Abstract
BRCC36 is a JAMM (JAB1/MPN/Mov34 metalloenzyme) domain, lysine 63-ubiquitin (K63-Ub)-specific deubiquitinating enzyme (DUB) and a member of two protein complexes: the DNA damage-responsive BRCA1-RAP80 complex, and the cytoplasmic BRCC36 isopeptidase complex (BRISC). The presence of several identical constituents in both complexes suggests common regulatory mechanisms and potential competition between K63-Ub-related signaling in cytoplasmic and nuclear compartments. Surprisingly, we discover that BRCC36 DUB activity requires different interactions within the context of each complex. Abraxas and BRCC45 were essential for BRCC36 DUB activity within the RAP80 complex, whereas KIAA0157/Abro was the only interaction required for DUB activity within the BRISC. Poh1 also required protein interactions for activity, suggesting a common regulatory mechanism for JAMM domain DUBs. Finally, BRISC deficiency enhanced formation of the BRCA1-RAP80 complex in vivo, increasing BRCA1 levels at DNA double strand breaks. These findings reveal that JAMM domain DUB activity and K63-Ub levels are regulated by multiple mechanisms within the cell.
Keywords: BRCA BRCB, Deubiquitination, DNA Damage, DNA Repair, Ubiquitination, BRCA1, BRCC36, DUB, JAMM Domain, RAP80
Introduction
The mammalian genome is remarkably stable despite an estimated 105 mutagenic events/cell cycle (1). This exquisite fidelity can be attributed to the multiple and varied activities of the DNA damage response (DDR).4 To maintain genome integrity, eukaryotic cells activate the DDR, a complex signaling network that integrates and coordinates DNA damage recognition, cell cycle checkpoints, and DNA repair (2, 3). Recent evidence implicates ubiquitin chain formation, recognition, and breakdown at the site of the genomic lesion as an essential component of the DDR.
Ubiquitin is a 76-amino acid protein that can be attached to a target protein via an isopeptide linkage between the ϵ-amino lysine residue of a target protein and the C-terminal glycine residue within ubiquitin. Protein ubiquitination can have quite complex outcomes resulting from the considerable structural information embedded within ubiquitin polymers. Specifically, a single ubiquitin monomer can be extended through the ubiquitination of any one of seven lysines or through the N terminus, creating polyubiquitin chains (4). These different ubiquitin topologies result in the formation of diverse structures resulting in vastly different biological outcomes. The canonical Lys48-linked polyubiquitination of proteins signals for proteasomal degradation (5); conversely, Lys63-linked polyubiquitin has been implicated in non-degradative signals in response to both cytoplasmic and nuclear cues. Specifically, Lys63-linked polyubiquitin is involved in both the recruitment and retention of DNA repair factors at sites of DNA damage (6–8).
BRCA1 is central to the DDR and forms a number of mutually exclusive macromolecular complexes, each with discrete activities (9, 10). Recently, it has been shown that the core RAP80 complex plays an important role in facilitating BRCA1 localization to DNA double strand breaks (DSBs). The RAP80 complex is a five-member stoichiometric complex consisting of RAP80, BRCC36, BRCC45, Abraxas, and MERIT40 (11–13). RAP80, in turn, is recruited to DSBs through its tandem ubiquitin-interacting motifs (UIMs) (6, 14–16), which specifically recognize K63-Ub chains (6) that are synthesized by the combined actions of several different E3 ligases (7, 8, 17–22).
BRCC36 is a member of a unique class of zinc-dependent metalloproteases, termed JAMM (JAB1/MPN/Mov34 metalloenzyme) DUBs (23–25). BRCC36 possesses DUB activity specifically for K63-Ub polyubiquitin chains (26–28). This activity parallels the ubiquitin binding preference of the RAP80 UIM domains (6, 26, 27) and supports the model that parallel and opposing Lys63-ubiquitin synthesis and hydrolysis at DSBs regulate the recruitment and retention of DNA repair factors (27, 29).
Recently, BRCC36 was also identified as a member of the four-subunit BRCC36 isopeptidase complex (BRISC) (26). This complex contains three protein components in common with the core RAP80 complex, BRCC36, BRCC45, and MERIT40, whereas the fourth component of the BRISC is the Abraxas paralog, KIAA0157/Abro1. The presence of multiple, common components of each DUB complex raises several possibilities. First, protein interactions within each core protein complex could regulate either the efficiency or the ubiquitin chain specificity of BRCC36 DUB activity. Second, a competition for assembly of the BRCA1-RAP80 and BRISC complexes may exist in vivo, with the relative levels of each complex potentially influencing K63-Ub chain abundance in either the nucleus or cytoplasm. In principle, altering the balance between these two BRCC36-containing complexes could affect the proposed ubiquitin landscape at DSBs to influence the DDR (30).
In an attempt to answer these questions, we reconstituted and characterized the DUB activity of the RAP80 and BRISC complexes in vitro and examined the in vivo consequences of altering the balance between these BRCC36-containing complexes on DSB recruitment events. Here, we show that BRCC36 DUB activity requires different interactions within the context of each protein complex. Abraxas and BRCC45 are essential for BRCC36 DUB activity in the RAP80 complex, whereas KIAA0157 is the only essential interaction necessary for BRCC36 DUB activity in the BRISC. Protein-protein interaction with MPN− (Mpr-1/Pad-1 N-terminal) domain-containing proteins was also required for the JAMM domain DUB Poh1, a component of the 19 S proteasome lid complex, suggesting a common mode of regulation for this class of DUBs. A second level of regulation was exposed by in vivo experiments. BRISC deficiency strongly increased BRCA1-RAP80 complex formation at sites of damage in vivo, whereas RAP80 deficiency produced the opposite effect, thereby increasing K63-Ub levels at DSBs.
EXPERIMENTAL PROCEDURES
Cell Lines
HeLa, HeLa S3, and U2OS cells were cultured in DMEM (Invitrogen) with 10% calf serum. Phoenix A retroviral packaging lines were used to produce FLAG-HA-tagged versions (POZ-FH), (31) of Abraxas, KIAA0157, BRCC36, BRCC36 ΔN13, BRCC45, and MERIT40 retrovirus. Cell lines infected with retrovirus were selected on anti-IL2-receptor antibody (Upstate Biotechnology)-coated Dynabeads® (Invitrogen). Transient transfections were performed on 293T cells using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen).
Antibodies
The following antibodies were employed. Abraxas was detected by immunoblot (IB) at 1:500 with a rabbit polyclonal antibody. BRCC36 was detected by IB with a rabbit polyclonal antibody. BRCC45 was detected by IB by a rabbit polyclonal antibody raised against human BRCC45 (Zymed Laboratories Inc.). HA-tagged proteins were detected by IB at 1:1000 and by immunofluorescence (IF) at 1:1000 dilutions using the mouse monoclonal antibody HA.11 (Covance). KIAA0157 was detected by IB at 1:500 using a rabbit polyclonal antibody raised against a full-length recombinant KIAA0157 protein. Lys63-linked ubiquitin was detected by IF at 1:1500 using a humanized monoclonal antibody (Genentech). MERIT40 was detected by IB using a rabbit polyclonal antibody at 1:500 and IF at 1:50. γH2AX was detected by IF with a mouse monoclonal antibody (clone JBW301, Upstate Biotechnology) at 1:2500. 53BP1 was detected by IF with a rabbit polyclonal antibody (Novus) at 1:250. BRCA1 was detected by IB with the mouse monoclonal antibody MS110 at a 1:5 dilution (32) and by IF with the mouse monoclonal antibody SC-6954 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 1:25. Total ubiquitin was detected for IB with the mouse monoclonal antibody P4D1 (Santa Cruz Biotechnology, Inc.) at 1:200. A RAP80 rabbit polyclonal antibody was used for IB at 1:500.
DNA Damage Induction
Cells were exposed to 10 Gy of radiation using an MDS Nordion Gammacell fixed source Cs-137 irradiator. Cells were incubated for 4 h at 37 °C prior to fixation. IF was performed as described previously (32).
Microirradiation-induced DSBs were induced by a PALM MicroBeam laser microdissection system (Carl Zeiss MicroImaging, Inc.) as described previously (6, 33, 34). Briefly, cells were cultured on coverslips for 36 h in DMEM with 10% calf serum, supplemented with 10 μm BrdU (Sigma). Laser stripes were performed on at least 50 cells/coverslip with a 337-nm wavelength laser set to 62% power using a ×40 objective. Cells were incubated for 30 min at 37 °C, and fixation and IF were performed as described above.
DUB Activity Assay
Equal amounts of FLAG-purified recombinant proteins, as determined by Coomassie staining, were incubated with hexa-K48-Ub or hexa-K63-Ub (Boston Biochem) in DUB buffer (125 mm HEPES, pH 7.5, 25 mm MgCl2, 10 mm NaF, 10 nm okadaic acid, 500 mm NaCl, 2.5 mm DTT, 0.5 mg/ml BSA) for 1 h at 25 °C. Reaction products were separated on a 4–12% gradient bis-tris gel (Invitrogen) and transferred to a nitrocellulose membrane. The membrane was denatured in a 6 m guanidine HCl, 25 mm Tris-HCl, pH 7.5, 0.25 mm PMSF, 20 mm β-mercaptoethanol solution for 30 min at 4 °C and then washed extensively in PBST to facilitate detection of ubiquitin by blotting with P4D1 antibody.
Immunofluorescent Microscopy and Image Analysis
8-Bit gray scale images of ∼100 cells/condition were captured. A QImaging RETIGA-SRV camera connected to a Nikon Eclipse 80i microscope driven by ImagePro 6.2 software was employed. Image analyses were performed using ImageJ software from the National Institutes of Health. For intensity analyses, a region of interest was selected, and the mean fluorescence intensity with subtraction of background fluorescence was measured on unprocessed images. Gray scale images were pseudocolored in Adobe Photoshop CS4.
Immunoprecipitation
FLAG immunoprecipitation was performed in NETN150 buffer (150 mm Tris-HCl, pH 7.4, 1 mm EDTA, 0.05% Nonidet P-40, 0.5 mm PMSF, and 5 mm β-mercaptoethanol) using anti-FLAG M2-agarose beads (Sigma). Immunoprecipitated proteins were eluted with either glycine or FLAG peptide (Sigma).
Mass Spectrometry
Catalytically inactive BRCC36 complexes were purified from nuclear extracts of HeLa S3 cells stably expressing N-terminal FLAG-HA-tagged BRCC36-QSQ by tandem affinity purification (Nakatani and Ogryzko 2003). Purified material was electrophoresed on a 4–12% bis-tris SDS gel (Invitrogen) and Coomassie-stained. The RAP80-Ub2 band was excised and submitted to the Taplin Biological Mass Spectrometry Facility (Harvard Medical School) for analysis.
The RAP80-Ub2 band was in-gel trypsin-digested and subjected to LC-MS/MS using an LTQ Orbitrap XL hybrid mass spectrometer (Thermo Finnigan) as described previously (35). In brief, a branched signature peptide sequence is produced at sites of ubiquitination following trypsin digestion of ubiquitinated proteins. This unique signature results in both a mass shift and a missed proteolytic cleavage as the lysine is protected by its modification with ubiquitin. Using data base-searching algorithms, the branched signature peptides can be identified, and the sequence can be determined by an additional mass of 114 daltons, representing an additional G-G dipeptide conjugated to the ϵ-amino group of a ubiquitinated lysine residue.
Plasmids
FLAG-HA-tagged versions of Abraxas, KIAA0157, BRCC36, BRCC36 ΔN13, BRCC45, MERIT40, and Poh1 were created using either the N-terminal or C-terminal versions of POZ-FH (31). FLAG-tagged versions of BRCC45 and BRCC45 mutants were created using the N-terminally tagged pCMV-Tag2B vector (Stratagene). FLAG-HA-tagged versions of each of these cDNAs were subcloned into pVL1392 and pVL1392 vectors for protein expression in Sf9 cells.
Purification of Recombinant Proteins
FLAG-HA-tagged Abraxas, BRCC36, BRCC45, KIAA0157, MERIT40, and RAP80 were cloned into either pVL1392 or pVL1392 vectors, and high titer recombinant virus was generated and used to infect Sf9 cells using standard methods (BD Biosciences). Two days postinfection, cells were harvested and lysed in a buffer containing 50 mm HEPES, pH 8.0, 120 mm NaCl, 0.5% Nonidet P-40, 0.5 mm PMSF, and 5 mm β-mercaptoethanol. The recombinant protein was purified using anti-FLAG M2-agarose beads (Sigma) and FLAG peptide (Sigma)-eluted. Purity was estimated to be ∼95% by Coomassie staining. Recombinant protein was quantified using the Coomassie Plus reagent (Pierce).
RNA Interference
Transfections were performed using either oligofectamine or Lipofectamine RNAiMax (Invitrogen) per the manufacturer's instructions and analyzed 72 or 48 h post-transfection, respectively. The following siRNA target sequences were employed: Abraxas, CGUUUAGAGAGAGGCUGCUUCACAA; BRCC36, AACAUCAACAUGUGAAGGCCTT; BRCC45, GGUGCAGUACGUGAUUCAA; Ct, UCGAAGUAUUCCGCGUACGTT; KIAA0157, GCAACACAGAAUUUCUGCAAGUAAU; MERIT40, CCCAUAUUUCUUCUUUGACGUUGUU; RAP80, CCAGUUGGAGGUUUAUCAA; Rpn8, CCUACAGAAGCGUACAUUUTT.
Statistical Analysis
GraphPad Prism software was used to create graphs and statistically analyze the data using Student's unpaired two-tailed t test without assuming equal variances. Enzyme kinetics were determined using nonlinear regression toward Michaelis-Menten enzyme kinetics equations as provided by the GraphPad Prism software.
RESULTS
BRCC36 DUB Activity Requires Interactions within the RAP80 Complex
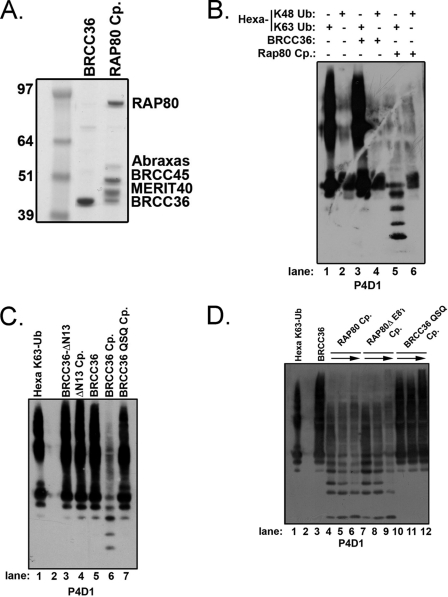
To determine if interactions within the RAP80 complex modify BRCC36 Lys63-specific DUB activity, the entire core RAP80 complex was reconstituted using a baculovirus Sf9 system and purified to homogeneity (Fig. 1A) (11). The RAP80 complex failed to display activity using ubiquitin conjugated to 7-amino-4-methylcoumarin as a substrate (data not shown); thus, we turned to non-fluorescence-based approaches using defined polyubiquitin substrates. Purified BRCC36 alone or in the context of the RAP80 complex was incubated with either hexa-K48-Ub or hexa-K63-Ub (Fig. 1B). DUB activity was detected by the production of ubiquitin cleavage products on IB. BRCC36 lacked DUB activity to K48-Ub substrates when expressed alone or in the RAP80 complex as anticipated (lanes 4 and 6). Strikingly, BRCC36 alone completely lacked K63-Ub DUB activity (lane 3), whereas BRCC36 DUB activity was easily detected in the context of the RAP80 complex (lane 5). To confirm that BRCC36 required interactions with the other members of the RAP80 complex in order to have DUB activity, we employed an interaction-deficient BRCC36 mutant (supplemental Fig. S1). This BRCC36 mutant lacks the first 13 N-terminal residues and no longer localizes to ionizing radiation-induced foci (IRIF); nor does it interact with any of the RAP80 complex members (supplemental Fig. S1, B and C). As predicted, the BRCC36 ΔN13 mutant either alone or in the context of the RAP80 complex lacked DUB activity (Fig. 1C, lanes 3 and 4, and supplemental Fig. S1D). The specificity of the K63-Ub-specific DUB activity of BRCC36 was confirmed by the absence of K63-Ub hydrolysis by a RAP80 complex assembled with a BRCC36 mutant in which the active site Zn2+-binding histidine residues were replaced with glutamines (BRCC36H122H124 → BRCC36Q122Q124; hereafter denoted as QSQ). These residues are required for JAMM domain DUB activity (6, 23, 25) but are not required for BRCC36 association with RAP80 or DSB recognition (27). The QSQ complex did not possess in vitro DUB activity (Fig. 1C, lane 7). These results indicate that one or more protein interactions within the RAP80 complex are required for BRCC36-dependent DUB activity.
FIGURE 1.
BRCC36 requires interactions within the RAP80 complex for DUB activity. A, Coomassie-stained gel representing purified proteins used for in vitro DUB assays. Recombinant Abraxas, BRCC36, BRCC45, MERIT40, and RAP80 (all of which are FLAG-HA-tagged) were purified from baculovirus-infected Sf9 cells by FLAG-IP and FLAG peptide elution and are estimated to be ∼95% pure. Cp, complex. A full-length gel is included in supplemental Fig. S1. B, BRCC36 Lys63-specific DUB activity requires interactions within the RAP80 complex. FLAG-HA-tagged BRCC36 or the entire RAP80 complex was purified from baculovirus-infected Sf9 cells 48 h postinfection. FLAG-purified complexes were incubated with either hexa-K48-Ub or hexa-K63-Ub for 1 h, and the products were detected by IB with the P4D1 antibody to ubiquitin. The figure is representative of three independent experiments. C, an interaction-deficient BRCC36 mutant (ΔN13) lacks DUB activity. FLAG-HA-tagged BRCC36-ΔN13, and RAP80 complexes containing either BRCC36 (BRCC36 complex) or BRCC36 mutants (ΔN13 complex and QSQ complex) were incubated with hexa-K63-Ub for 1 h, and the products were detected by IB with the P4D1 antibody to detect ubiquitin. The figure is representative of three independent experiments. See supplemental Fig. S1 for a Coomassie-stained gel representing purified proteins. D, the RAP80 complex does not require ubiquitin targeting for BRCC36 DUB activity. FLAG-HA-tagged BRCC36, RAP80 complex, RAP80ΔE81 complex, or QSQ complex were purified from baculovirus-infected Sf9 cells 48 h postinfection. FLAG-purified complexes were incubated with hexa-K63-Ub for 1 h, and the products were detected by IB with the P4D1 antibody to ubiquitin. Increasing amounts of the RAP80, RAP80ΔE81, and QSQ complexes were employed as denoted by the arrow. The figure is representative of three independent experiments.
The RAP80 UIM domains specifically recognize K63-Ub (6), thus raising the possibility that BRCC36 DUB activity requires the ubiquitin-targeting action of RAP80. To test this option, we employed a human breast cancer-associated RAP80 mutant with an in-frame deletion at residue Glu81 in a highly conserved region of the first UIM (36). This RAP80ΔE81 protein displays significantly reduced ubiquitin binding and DSB localization, leading to impairments in the DDR. No clear change in K63-Ub DUB activity was observed between complexes containing either RAP80 WT (lanes 4–6) or RAP80ΔE81 (lanes 7–9), indicating that RAP80-dependent ubiquitin recognition is not required for DUB activity or specificity (Fig. 1D).
Abraxas and BRCC45 Interact with BRCC36 through MPN− and Ubiquitin E2 Variant (UEV) Domains, Respectively
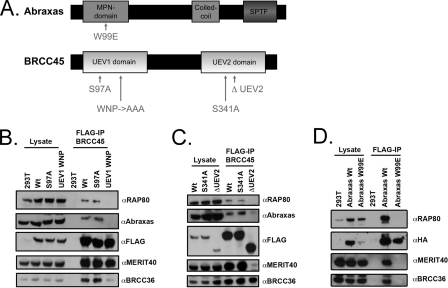
These data suggest that BRCC36 DUB activity requires one or more interactions within the RAP80 complex. To gain further insight into these observations, we mutated the putative ubiquitin binding domains of Abraxas and BRCC45 (Fig. 2A). BRCC45 contains two UEV domains that lack a cysteine necessary for the formation of a thioester-ubiquitin intermediate. Abraxas contains a JAMM/MPN− domain but lacks conserved residues that coordinate Zn2+ binding required for enzymatic activity. Point mutations or deletions were made within highly conserved regions in the first or second UEV of BRCC45 (Fig. 2, A–C).
FIGURE 2.
Abraxas and BRCC45 interact with BRCC36 through MPN- and UEV domains respectively. A, schematic diagram of the domains and mutations used for Abraxas (top) and BRCC45 (bottom) interaction studies with BRCC36. Abraxas contains an MPN− domain, a coiled coil domain, and a phosphoserine motif. BRCC45 contains two UEV domains, labeled UEV1 and UEV2 for ease of communication. B, interaction profile of BRCC45 UEV1 mutants. FLAG-tagged plasmids encoding wild type BRCC45 (Wt), BRCC45 S97A, and BRCC45 UEV1 WNP were transiently transfected into 293T cells, and FLAG-IP was performed 48 h later. RAP80, Abraxas, MERIT40, and BRCC36 were detected as BRCC45-associated proteins for both wild type BRCC45 and mutant BRCC45 S97A. BRCC45 UEV1 WNP demonstrated reduced interaction with RAP80, Abraxas, and BRCC36. C, interaction profile of BRCC45 UEV2 mutants. FLAG-tagged plasmids encoding WT BRCC45, BRCC45 S341A, and BRCC45 ΔUEV2 were transiently transfected into 293T cells, and IP was performed 48 h later. RAP80, Abraxas, MERIT40, and BRCC36 were detected as BRCC45-associated proteins for both WT BRCC45 and mutant BRCC45 S341A. BRCC45 ΔUEV2 demonstrated reduced interaction with RAP80, Abraxas, and MERIT40 while maintaining interaction with BRCC36. D, interaction profile of an Abraxas MPN domain mutant. FLAG-HA-tagged plasmids encoding wild type Abraxas and Abraxas W99E were transiently transfected into 293T cells, and IP was performed 48 h later. The Abraxas W99E mutant demonstrated loss of interaction with RAP80, MERIT40, and BRCC36.
An alignment of BRCC45 and other UEV domains was created using ClustalW (37). Highly conserved residues were mapped onto the structure of Ubc13/Mms2 dimer bound to ubiquitin (Protein Data Bank code 2GMI) and Tsg101 UEV domain bound to ubiquitin (Protein Data Bank code 1S1Q) (38, 39). Ser97 and Ser341 are serine residues that align with the catalytically active cysteine of E2 enzymes and should minimally disrupt the overall structure. To identify protein-protein interactions with BRCC45, we mutated the highly conserved W105N106P107 to AAA within the UEV1 domain and deleted UEV2.
Abraxas has a predicted MPN− domain, similar to Mov34/MPN/Pad-1 but enzymatically inactive. Abraxas was aligned with other members of the MPN family. Highly conserved residues were mapped onto the structure of AMSH-like protease (AMSH-LP) bound to diubiquitin (Protein Data Bank code 2ZNV) (40). Trp99 is highly conserved among MPN family members and, based on structural homology, is predicted to interact with the C terminus of the distal ubiquitin in Lys63-linked diubiquitin. To assess the importance of ubiquitin binding of Abraxas, we mutated Trp99 to a bulky negatively charged residue, glutamate.
We observed that UEV1 and UEV2 were required for protein interactions with other members of the RAP80 complex. Mutation of 3 residues to alanine within UEV1 (UEV1 WNP to AAA) disrupted interaction with RAP80, Abraxas, and BRCC36, whereas complete deletion of UEV2 disrupted interaction with RAP80, Abraxas, and MERIT40 (Fig. 2, B and C). Similarly, a point mutation in the Abraxas MPN− domain (W99E) and a corresponding mutation in the KIAA0157 MPN− domain (W98E) resulted in the loss of interaction with all members of the RAP80 complex (Fig. 2, A and D, and supplemental Fig. S2A). More subtle point mutations to BRCC45 UEV1 and UEV2 did not disrupt interaction with the other RAP80 core components. BRCC45 UEV1 mutant S97A and UEV2 mutant S341A maintained WT levels of interaction with each of the other RAP80 core constituents (Fig. 2, B and C), enabling an assessment of their contribution to DUB activity.
Abraxas and BRCC45 Are Essential for BRCC36 DUB Activity
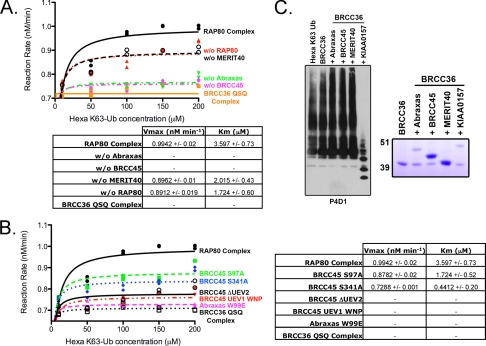
To ascertain what effect loss of these interactions would have on BRCC36 DUB activity compared with loss of the complete protein itself, we first reconstituted RAP80 complexes lacking a single component and compared BRCC36 DUB activity in each of these singularly deficient complexes with that of the complete RAP80 complex. FLAG-purified complexes were incubated with increasing concentrations of hexa-K63-Ub, and the appearance of diubiquitin was quantified to calculate DUB activity as a function of input hexa-K63-Ub (Fig. 3A). MERIT40 or RAP80 deficiency resulted in a reproducibly modest reduction in DUB activity. In comparison, a more dramatic impact on DUB activity was observed upon Abraxas or BRCC45 deficiency, either of which essentially eliminated detectable hydrolysis products (Fig. 3A).
FIGURE 3.
Abraxas and BRCC45 are essential for BRCC36 DUB activity within the RAP80 complex. A, assessment of the contribution of each core RAP80 complex constituent to BRCC36 DUB activity. FLAG-HA-tagged RAP80 complexes lacking a single component of the core RAP80 complex (Abraxas, BRCC45, MERIT40, or RAP80) were FLAG-purified from baculovirus-infected Sf9 cells 48 h postinfection. Complexes were incubated with increasing concentrations of hexa-K63-Ub for 1 h, as indicated, and the products were detected by IB with the P4D1 antibody. The appearance of diubiquitin was quantified using NIH Image J software, and DUB activity was calculated as a function of input hexa-K63-Ub. The figure is an average of two independent experiments. Enzyme kinetics were determined using nonlinear regression (Michaelis-Menten enzyme kinetics). The representative graph is displayed alongside the calculated Vmax and Km values. B, full BRCC36 DUB activity requires protein-protein interactions and WT BRCC45 UEV domains. RAP80 complexes, of which all components are FLAG-HA-tagged, containing wild type Abraxas/BRCC45 or mutants were purified from baculovirus-infected Sf9 cells 48 h postinfection. FLAG-purified complexes were incubated with hexa-K63-Ub for 1 h, and the products were detected by IB with the P4D1 antibody to ubiquitin. The appearance of diubiquitin was quantified using NIH Image J software, and DUB activity was calculated as a function of input hexa-K63-Ub. The figure is an average of two independent experiments. Enzyme kinetics were determined using nonlinear regression (Michaelis-Menten enzyme kinetics). The representative graph is displayed alongside the calculated Vmax and Km values. C, KIAA0157, but not Abraxas, is sufficient to impart K63-Ub DUB activity on BRCC36 in vitro. FLAG-purified complexes (right, Coomassie-stained gel representing FLAG-purified complexes) were incubated with hexa-K63-Ub for 1 h, and the products were detected by IB with the P4D1 antibody (left). The figure is representative of three independent experiments.
In agreement with the requirement for multiple protein interactions, UEV deletions or the UEV1 WNP mutation strongly reduced DUB activity in complexes that had been reconstituted in Sf9 cells (Fig. 3B). More subtle changes to UEV1 (S97A) and UEV2 (S341A) did not detectably alter protein interactions for BRCC45 and other members of the RAP80 complex (Fig. 2, B and C) yet resulted in a discernable decrease in DUB activity (Fig. 3B), suggesting that the BRCC45 UEVs contribute to DUB activation by mechanisms beyond the maintenance of protein-protein interactions. Although it is unclear why BRCC45 UEV mutations reduce DUB activity, this could be due to an influence on the BRCC36 active site residues or access to ubiquitin chain substrates. Definitive evidence of these possible mechanisms awaits higher resolution structural analysis. Similarly, the Abraxas MPN− domain point mutation (W99E) strongly reduced BRCC36 dependent DUB activity in vitro (Fig. 3B). Similarly, the corresponding KIAA0157 mutation in the MPN− domain (W98E) resulted in decreased BRCC36-dependent DUB activity (supplemental Fig. S2B). These observations confirm the need for both Abraxas and BRCC45 interaction with BRCC36 and demonstrate that the putative ubiquitin binding domains also play a role in maintaining protein-protein interactions necessary for BRCC36 DUB activity.
The RAP80 and BRISC complexes differ by the unique presence of the paralogs Abraxas and KIAA0157, respectively. Our previous findings (Fig. 3) demonstrated that both Abraxas and BRCC45 were essential for BRCC36 DUB activity within the RAP80 complex, whereas recent reports indicate that BRCC36 and KIAA0157 form a minimal heterodimer for BRCC36 DUB activity (28). Producing recombinant two-member complexes of BRCC36 plus Abraxas, BRCC45, MERIT40, or KIAA0157 (Fig. 3C, right), we observe that the KIAA0157 was sufficient to activate BRCC36 DUB activity, whereas Abraxas failed to do so (Fig. 3C, left). Thus, despite a high degree of homology and similar domain structures, it appears that KIAA0157 imparts a more potent DUB-stimulatory activity compared with Abraxas. In addition, we observed increased DUB activity within the BRISC as compared with the core RAP80 complex (supplemental Fig. S3, A–C). These results reveal differential requirements for Lys63-specific DUB activity between the RAP80 and BRISC complexes, implicating BRCC45 as an essential component of the RAP80 complex.
Interactions with MPN− Domain-containing Proteins Are a Common Mechanism of Regulating JAMM Domain DUB Activity
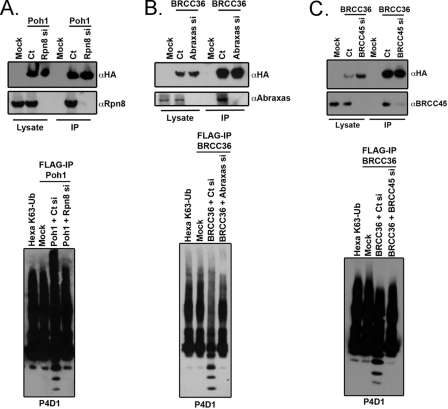
Based on the necessity of BRCC36 interaction with Abraxas, an MPN− domain-containing protein, for DUB activity, we asked if other JAMM domain DUBs also required interaction with MPN− domain proteins for K63-Ub-specific DUB activity. The JAMM domain family of DUBs consists of seven members (41). One of these members, Poh1 is responsible for Lys63-Ub DUB activity within the 19 S subunit of the 26 S proteasome. Computational analysis of the RAP80 complex suggested weak similarity to other components of the 19 S subunit (13), raising the possibility that protein-protein interactions might be a common mechanism of regulating JAMM domain DUBs. Rpn8 contains an MPN-/JAMM- domain, like Abraxas, and makes direct contact with Poh1 within the 19 S proteasome lid (42). These similarities suggest the hypothesis that MPN−/JAMM+ pairs may allow for JAMM domain DUB activity. To test this hypothesis, we purified a FLAG-HA-tagged version of Poh1 from 293T cells transfected with siRNA targeting either a control or Rpn8 and assessed DUB activity in vitro (Fig. 4A). Poh1 complexes exhibited Lys63-specific DUB activity that was abrogated by Rpn8 knockdown (Fig. 4A, bottom). Similarly, either Abraxas or BRCC45 knockdown strongly reduced DUB activity from BRCC36 that had been purified from nuclear extracts to eliminate the possibility of contaminating DUB activity from the BRISC (Fig. 4, B and C, bottom). Consequently, protein-protein interactions with MPN− or UEV domain-containing proteins can regulate JAMM domain DUB activity. Furthermore, these data support a model in which protein-protein interactions can regulate JAMM domain DUB activity, as suggested in Figs. 1–3.
FIGURE 4.
Protein-protein interactions with MPN− domain proteins are a common mechanism of regulating JAMM/JAB DUB activity. A, IB of ectopic Poh1 complexes after FLAG-IP from 293T cells. FLAG-HA-tagged plasmids encoding Poh1 were co-transfected with control (Ct) or Rpn8 siRNA, and IP was performed 48 h later (top). Depletion of Rpn8 from Poh1 complexes abrogates Poh1 DUB activity (bottom). Equal amounts of FLAG peptide-eluted Poh1 complexes were incubated with hexa-K63-Ub for 1 h, and the products were detected by IB with the P4D1 antibody to ubiquitin. B, IB of ectopic BRCC36 complexes after FLAG-IP from HeLa S3 cell nuclear extracts. Cells were treated with control or Abraxas siRNA, and IP was performed on nuclear extracts 48 h later (top). Abraxas deficiency abrogates BRCC36 DUB activity from nuclear extracts (bottom). Equal amounts FLAG peptide-eluted BRCC36 protein were incubated with hexa-K63-Ub for 1 h, and the products were detected by IB with the P4D1 antibody. C, IB of ectopic BRCC36 complexes after FLAG-IP from HeLa S3 cell nuclear extracts. Cells were treated with control or BRCC45 siRNA, and IP was performed on nuclear extracts 48 h later (top). BRCC45 deficiency abrogates BRCC36 DUB activity from nuclear extracts (bottom). Equal amounts of FLAG peptide-eluted BRCC36 protein were incubated with hexa-K63-Ub for 1 h, and the products were detected by IB with the P4D1 antibody.
RAP80 Is an in Vivo Target of BRCC36 Lys63-specific DUB Activity
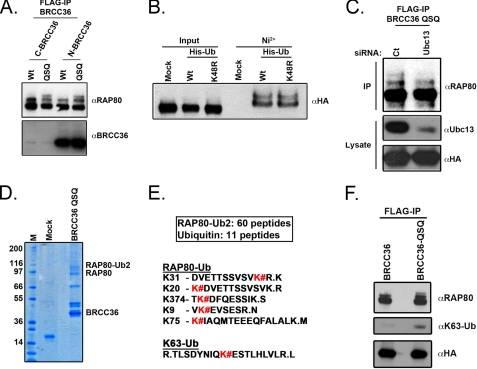
BRCC36 Lys63-specific DUB activity has been demonstrated in vitro, paralleling the ubiquitin binding preference of the RAP80 UIM domains (6, 11, 27). RAP80, like many ubiquitin-binding proteins, had been reported to be ubiquitinated in a manner dependent on its UIM domains (43). Given this observation and the intimate relationship between RAP80 and BRCC36, we tested if RAP80 was itself a target of BRCC36 DUB activity. FLAG immunoprecipitation of epitope-tagged BRCC36 wild type (BRCC36 WT) versus epitope-tagged BRCC36 QSQ (BRCC36 QSQ) was performed to determine if BRCC36 DUB targets could be revealed among the BRCC36-interacting partners. The RAP80 species that associated with DUB-inactive BRCC36 QSQ demonstrated higher migrating forms consistent with mono- and diubiquitination (Fig. 5A). Indeed, RAP80 was diubiquitinated equally well by both WT and K48R mutated ubiquitin (Fig. 5B). RAP80-BRCC36 complexes exhibit an in vitro preference for K63-Ub hydrolysis. Therefore, we examined the dependence of these higher migrating forms on the Lys63-specific E2 enzyme, Ubc13. Ubc13 knockdown strongly decreased the polyubiquitinated form of RAP80 associated with BRCC36 QSQ (Fig. 5C).
FIGURE 5.
RAP80 is an in vivo target of BRCC36 Lys63-specific DUB activity. A, FLAG-IP of ectopic BRCC36 complexes was performed and probed with anti-BRCC36 and anti-RAP80 antibodies as indicated. The anti-BRCC36 antibody (J86) recognizes the C-terminal 20 amino acids of BRCC36. C-terminal epitope tags strongly reduce J86 recognition on Western blot, accounting for the weak signal of BRCC36 species with C-terminal FLAG-HA tags (C-B36 and C-QSQ) compared with N-terminal FLAG-HA-tagged BRCC36 (N-B36 and N-QSQ). B, RAP80 is ubiquitinated by both WT and K48R mutated ubiquitin. 293T cells expressing FLAG-HA-RAP80 (RAP80) were mock-transfected or transfected with either WT ubiquitin or ubiquitin K48R mutant containing an N-terminal 6-histidine sequence. Ubiquitinated proteins were purified over a Ni2+-agarose column and eluted with imidazole-containing buffer. IB was performed with an anti-HA antibody. C, Ubc13 knockdown strongly decreases the polyubiquitinated form of RAP80 associated with BRCC36 QSQ. FLAG-IP of ectopic BRCC36 QSQ complex was performed following control (Ct) or Ubc13 siRNA-mediated knockdown. IB was performed on FLAG-purified BRCC36 complexes as indicated. D, FLAG and HA tandem immunoaffinity purification was performed for ectopic BRCC36 QSQ from HeLa S3 nuclear extracts. The RAP80-Ub2 Coomassie-stained band was excised and trypsin-digested prior to mass spectrometry analysis. M, molecular weight marker; Mock, mock-transfected cell line. E, peptide sequences obtained by mass spectrometry of tryptic digests from the RAP80-Ub2 band. K#, RAP80 lysine residues that are conjugated to ubiquitin. A peptide sequence indicating the presence of Lys63-linked polyubiquitin was also detected by mass spectrometry from tryptic digests of the RAP80-Ub2 band. F, IB of ectopically expressed FLAG-HA BRCC36 or FLAG-HA BRCC36-QSQ after FLAG-IP. Increased RAP80 polyubiquination, specifically Lys63-linked ubiquitin, is observed in association with BRCC36-QSQ compared with BRCC36.
These results are consistent with Ubc13 and BRCC36 playing opposing roles on the same K63-Ub substrates (27). Supporting this assertion, mass spectrometry of RAP80 ubiquitinated bands (RAP80-Ub2) that co-precipitated with BRCC36 QSQ mutant after tandem affinity purification revealed five ubiquitinated lysine residues on RAP80 and the presence of K63-Ub (Fig. 5, D–F). These results confirm that BRCC36 acts on Lys63-ubiquitinated proteins in vivo and that RAP80 is one substrate common to both BRCC36 K63-Ub DUB and Ubc13 ubiquitin ligase activities in vivo.
BRISC Deficiency Enhances the DNA Damage Association of the BRCA1-RAP80 Complex
KIAA0157 and Abraxas are unique members of the BRISC and BRCA1-RAP80 complexes, respectively (supplemental Fig. S4A), enabling each complex to be individually manipulated by targeting either protein. We have detected KIAA0157 both in the cytoplasm and nucleus (supplemental Fig. S4B). Consequently, we tested whether the BRISC complex (containing KIAA0157) might play a role in the DDR.
We performed IF of eAbraxas and eKIAA0157 in HeLa S3 cells at 4 h following 10 grays of ionizing radiation, demonstrating that although Abraxas colocalizes with the DNA damage marker, 53BP1, at DSBs, KIAA0157 does not (supplemental Fig. S5A). Along these lines, knockdown of endogenous KIAA0157 in U2OS cells did not result in increased ionizing radiation sensitivity in a clonogenic assay, whereas, as previously reported, knockdown of Abraxas did (supplemental Fig. S5B) (18). BRCC36 and BRCA1 displayed reduced ionizing radiation-induced foci (IRIF) at 4 h after 10 grays of ionizing radiation following Abraxas knockdown (supplemental Fig. S5, C and D), whereas KIAA0157 knockdown did not impair either at IRIF. Interestingly, both BRCC36 and BRCA1 qualitatively formed more robust IRIF in KIAA0157-depleted cells, hinting at the possibility of enhanced DSB recruitment of the BRCA1-RAP80 complex in the context of BRISC deficiency.
These aforementioned results raise the possibility that the steady state levels of common constituents within RAP80 and BRISC complexes are in balance, with formation of one limiting the abundance of the other complex. Moreover, it is conceivable that the dominant complex would direct increased quantities of K63-Ub DUB activity to its cognate cellular compartments.
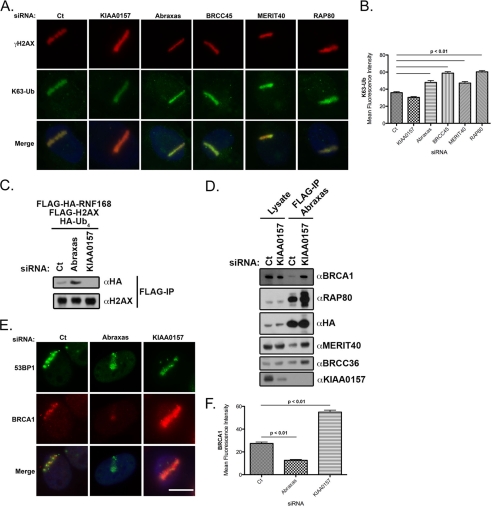
To test this hypothesis, we examined K63-Ub intensity at IRIF and laser-microirradiated DSBs as an in vivo assay of BRCC36 DUB activity following knockdown of different members of each complex. We performed laser microirradiation of U2OS cells following knockdown of RAP80 complex members or KIAA0157 and assayed the fluorescence intensity of K63-Ub (Fig. 6, A and B). As predicted from the K63-Ub foci formation data, knockdown of Abraxas, BRCC45, MERIT40, and RAP80 increased K63-Ub fluorescence. Conversely, knockdown of the KIAA0157 resulted in a slight decrease in K63-Ub fluorescence (Fig. 6, A and B). Moreover, knockdown of Abraxas, BRCC45, MERIT40, and RAP80 increased the percentage of cells with 10 or more K63-Ub foci colocalizing with γH2AX (supplemental Fig. S6, B and C), whereas knockdown of KIAA0157 did not increase K63-Ub foci formation (supplemental Fig. S6, B and C). These results were replicated at the chromatin level, where ubiquitination of γH2AX was increased following Abraxas depletion and decreased following KIAA0157 depletion (Fig. 6C).
FIGURE 6.
Relative levels of KIAA0157 influence the abundance and localization of the BRCA1-RAP80 complex. A, differential influence of BRISC and RAP80 complexes on K63-Ub levels at DSBs. IF was performed for γH2AX and K63-Ub following control, KIAA0157, Abraxas, BRCC45, MERIT40, and RAP80 siRNA treatment in laser-microirradiated U2OS cells. Knockdown of Abraxas, BRCC45, MERIT40, and RAP80 increased K63-Ub intensity at DSBs. Bars, 10 μm. B, quantification of K63-Ub stripe intensity displayed graphically from A. At least 100 cells were counted in triplicate for the analysis. Error bars, S.D. p values were calculated by Student's t test. C, IB of H2AX-Ub following siRNA knockdown of either Abraxas or KIAA0157. Compared with control knockdown cells (Ct), H2AX-Ub levels are increased following Abraxas knockdown and decreased following KIAA0157 knockdown. IB is representative of three independent experiments. D, IB of ectopic Abraxas following KIAA0157 siRNA-mediated depletion. FLAG-HA-tagged Abraxas was FLAG-immunoprecipitated and blotted as indicated. Knockdown of KIAA0157 increased interaction between Abraxas and BRCA1, RAP80, MERIT40, and BRCC36. E, KIAA0157 depletion increases BRCA1 localization to DSBs. IF was performed for 53BP1 and BRCA1 following control and KIAA0157 siRNA treatment in laser-microirradiated U2OS cells. Knockdown of KIAA0157 increased BRCA1 intensity compared with control. Bars, 10 μm. F, quantification of BRCA1 stripe intensity displayed graphically from E. At least 100 cells were counted in triplicate for the analysis. Error bars, S.D. p values were calculated by Student's t test.
Together, these observations support a model whereby RAP80 complex targeted BRCC36 hydrolyzes K63-Ub chains at DSBs, resulting in a decreased K63-Ub signal. Consistent with a model of a limited pool of common subunits, knockdown of KIAA0157 strongly increased the association of Abraxas with BRCC36, MERIT40, RAP80, and BRCA1 (Fig. 6D). Given that the RAP80 complex targets BRCA1 to DSBs and that BRCA1 association with Abraxas is increased in the face of BRISC deficiency, we asked if KIAA0157 knockdown would ultimately lead to increased BRCA1 at DSBs. BRCA1 localization to laser-microirradiated stripes of DNA damage was reduced following Abraxas knockdown, as reported previously (Fig. 6, E and F) (38). Conversely, BRCA1 localization was increased following KIAA0157 knockdown (Fig. 6, E and F).
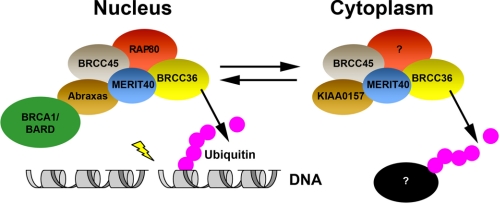
Thus, BRCA1 DSB recruitment is modulated by the relative levels of BRISC and RAP80 complexes, revealing a competition between cytoplasmic and nuclear DUB complexes that influences events at DSBs (Fig. 7). Formation of each complex is limited in part by the abundance of the other complex. In the nucleus, the ubiquitin binding of RAP80 targets the RAP80 complex to DSBs. We postulate that a similar mechanism exists in the cytoplasm, whereby one or more currently unknown BRISC-interacting partners targets the BRCC36 DUB activity to act on K63-Ub substrates in different locales within the cell (Fig. 7).
FIGURE 7.
Model for Lys63-specific DUB regulation in different cellular compartments. The BRISC and BRCA1-RAP80 complexes are differentially regulated by protein-protein interactions in the cytoplasm and nucleus, respectively. Formation of each complex is limited in part by the abundance of the other complex. In the nucleus, the ubiquitin binding of RAP80 targets the RAP80 complex to DSBs. We theorize that the same mechanism exists in the cytoplasm, where an unknown protein targets DUB activity within the BRISC to act on yet to be identified K63-Ub targets.
DISCUSSION
A network of enzymes that synthesize and degrade ubiquitin chains stringently regulates ubiquitination and deubiquitination reactions. This is evident by the kinetics of appearance and disappearance of ubiquitin chains at DSBs. A rapid response of at least four different E3 ligases synthesizes a ubiquitin platform for recruitment of DNA repair complexes at DSBs within minutes of DDR induction. Although considerable speculation exists, it is still unknown if the ubiquitin chain topologies are remodeled or edited during the course of the damage response. What is clear, however, is that the kinetics of disappearance of ubiquitin closely follows the resolution of ATM-dependent H2AX phosphorylation on chromatin adjacent to DSBs (19), revealing a tight link between DDR phosphorylation and ubiquitination.
It has been well established that ubiquitin chain synthesis is regulated by numerous protein-protein interactions. E3 ligases must pair with an E2 enzyme-ubiquitin thioester intermediate in order to transfer ubiquitin to a target substrate. Moreover, many E3 ligases require additional regulatory mechanisms to target them to the site of action. For example, RNF8 utilizes a FHA domain to bind phosphorylated MDC at DSBs, whereas RNF168 employs ubiquitin-binding MIU (motif interacting with ubiquitin) domains to target its E3 ligase activity to sites of damage. The RING domain containing E3 ligase, BRCA1 employs a similar strategy via interaction with RAP80.
Emerging evidence suggests that DDR-related DUB activity is also tightly controlled. In this report, we document that BRCC36 requires one or more protein interactions for enzymatic activity. We provide evidence that such regulation is not unique to BRCC36 but also applies to a second JAMM domain DUB, Poh1. Notably, each required interaction with an MPN− domain protein for activity on K63-Ub. It is interesting to note that, although interaction with MPN− domain proteins may be necessary for some JAMM domain DUBs, it is not an absolute requirement for all DUBs of this class. For example, AMSH-LP is also a Lys63-specific, JAMM domain DUB, and it displays DUB activity in the absence of other protein interactions. High resolution structural information exists for AMSH-LP (40). It will be interesting to compare structures of BRCC36 and Poh1 with AMSH-LP when they become available. Such information may be necessary to fully understand the switch to active enzyme upon association with other members of the RAP80 and BRISC complexes. Differences should be anticipated, given the ability of AMSH-LP to be active as a single polypeptide and to display activity toward artificial substrates not recognized by either the RAP80 or BRISC complexes. In line with this reasoning, sequence alignments reveal a structure of the BRCC36 JAMM domain more similar to that of Poh1 than to that of AMSH-LP (39% similarity between BRCC36 and Poh1 compared with 22% similarity between BRCC36 and AMSH-LP) (supplemental Fig. S7).
Although a member of a different class of DUB enzyme than BRCC36, USP1 also requires protein interactions for DUB activity. USP1 utilizes an active site cysteine nucleophile to hydrolyze monoubiquitinated proliferating cell nuclear antigen and FANCD2 (44, 45). USP1-dependent DUB activity relies on an interaction with a stoichiometric binding partner, UAF1 (46). Another example of protein interactions modifying DUB activity is the USP14 interaction with UBP6. The activity of USP14/UBP6 is stimulated more than 300-fold upon binding to the 19 S regulatory particle. In the case of USP14, two loops (BL1 and BL2) that occlude the active site in the apostructure become ordered when bound to ubiquitin (47). Exactly how the 19 S regulatory particle relieves the self-inhibition of USP14 is still under investigation. We note, however, that in the alignment of BRCC36 with Poh1 and AMSH-LP, BRCC36 contains two insertions in loop regions before and after the Ins-1 region of AMSH-LP, which in the crystal structure make critical contacts to the C-terminal tail of the distal ubiquitin (supplemental Fig. S7). It remains to be seen whether these insertions may be involved in the regulation of the activity of BRCC36.
A second interesting feature of BRCC36 DUB regulation unique to the RAP80 complex is the requirement of BRCC45. The RAP80 complex lacked detectable DUB activity in the absence of BRCC45. The dominant contribution appeared to be through BRCC45 UEV domain-mediated protein interactions. However, UEV1 and UEV2 mutations, which did not produce a discernable change in protein associations, also reduced DUB activity, suggesting an additional contribution to DUB activity. It is interesting to note that KIAA0157 was sufficient to activate BRCC36 DUB activity, revealing previously unappreciated differences between Abraxas and KIAA0157. Thus, BRCC45 UEV domains are not an absolute requirement for stimulating BRCC36 DUB activity within the BRISC. We had previously demonstrated that cooperative interactions within the RAP80 complex were necessary to maintain protein stability in vivo. Our current findings related to DUB activity provide an additional explanation for the necessity of maintaining multiple different partners within the same protein complex. Each of these proteins appears to contribute to DUB activity in addition to their well documented in vivo roles.
This study also suggests that a third mode of Lys63-specific DUB regulation exists in the cell: the partitioning of common components of each complex to either nuclear or cytoplasmic compartments based on the availability of unique members of each complex (e.g. Abraxas and KIAA0157). BRISC deficiency, created by KIAA0157 knockdown, resulted in an increased abundance of the BRCA1-RAP80 complex commensurate with increased BRCA1-RAP80 complex at damage sites. Together, these findings reveal a means by which KIAA0157 protein levels can indirectly influence ubiquitin landscapes within cellular compartments in which it is not present.
Interestingly, along with γH2AX (27), RAP80 is the second potential target of BRCC36 Lys63-specific DUB activity, as revealed by the presence of higher migrating forms associated with BRCC36 QSQ in comparison with BRCC36 WT. These higher forms of RAP80 were Ubc13-dependent and were also definitively identified as modified with K63-Ub by mass spectrometry. We postulate that RAP80 will be one of several Lys63-ubiquitinated proteins at DSBs. In line with this reasoning, RAP80 deficiency decreases BRCC36 DUB activity at DSBs and actually increases K63-Ub present at laser stripes, indicating that RAP80 itself is not responsible for the majority of the K63-Ub signal at DSBs. We propose a varied ubiquitin landscape at DSBs containing more than one major target of K63-Ub, enabling parallel signaling events to occur simultaneously during the DDR (30, 48).
Regulation of DUB activity is therefore contained within multiple distinct elements, including protein-protein interactions and a competition between limiting components that shuttle between the cytoplasm and nucleus (Fig. 7). These elements also point to DUB inhibition strategies that disrupt non-catalytic domains of proteins associated with BRCC36 or Poh1 within the 19 S proteasome. Conceivably, such approaches may offer an opportunity to achieve more specific DUB inhibition among JAMM domain DUB enzymes. Furthermore, it may be possible to selectively inhibit DUB activity within the BRISC or BRCA1-RAP80 complexes, given their differential reliance on protein associations for DUB activity. Together, they suggest a multileveled regulation of DUBs that includes essential biochemical interactions and higher order assembly of protein complexes in different cellular compartments.
Supplementary Material
Acknowledgments
Results from these studies were shared with J. Chen (M. D. Anderson Cancer Center) prior to publication. We thank R. Tomaino and S. Gygi (Harvard Medical School) for discussion of mass spectrometry data. We also acknowledge, from the University of Pennsylvania, members of the Greenberg laboratory and B. Black, J. A. Diehl, S. Y. Fuchs, and B. Johnson for helpful discussions and L. Pontano and M. Romero for advice on baculoviral transduction and protein purification in Sf9 cells.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grant 1R01CA138835-01 and K08 awards 1K08CA106597-01 and 3K08CA106597-04S1 (to R. A. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
- DDR
- DNA damage response
- BRISC
- BRCC36 isopeptidase complex
- DSB
- double strand break
- DUB
- deubiquitinating enzyme
- IRIF
- ionizing radiation-induced foci
- JAMM
- JAB1/MPN/Mov34 metalloenzyme
- K63-Ub and K48-Ub
- lysine 63- and lysine 48-ubiquitin, respectively
- MPN
- Mpr-1/Pad-1 N-terminal
- UEV
- ubiquitin E2 variant
- UIM
- ubiquitin-interacting motif
- IB
- immunoblot
- IF
- immunofluorescence
- IP
- immunoprecipitation
- bis-tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Aguilera A., Gómez-González B. (2008) Nat. Rev. Genet. 9, 204–217 [DOI] [PubMed] [Google Scholar]
- 2.Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 3.Bartek J., Lukas J. (2007) Curr. Opin. Cell Biol. 19, 238–245 [DOI] [PubMed] [Google Scholar]
- 4.Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 5.Pickart C. M., Cohen R. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 6.Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., Greenberg R. A. (2007) Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 8.Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 9.Greenberg R. A., Sobhian B., Pathania S., Cantor S. B., Nakatani Y., Livingston D. M. (2006) Genes Dev. 20, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X., Chen J. (2004) Mol. Cell. Biol. 24, 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao G., Patterson-Fortin J., Messick T. E., Feng D., Shanbhag N., Wang Y., Greenberg R. A. (2009) Genes Dev. 23, 740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng L., Huang J., Chen J. (2009) Genes Dev. 23, 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B., Hurov K., Hofmann K., Elledge S. J. (2009) Genes Dev. 23, 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., Elledge S. J. (2007) Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H., Chen J., Yu X. (2007) Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 16.Yan J., Kim Y. S., Yang X. P., Li L. P., Liao G., Xia F., Jetten A. M. (2007) Cancer Res. 67, 6647–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B., Elledge S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 20.Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao G. Y., Sonoda E., Barber L. J., Oka H., Murakawa Y., Yamada K., Ikura T., Wang X., Kobayashi M., Yamamoto K., Boulton S. J., Takeda S. (2007) Mol. Cell 25, 663–675 [DOI] [PubMed] [Google Scholar]
- 22.Bekker-Jensen S., Rendtlew Danielsen J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. (2010) Nat. Cell Biol. 12, 80–86 [DOI] [PubMed] [Google Scholar]
- 23.Yao T., Cohen R. E. (2002) Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 24.Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 25.Ambroggio X. I., Rees D. C., Deshaies R. J. (2004) PLoS Biol. 2, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper E. M., Cutcliffe C., Kristiansen T. Z., Pandey A., Pickart C. M., Cohen R. E. (2009) EMBO J. 28, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao G., Lilli D. R., Patterson-Fortin J., Coleman K. A., Morrissey D. E., Greenberg R. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3166–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper E. M., Boeke J. D., Cohen R. E. (2010) J. Biol. Chem. 285, 10344–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenberg R. A. (2008) Chromosoma 117, 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messick T. E., Greenberg R. A. (2009) J. Cell Biol. 187, 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakatani Y., Ogryzko V. (2003) Methods Enzymol. 370, 430–444 [DOI] [PubMed] [Google Scholar]
- 32.Scully R., Chen J., Ochs R. L., Keegan K., Hoekstra M., Feunteun J., Livingston D. M. (1997) Cell 90, 425–435 [DOI] [PubMed] [Google Scholar]
- 33.Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M. B., Bartek J., Lukas J. (2006) J. Cell Biol. 173, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haas W., Faherty B. K., Gerber S. A., Elias J. E., Beausoleil S. A., Bakalarski C. E., Li X., Villén J., Gygi S. P. (2006) Mol. Cell Proteomics 5, 1326–1337 [DOI] [PubMed] [Google Scholar]
- 36.Nikkilä J., Coleman K. A., Morrissey D., Pylkäs K., Erkko H., Messick T. E., Karppinen S. M., Amelina A., Winqvist R., Greenberg R. A. (2009) Oncogene 28, 1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 38.Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 39.Sundquist W. I., Schubert H. L., Kelly B. N., Hill G. C., Holton J. M., Hill C. P. (2004) Mol. Cell. 13, 783–789 [DOI] [PubMed] [Google Scholar]
- 40.Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., Fukai S. (2008) Nature 455, 358–362 [DOI] [PubMed] [Google Scholar]
- 41.Komander D., Clague M. J., Urbé S. (2009) Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 42.Sharon M., Taverner T., Ambroggio X. I., Deshaies R. J., Robinson C. V. (2006) PLoS Biol. 4, e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan J., Kim Y. S., Yang X. P., Albers M., Koegl M., Jetten A. M. (2007) Nucleic Acids Res. 35, 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nijman S. M., Huang T. T., Dirac A. M., Brummelkamp T. R., Kerkhoven R. M., D'Andrea A. D., Bernards R. (2005) Mol. Cell 17, 331–339 [DOI] [PubMed] [Google Scholar]
- 45.Huang T. T., Nijman S. M., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D'Andrea A. D. (2006) Nat. Cell Biol. 8, 339–347 [DOI] [PubMed] [Google Scholar]
- 46.Cohn M. A., Kowal P., Yang K., Haas W., Huang T. T., Gygi S. P., D'Andrea A. D. (2007) Mol. Cell 28, 786–797 [DOI] [PubMed] [Google Scholar]
- 47.Hu M., Li P., Song L., Jeffrey P. D., Chenova T. A., Wilkinson K. D., Cohen R. E., Shi Y. (2005) EMBO J. 24, 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A. (2010) Cell 141, 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.