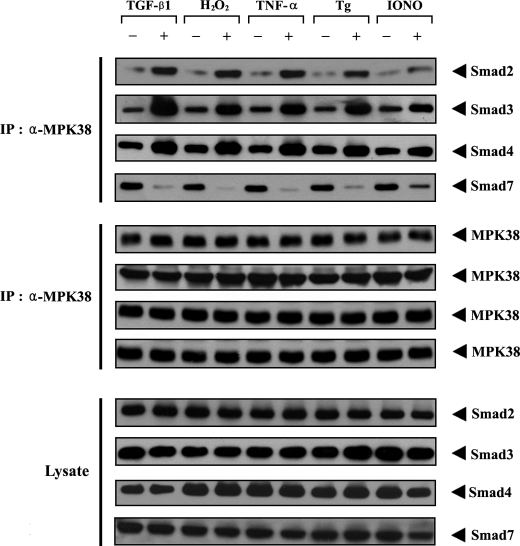
In addition to binding to Smad proteins, MPK38 can stimulate ASK1-mediated signaling (17). We therefore investigated whether TGF-β and ASK1 stimuli, including H2O2, TNF-α, endoplasmic reticulum stress (thapsigargin), and calcium overload (ionomycin), can influence MPK38-Smads complex formation. Upon TGF-β1, H2O2, TNF-α, thapsigargin, or ionomycin treatment, the association between MPK38 and Smad2, -3, and -4 was considerably increased compared with control untreated HEK293 cells (Fig. 4, 1st to 3rd panels). However, exposure of cells to TGF-β1, H2O2, TNF-α, thapsigargin, and ionomycin resulted in a significant decrease in MPK38-Smad7 complex formation (Fig. 4, 4th panel). These data indicate that the endogenous interaction between MPK38 and Smad proteins (Smad2, -3, -4, and -7) appears to be differentially regulated by TGF-β and ASK1 stimulation, suggesting that a functional link between the MPK38 and TGF-β signaling pathways may occur via Smad proteins in cells.
FIGURE 4.
Regulation of MPK38-Smads association by TGF-β and ASK1 stimulation. HEK293 cell lysates were treated with or without the following stimuli: TGF-β1 (100 pm, 20 h), H2O2 (2 mm, 30 min), TNF-α (500 ng/ml, 30 min), thapsigargin (Tg: 20 μm, 30 min), or ionomycin (IONO: 1 μm, 24 h). They were then immunoprecipitated with an anti-MPK38 antibody (α-MPK38), followed by immunoblotting with anti-Smad2, -3, -4, and -7 antibodies to determine the endogenous association between MPK38 and Smad proteins (top panels). The expression level of MPK38 in the immunoprecipitates was determined by immunoblot using an anti-MPK38 antibody (middle panels).
MPK38 Stimulates TGF-β-induced Transcription in a Kinase-dependent Manner
Given that MPK38 forms a complex with and phosphorylates Smad proteins (see Figs. 1–3), we next examined whether MPK38 could have an effect on TGF-β-induced transcription. To examine the effect of MPK38 on TGF-β-induced transcription, we transfected HepG2 cells with increasing amounts of wild-type or kinase-dead MPK38, together with the p3TP-Lux reporter or the p21-Luc reporter plasmid (20), in the presence or absence of TGF-β1. Wild-type MPK38 significantly increased TGF-β-induced transcription in a dose-dependent manner (Fig. 5A, lane 2 versus lanes 3–5). However, the expression of kinase-dead MPK38 had no effect on TGF-β-induced transcription (Fig. 5A, lane 2 versus lanes 6–8). These data indicated that the stimulation of TGF-β-induced transcription by MPK38 is dependent on its kinase activity. Consistently, the knockdown of endogenous MPK38 by MPK38-specific siRNA resulted in a dose-dependent decrease in TGF-β-induced transcriptional activity in the presence of both the p3TP-Lux and the p21-Luc reporter systems, whereas the control scrambled siRNA had no effect (Fig. 5B). To further investigate the role of endogenous MPK38 in TGF-β signaling, we employed an inducible MPK38 shRNA system to deplete the expression of endogenous MPK38 in NIH 3T3 cells. The knockdown of endogenous MPK38 resulted in a significant down-regulation of TGF-β targets, such as plasminogen activator inhibitor-1 (PAI-1), the cyclin-dependent kinase inhibitor p21Cip1, and Smad7, as well as up-regulation of CDK4 and Cyclin D1, which are involved in TGF-β-induced G1 arrest, compared with either parental NIH 3T3 cells or a control expressing the empty vector alone (Fig. 5C, 1st to 5th panels). NIH 3T3 cells (inducible MPK38 shRNA) stably expressing the pSingle-tTS-shRNA vector harboring MPK38-specific shRNA showed a doxycycline-dependent RNA interference effect on endogenous MPK38 silencing, whereas stable NIH 3T3 cells containing the empty vector alone (Vector) or parental NIH 3T3 cells showed no effect in the presence of doxycycline (Fig. 5C, 6th panel). These findings clearly suggest that MPK38 physically associates with Smad proteins and stimulates TGF-β-induced transcription.
FIGURE 5.
Stimulation of TGF-β-induced transcription by MPK38. A and B, HepG2 cells were transfected as described under “Materials and Methods” with increasing amounts of MPK38 (WT, wild-type; K40R, kinase-dead), MPK38-specific siRNA, or control scrambled siRNA, as indicated, and 0.3 μg of p3TP-Lux or 0.3 μg of p21-Luc reporter plasmid, and incubated in the presence or absence of 100 pm TGF-β1. Luciferase activity was measured 48 h after transfection. C, effect of MPK38 on TGF-β targets. NIH 3T3 cells harboring stably integrated pSingle-tTS-shRNA empty vector (Vector) or pSingle-tTS-shRNA vector containing MPK38-specific shRNA (inducible MPK38 shRNA), as well as Parental NIH 3T3 cells (NIH 3T3), were lysed and subjected to immunoblot analyses using antibodies for PAI-1, p21, Smad7, CDK4, Cyclin D1, MPK38, and β-actin. Inducible silencing of endogenous MPK38 expression by doxycycline (Dox: 1 μg/ml, 72 h) was assessed by immunoblotting using an anti-MPK38 antibody. β-Actin was used as a loading control. The relative expression level of TGF-β target genes was quantified by densitometry, and -fold increase relative to the untreated control in parental NIH 3T3 cells was calculated.
MPK38-mediated Phosphorylation of Smad Proteins Is Required for its Positive Regulation of TGF-β-induced Transcription
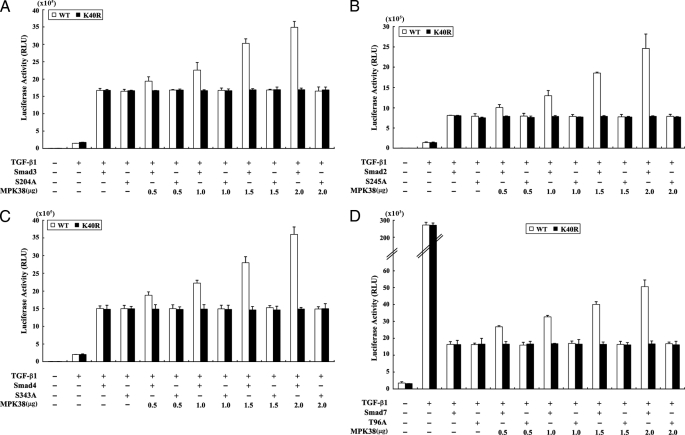
To investigate whether MPK38 plays an important role in the regulation of TGF-β-mediated transcriptional activity through direct phosphorylation of Smad proteins (Smad2, -3, -4, and -7), we compared the effect of MPK38 on TGF-β-induced transcription in the presence of wild-type Smad proteins with its effect in the presence of the four MPK38-mediated phosphorylation-defective Smad substitution mutants (S245A of Smad2, S204A of Smad3, S343A of Smad4, and T96A of Smad7) (see Fig. 3B). Smad3(S204A) had no effect on MPK38-mediated stimulation of TGF-β-induced transcription compared with the control expressing wild-type Smad3 (Fig. 6A). Similar results were also obtained for Smad2 and -4, in which both Smad2(S245A) and Smad4(S343A) mutants, like the Smad3(S204A) mutant, did not alter the MPK38-mediated stimulation of TGF-β-induced transcriptional activity (Fig. 6, B and C). We also compared the effect of MPK38 on TGF-β-mediated transcriptional activity in the presence of wild-type Smad7, an inhibitory Smad, with its effect in the presence of the Smad7(T96A) mutant. Likewise, expression of the Smad7(T96A) mutant did not alleviate the Smad7-mediated suppression of TGF-β-induced transcription in the presence of wild-type MPK38 (Fig. 6D). These findings suggest that the phosphorylation of Smad proteins by MPK38 is apparently important for MPK38-mediated regulation of TGF-β signaling.
FIGURE 6.
Effect of MPK38-mediated phosphorylation of Smad proteins on TGF-β-induced transcriptional activity. A–D, HepG2 cells were transfected with 0.2 μg of p3TP-Lux plasmid, increasing amounts of wild-type (WT), and kinase-dead (K40R) MPK38, and wild-type and mutant forms (S245A of Smad2, S204A of Smad3, S343A of Smad4, and T96A of Smad7) of Smad proteins (each 6 μg), as indicated, in the presence or absence of TGF-β1 (100 pm).
MPK38 Differentially Modulates the Association between Type I TGF-β Receptor and Smad Proteins
We next examined whether MPK38 could have an effect on the association between TβR1(TD), an activated type 1 TGF-β receptor, and Smad3 and -7, because we reasoned that MPK38 could contribute to the alteration of the association between TβR1(TD) and the Smad proteins that is crucial for TGF-β signaling. To this end, FLAG-Smad proteins (Smad3 and -7) were cotransfected with HA-TβR1(TD) into HEK293 cells in the presence or absence of wild-type and kinase-dead MPK38. Compared with control cells that did not express MPK38, the coexpression of wild-type MPK38 significantly increased the association between TβR1(TD) and Smad3 (Fig. 7A, left panel), or decreased the association between TβR1(TD) and Smad7 (Fig. 7B, left panel), whereas the kinase-dead MPK38 had no effect on the association of the proteins. To confirm whether the association between TβR1(TD) and Smad proteins (Smad3 or -7) is dependent on endogenous MPK38, TβR1(TD)-Smads complex formation was examined in cells using MPK38-specific siRNA. Consistently, the knockdown of endogenous MPK38 had an opposite effect on the association (Fig. 7, A and B, right panels). To further investigate whether the MPK38-mediated phosphorylation of Smad proteins (Smad3 and -7) could influence the TβR1(TD)-Smads complex formation, we also analyzed the effect of Smad substitution mutants (S204A of Smad3 and T96A of Smad7) on the association between TβR1(TD) and Smad3 and -7 using a binding assay. The association between TβR1(TD) and Smad3 was significantly decreased in the presence of the Smad3(S204A) mutant when compared with wild-type Smad3, whereas the Smad7(T96A) mutant resulted in a considerable increase in TβR1(TD)-Smad7 complex formation, leading to the suppression of TGF-β signaling (Fig. 7C). These data provide evidence that MPK38 positively regulates TGF-β signaling through modulation of the physical interaction between the TGF-β receptor and Smad3 and -7.
FIGURE 7.
Modulation of the association between activated type I TGF-β receptor and Smad proteins by MPK38. A and B, HEK293 cells were transfected with the indicated combinations of plasmid vectors expressing TβR1(TD), an activated type I TGF-β receptor, FLAG-Smad3 or -7, and GST-tagged wild-type (WT) or kinase-dead (K40R) MPK38, and the cell lysates were subjected to immunoprecipitation with an anti-HA antibody (α-HA). Complex formation between TβR1(TD) and Smad3 (A, left, top panel) or Smad7 (B, left, top panel) was determined by anti-FLAG antibody immunoblot. HEK293 cells were transfected with the indicated siRNA duplexes (MPK38-specific siRNA or control scrambled siRNA) together with plasmid vectors expressing TβR1(TD) and FLAG-Smad3 or -7. Complex formation between TβR1(TD) and Smad3 (A, right, top panel) or Smad7 (B, right, top panel) was determined by anti-FLAG antibody immunoblot. The expression level of endogenous MPK38 was determined by anti-MPK38 immunoblotting (A and B, right, bottom panels). C, effect of MPK38-mediated phosphorylation of Smad proteins on TβR1(TD)-Smads association. FLAG-tagged wild-type Smad proteins (Smad3 and -7) or their substitution mutants (S204A of Smad3 and T96A of Smad7) were cotransfected with HA-TβR1(TD) into HEK293 cells. The cell lysates were subjected to immunoprecipitation with an anti-HA antibody (α-HA), and complex formation between TβR1(TD) and Smad proteins was determined by an anti-FLAG antibody immunoblot (top panel). The relative level of complex formation between TβR1(TD) and Smad3 (A and C) or Smad7 (B and C) was quantitated by densitometric analysis, and -fold increase relative to control samples expressing TβR1(TD) and Smad3 or -7 alone was calculated. Sc, scrambled.
MPK38 Modifies the Subcellular Localization of Smad Proteins
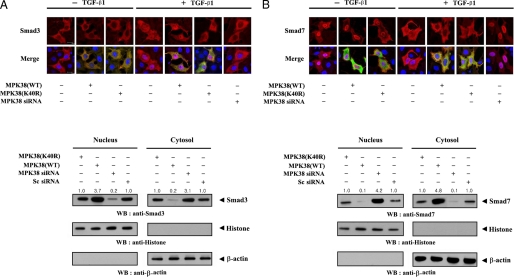
As nuclear translocation of R-Smads, such as Smad3, has been shown to be crucial for TGF-β signaling, we investigated whether MPK38 affects the intracellular localization of Smad3. To this end, we monitored the nuclear translocation of Smad3 by immunofluorescence microscopy using Hep3B cells transfected with Smad3 alone or together with wild-type or kinase-dead MPK38 in the presence or absence of TGF-β1. As expected, TGF-β1 treatment significantly increased the nuclear localization of Smad3 in cells, leading to the stimulation of TGF-β signaling (Fig. 8A, upper panel, lane 1 versus lane 4). In the presence of wild-type MPK38, a further increase in Smad3 nuclear translocation was observed, but no such increase was observed for kinase-dead MPK38 (Fig. 8A, upper panel, lane 4 versus lanes 5 and 6). For Smad7, in contrast, TGF-β1 stimulated translocation in the opposite direction, from the nucleus to the cytoplasm (Fig. 8B, upper panel, lane 1 versus lane 4). Coexpression of wild-type MPK38 significantly increased the translocation of Smad7 from the nucleus into the cytoplasm, whereas the coexpression of kinase-dead MPK38 had no effect (Fig. 8B, upper panel, lane 4 versus lanes 5 and 6), consistent with the data for Smad3. To confirm that MPK38 is physiologically associated with the modulation of intracellular localization of Smad proteins, we also performed knockdown experiments using MPK38-specific siRNA. Our aim was to determine whether the knockdown of endogenous MPK38 could alter the subcellular localization of Smad proteins. Using siRNA to reduce the amount of endogenous MPK38 suppressed the normal translocation of Smad proteins (Smad3 and -7) in the presence of TGF-β1 (Fig. 8, A and B, upper panels, lanes 4 versus lanes 5 and 7). To further verify the effect of MPK38 on the subcellular localization of Smad proteins, Hep3B cells transfected with plasmid vectors expressing wild-type or kinase-dead MPK38, an MPK38-specific siRNA, or a nonspecific scrambled siRNA were treated with TGF-β1 and separated into cytoplasmic and nuclear fractions. Each fraction was analyzed by Western blot analysis. The accumulation of Smad3 in the nuclear fraction was significantly decreased in MPK38-knockdown cells compared with control cells expressing a nonspecific scrambled siRNA (Fig. 8A, lower left, lane 1 versus lanes 3 and 4), whereas the cytoplasmic accumulation of Smad3 was markedly increased. In addition, the opposite trend was observed for translocation of Smad7 in response to TGF-β1 under the same conditions (Fig. 8B, lower panel). These results indicate that MPK38 stimulates the normal translocation of Smad proteins in response to TGF-β1.
FIGURE 8.
Modulation of Smads localization by MPK38. Hep3B cells were transiently transfected with plasmid vectors expressing FLAG-Smad3 (A, upper) or FLAG-Smad7 (B, upper), together with GST-tagged wild-type (WT) and kinase-dead (K40R) MPK38, or an MPK38-specific siRNA, and incubated in the presence or absence of 100 pm TGF-β1. Cells were immunostained with anti-FLAG or anti-MPK38 antibodies, followed by Alexa Fluor-594 anti-mouse secondary antibodies (red, for Smad3 and Smad7) or Alexa Fluor-488 anti-rabbit secondary antibodies (green, for MPK38), and analyzed by confocal microscopy. Cytoplasmic and nuclear fractions from extracts of Hep3B cells transfected with the indicated plasmid vectors expressing wild-type or kinase-dead MPK38, or siRNA duplexes (MPK38-specific siRNA and control scrambled siRNA), in the presence of TGF-β1 were separated from each other and used for Western blot analysis using the indicated antibodies (A and B, lower panels). Nucleus and Cytosol indicate the nuclear and cytoplasmic fractions of cell extracts, respectively. The relative level of Smads expression (Smad3 and -7) was quantified by densitometry. Sc, scrambled.
MPK38 Stimulates TGF-β-induced Apoptosis and Growth Arrest
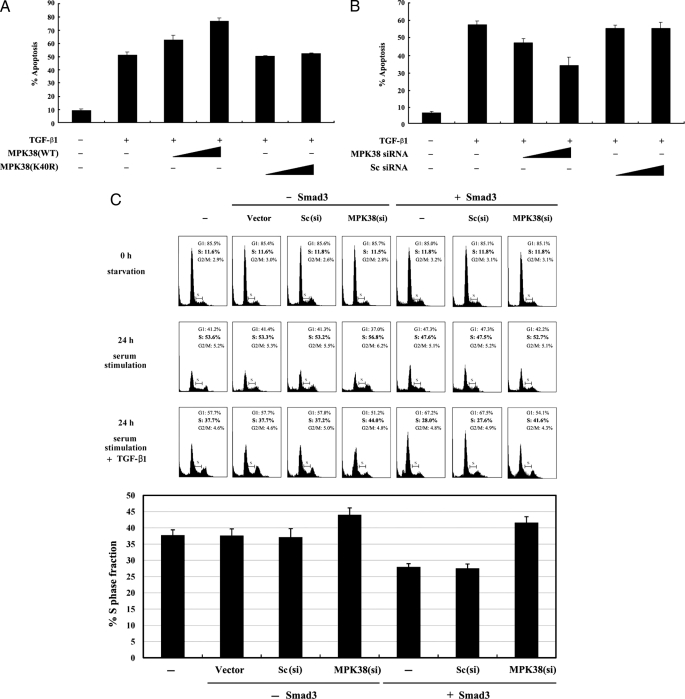
To explore the functional significance of the MPK38-Smads association, we examined the effect of wild-type and kinase-dead MPK38 on TGF-β-induced apoptosis using a GFP assay system (27). HaCaT cells were transfected with an expression plasmid encoding GFP, together with wild-type or kinase-dead MPK38, and incubated in the presence or absence of TGF-β1. Approximately 51% of the HaCaT cells were apoptotic following TGF-β1 treatment (Fig. 9A, 2nd lane). Cells transfected with wild-type MPK38 showed higher apoptotic function (∼52% increase) than cells treated with TGF-β1 alone, and the effect was dose-dependent (Fig. 9A, lane 2 versus lanes 3 and 4). However, the stimulatory effect on TGF-β-induced apoptosis was not observed in cells transfected with kinase-dead MPK38 (Fig. 9A, lane 2 versus lanes 5 and 6). These data are consistent with the results obtained from luciferase assays (Fig. 5) and indirect immunofluorescence studies (Fig. 8). Furthermore, the knockdown of endogenous MPK38 with MPK38-specific siRNA resulted in a dose-dependent decrease in TGF-β-induced apoptosis, whereas a control scrambled siRNA had no effect (Fig. 9B). Next, to examine whether TGF-β-induced cell cycle arrest was also affected by knockdown of endogenous MPK38, we performed flow cytometry analysis using HaCaT cells expressing a vector alone, control scrambled siRNA [Sc(si)], or MPK38 siRNA [MPK38(si)] in the presence or absence of Smad3. A much higher number of cells (∼42% versus ∼28%) was found to be in S phase in the presence of MPK38 siRNA after 24 h of serum stimulation in the presence of TGF-β1, compared with controls expressing vector alone or control scrambled siRNA (Fig. 9C, lower, lane 5 versus lanes 6 and 7), indicating that MPK38 stimulates TGF-β-induced growth arrest. In addition, we found that MPK38 has a similar effect on TGF-β-induced epithelial-mesenchymal transition (supplemental Fig. S2). Together, these results suggest that TGF-β-mediated biological functions are positively regulated by MPK38.
FIGURE 9.
Effect of MPK38 on TGF-β-induced apoptosis and growth inhibition. A and B, stimulation of TGF-β-induced apoptosis by MPK38. HaCaT cells were transiently transfected with increasing amounts of wild-type and kinase-dead MPK38 (1 and 2 μg) or MPK38-specific siRNA (100 and 200 nm) as indicated, together with an expression vector encoding GFP (3 μg). As a control, a control scrambled siRNA (100 and 200 nm) was transfected into cells. After treatment of the transfected cells with TGF-β1 (2 ng/ml, 20 h), apoptotic cell death was determined using the GFP expression system (17, 27). C, effect of MPK38 knockdown on TGF-β-induced cell cycle arrest. HaCaT cells (∼2 × 105/dish) transfected with the indicated siRNA duplexes (MPK38-specific siRNA and control scrambled siRNA) in the presence or absence of Smad3 were synchronized in G0/G1 by hydroxyurea (2 mm) treatment for 20 h. Cells were collected before (0 h starvation) or after 10% serum treatment for 24 h in the absence (24 h serum stimulation) or presence (24 h serum stimulation + TGF-β1) of TGF-β1 (2 ng/ml), and the percentage of cells in the G1, S, or G2/M phases was analyzed by flow cytometry. The fraction of cells in S phase (24 h serum stimulation + TGF-β1) was quantitated and is presented in the bar graphs (lower panel). si, siRNA.
DISCUSSION
We have previously shown that PDK1 interacts with intracellular signaling mediators of the TGF-β signaling pathway, such as Smad proteins (Smad2, -3, -4, and -7) and serine-threonine kinase receptor-associated protein, and that this interaction is involved in the reciprocal regulation of PI3K/PDK1 and TGF-β signaling (20, 30), suggesting a functional link between the PI3K/PDK1 and TGF-β signaling pathways. Furthermore, cross-talk between p53 and TGF-β signaling has been reported (31). These data reveal the possibility of cross-talk between cell-survival and death-inducing signals, or between cell death signaling pathways. We have recently found that MPK38 participates in ASK1 activation through direct interaction and phosphorylation, resulting in the stimulation of ASK1-mediated apoptosis (17). These findings, together with TGF-β-induced stimulation of MPK38 kinase activity (Fig. 1A), raise the possibility that TGF-β signaling may be associated with MPK38-mediated signaling. Based on this, we speculated that the physical interaction between MPK38 and Smad proteins may occur in vivo.
In this study, we demonstrated that MPK38 physically interacts with Smad proteins (Smad2, -3, -4, and -7) (Fig. 1). Moreover, our data indicate that the kinase activity of MPK38 is required for its ability to stimulate TGF-β-induced transcription (Fig. 5) and apoptosis (Fig. 9). These findings are similar to those from recent studies showing that PDK1 and its downstream target Akt physically interact with Smad2, -3, -4, and -7, and inhibit TGF-β signaling in a kinase-dependent manner (20, 32). In addition, in our study, wild-type MPK38 stimulated the normal movement of Smad proteins (Smad3 and -7) in response to TGF-β1, but the kinase-dead MPK38 did not show such an effect (Fig. 8). In this context, our current evidence indicates that MPK38 is a potential positive regulator of the TGF-β signaling pathway. This is in contrast to PDK1, which acts as a negative regulator of TGF-β signaling. However, our present results do not support the possibility that the MPK38-mediated stimulation of TGF-β signaling is due only to the physical interaction between MPK38 and Smad proteins, because the kinase-dead MPK38, which lacks the ability to regulate TGF-β signaling, was shown to associate with Smad proteins at levels similar to those of wild-type MPK38 (data not shown). These data suggest that MPK38-mediated phosphorylation of Smad proteins (Smad2, -3, -4, and -7) through direct interaction plays a crucial role in the positive regulation of TGF-β signaling induced by MPK38.
Although C-terminal SXS phosphorylation of R-Smad by the type I TGF-β receptor is an important step in TGF-β signaling, additional phosphorylations by intracellular protein kinases are also involved in the regulation of Smad activities. Recent studies have shown that p38 mitogen-activated protein kinase (MAPK) and Rho-associated, coiled-coil containing protein kinase contribute to Smad3 phosphorylation at Ser204/208/213 in the linker region and stimulate its transcriptional activity (33). Similarly, our results showed that the MPK38-mediated phosphorylation of Smad3 at Ser204 plays a key role in the stimulation of TGF-β signaling (Fig. 6). However, extracellular signal-regulated kinase (ERK)-mediated phosphorylation of Smad3 at Ser204 and other sites (Ser208 and Thr179), and of Smad2 at Ser245/250/255 and Thr220, showed an inhibitory effect on Smad3 activity (34). These findings imply that MAPK-mediated phosphorylation of Smad2 and -3, in contrast to that of MPK38, appears to have a dual role in the regulation of Smad2 and -3. Growing evidence indicates that, in addition to these kinases, a variety of intracellular protein kinases (5, 35–38), including cyclin-dependent kinase 2/4 (CDK2/4), c-Jun N-terminal kinase (JNK), G protein-coupled receptor kinase 2 (GRK2), and calmodulin-dependent kinase II, contribute to the phosphorylation of the Smad2 and -3 linker region, although the exact mechanism(s) by which phosphorylation in the Smad linker region regulates Smad-mediated signaling remains unclear. On the other hand, Smad4 is known to be constitutively phosphorylated in cells, although the phosphorylation sites are yet to be discovered (5, 6). In this regard, the discovery of phosphorylation of Smad4 at Ser343 by MPK38 (Fig. 3B), together with a recent result showing that ERK-mediated phosphorylation of Smad4 at Thr277 is involved in the nuclear translocation of Smad4 (6), should aid in the understanding of TGF-β signal regulation. The inhibitory Smad proteins, Smad6 and Smad7, are phosphoproteins (7, 8, 39). It has been shown that protein kinase X, involved in macrophage differentiation, phosphorylates Smad6 at Ser435 (39). Our results showed that MPK38 is also involved in Smad6 phosphorylation at Thr176, and that this phosphorylation is important for the regulation of Smad6-mediated bone morphogenetic protein signaling (supplemental Fig. S3). In addition, Smad7 was found to be phosphorylated at Ser249 (8), although the intracellular kinases responsible for phosphorylating Smad7 have not yet been identified. Our results from this study have provided evidence that MPK38 stimulates TGF-β signaling by phosphorylating Smad7 at Thr96 (Fig. 6D), suggesting that MPK38 is a putative intracellular kinase for Smad7 phosphorylation in cells.
We previously observed that PDK1-mediated inhibition of TGF-β signaling is accompanied by the modulation of complex formation between the TGF-β receptor and Smad proteins (Smad3 and -7) (20). To explore the mechanism by which MPK38 stimulates TGF-β-induced transcriptional activation, we also investigated whether MPK38 can modulate the association between the TGF-β receptor and Smad proteins (Smad3 and -7). Our results showed that the ability of wild-type MPK38 to stimulate TGF-β-mediated transcriptional activation is dependent on MPK38-induced modulation of Smad(s) binding to the TGF-β receptor (Fig. 7). Wild-type MPK38 increased complex formation between the TGF-β receptor and Smad3, and simultaneously decreased complex formation between the TGF-β receptor and Smad7, leading to stimulation of TGF-β signaling. However, the association of TGF-β receptor with Smad proteins was not affected by the kinase-dead MPK38. These observations imply that the kinase activity of MPK38 is also required for modulation of complex formation between the TGF-β receptor and Smad proteins, in agreement with reporter (Figs. 5 and 6), localization (Fig. 8), and apoptosis (Fig. 9) studies.
In summary, our findings suggest that MPK38 positively regulates TGF-β signaling through direct interaction and phosphorylation of Smad proteins, and also that Smad proteins may function as linkers between TGF-β and MPK38-mediated signaling. The discovery of MPK38-mediated stimulation of TGF-β signaling should allow us to gain insight into how TGF-β signaling is regulated in vivo, since MPK38 directly interacts and phosphorylates all of the Smad proteins (Smad2, -3, -4, and -7) we tested.
Supplementary Material
This work was supported by Grant R0A-2007-000-20006-0 from the National Research Foundation of Korea and in part by Chungbuk National University Grant 2009.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S3.
- CDK
- cyclin-dependent kinase
- Smad
- Sma- and Mad-related protein
- I-Smad
- inhibitory Smad
- MH
- Mad homology
- TβR
- transforming growth factor-β receptor
- ZPR9
- zinc-finger-like protein 9
- MPK38
- murine protein serine/threonine kinase 38
- PDK1
- 3-phosphoinositide-dependent protein kinase 1.
REFERENCES
- 1.Abdollah S., Macías-Silva M., Tsukazaki T., Hayashi H., Attisano L., Wrana J. L. (1997) J. Biol. Chem. 272, 27678–27685 [DOI] [PubMed] [Google Scholar]
- 2.Macías-Silva M., Abdollah S., Hoodless P. A., Pirone R., Attisano L., Wrana J. L. (1996) Cell 87, 1215–1224 [DOI] [PubMed] [Google Scholar]
- 3.Souchelnytskyi S., Tamaki K., Engström U., Wernstedt C., ten Dijke P., Heldin C. H. (1997) J. Biol. Chem. 272, 28107–28115 [DOI] [PubMed] [Google Scholar]
- 4.Xu L. (2006) Biochim. Biophys. Acta 1759, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrighton K. H., Lin X., Feng X. H. (2009) Cell Res. 19, 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roelen B. A., Cohen O. S., Raychowdhury M. K., Chadee D. N., Zhang Y., Kyriakis J. M., Alessandrini A. A., Lin H. Y. (2003) Am. J. Physiol. Cell Physiol. 285, C823–C830 [DOI] [PubMed] [Google Scholar]
- 7.Imamura T., Takase M., Nishihara A., Oeda E., Hanai J., Kawabata M., Miyazono K. (1997) Nature 389, 622–626 [DOI] [PubMed] [Google Scholar]
- 8.Pulaski L., Landström M., Heldin C. H., Souchelnytskyi S. (2001) J. Biol. Chem. 276, 14344–14349 [DOI] [PubMed] [Google Scholar]
- 9.Gil M., Yang Y., Lee Y., Choi I., Ha H. (1997) Gene 195, 295–301 [DOI] [PubMed] [Google Scholar]
- 10.Heyer B. S., Warsowe J., Solter D., Knowles B. B., Ackerman S. L. (1997) Mol. Reprod. Dev. 47, 148–156 [DOI] [PubMed] [Google Scholar]
- 11.Bachvarova R., Cohen E. M., De Leon V., Tokunaga K., Sakiyama S., Paynton B. V. (1989) Development 106, 561–565 [DOI] [PubMed] [Google Scholar]
- 12.Ohsugi M., Hwang S. Y., Butz S., Knowles B. B., Solter D., Kemler R. (1996) Dev. Dyn. 206, 391–402 [DOI] [PubMed] [Google Scholar]
- 13.Nakano I., Paucar A. A., Bajpai R., Dougherty J. D., Zewail A., Kelly T. K., Kim K. J., Ou J., Groszer M., Imura T., Freije W. A., Nelson S. F., Sofroniew M. V., Wu H., Liu X., Terskikh A. V., Geschwind D. H., Kornblum H. I. (2005) J. Cell Biol. 170, 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heyer B. S., Kochanowski H., Solter D. (1999) Dev. Dyn. 215, 344–351 [DOI] [PubMed] [Google Scholar]
- 15.Saito R., Tabata Y., Muto A., Arai K., Watanabe S. (2005) Mol. Cell Biol. 25, 6682–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beullens M., Vancauwenbergh S., Morrice N., Derua R., Ceulemans H., Waelkens E., Bollen M. (2005) J. Biol. Chem. 280, 40003–40011 [DOI] [PubMed] [Google Scholar]
- 17.Jung H., Seong H. A., Ha H. (2008) J. Biol. Chem. 283, 34541–34553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray D., Jubb A. M., Hogue D., Dowd P., Kljavin N., Yi S., Bai W., Frantz G., Zhang Z., Koeppen H., de Sauvage F. J., Davis D. P. (2005) Cancer Res. 65, 9751–9761 [DOI] [PubMed] [Google Scholar]
- 19.Vulsteke V., Beullens M., Boudrez A., Keppens S., Van Eynde A., Rider M. H., Stalmans W., Bollen M. (2004) J. Biol. Chem. 279, 8642–8647 [DOI] [PubMed] [Google Scholar]
- 20.Seong H. A., Jung H., Kim K. T., Ha H. (2007) J. Biol. Chem. 282, 12272–12289 [DOI] [PubMed] [Google Scholar]
- 21.Seong H. A., Jung H., Ichijo H., Ha H. (2010) J. Biol. Chem. 285, 2397–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seong H. A., Jung H., Ha H. (2007) J. Biol. Chem. 282, 12075–12096 [DOI] [PubMed] [Google Scholar]
- 23.Yang Y., Gil M., Lee Y., Ha H. (1999) Appl. Biochem. Biotechnol. 80, 13–22 [DOI] [PubMed] [Google Scholar]
- 24.Jung H., Kim T., Chae H. Z., Kim K. T., Ha H. (2001) J. Biol. Chem. 276, 15504–15510 [DOI] [PubMed] [Google Scholar]
- 25.Jung H., Seong H. A., Manoharan R., Ha H. (2010) J. Biol. Chem. 285, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao K., Shida S., Selvakumaran M., Zimmerman R., Simon E., Schick J., Haas N. B., Balke M., Ross H., Johnson S. W., O'Dwyer P. J. (2005) Clin. Cancer Res. 11, 7264–7272 [DOI] [PubMed] [Google Scholar]
- 27.Jung H., Seong H. A., Ha H. (2007) J. Biol. Chem. 282, 35293–35307 [DOI] [PubMed] [Google Scholar]
- 28.Datta P. K., Moses H. L. (2000) Mol. Cell Biol. 20, 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakobsen S. N., Hardie D. G., Morrice N., Tornqvist H. E. (2001) J. Biol. Chem. 276, 46912–46916 [DOI] [PubMed] [Google Scholar]
- 30.Seong H. A., Jung H., Choi H. S., Kim K. T., Ha H. (2005) J. Biol. Chem. 280, 42897–42908 [DOI] [PubMed] [Google Scholar]
- 31.Cordenonsi M., Dupont S., Maretto S., Insinga A., Imbriano C., Piccolo S. (2003) Cell 113, 301–314 [DOI] [PubMed] [Google Scholar]
- 32.Song K., Wang H., Krebs T. L., Danielpour D. (2006) EMBO J. 25, 58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamaraju A. K., Roberts A. B. (2005) J. Biol. Chem. 280, 1024–1036 [DOI] [PubMed] [Google Scholar]
- 34.Kretzschmar M., Doody J., Timokhina I., Massagué J. (1999) Genes Dev. 13, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuura I., Denissova N. G., Wang G., He D., Long J., Liu F. (2004) Nature 430, 226–231 [DOI] [PubMed] [Google Scholar]
- 36.Engel M. E., McDonnell M. A., Law B. K., Moses H. L. (1999) J. Biol. Chem. 274, 37413–37420 [DOI] [PubMed] [Google Scholar]
- 37.Mori S., Matsuzaki K., Yoshida K., Furukawa F., Tahashi Y., Yamagata H., Sekimoto G., Seki T., Matsui H., Nishizawa M., Fujisawa J., Okazaki K. (2004) Oncogene 23, 7416–7429 [DOI] [PubMed] [Google Scholar]
- 38.Wrighton K. H., Feng X. H. (2008) Cell Signal. 20, 1579–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glesne D., Huberman E. (2006) Oncogene 25, 4086–4098 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.