Abstract
BRCC36 is a member of the JAMM/MPN+ family of zinc metalloproteases that specifically cleaves Lys 63-linked polyubiquitin chains in vitro. We and others showed previously that BRCC36 is a component of the BRCA1-A complex, which consists of RAP80, CCDC98/ABRAXAS, BRCC45/BRE, MERIT40/NBA1, BRCC36, and BRCA1. This complex participates in the regulation of BRCA1 localization in response to DNA damage. Here we provide evidence indicating that BRCC36 regulates the abundance of Lys63-linked ubiquitin chains at chromatin and that one of its substrates is diubiquitinated histone H2A. Moreover, besides interacting with CCDC98 within the BRCA1-A complex, BRCC36 also associates with another protein KIAA0157, which shares significant sequence homology with CCDC98. Interestingly, although CCDC98 functions as an adaptor of BRCC36 and regulates BRCC36 activity in the nucleus, KIAA0157 mainly localizes in cytosol and activates BRCC36 in the cytoplasm. Moreover, these two complexes appear to exist in fine balance in vivo because reduction of KIAA0157 expression led to an increase of the BRCA1-A complex in the nucleus. Together, these results suggest that scaffold proteins not only participate in the regulation of BRCC36 activity but also determine its subcellular localization and cellular functions.
Keywords: Chromatin Histone Modification, Deubiquitination, DNA Damage, Subcellular Fractionation, Ubiquitination
Introduction
Ubiquitin (Ub)2 is a protein of 76 residues that is highly conserved from yeast to humans. Conjugation of Ub needs a cascade of reactions that involve E1, E2, and E3 enzymes, which ultimately lead to the formation of an isopeptide bond between C-terminal Gly of Ub and a Lys residue on the substrate. Ubiquitin contains seven lysine residues (at positions 6, 11, 27, 29, 33, 48, and 63), and polyubiquitin chain assembly can occur at any of these lysine residues (1). Lys48-linked polyubiquitination of proteins is quite common and normally targets substrates for proteolysis by 26 S proteasome, whereas Lys63-linked polyubiquitination is not typically associated with protein degradation (2, 3). Instead, Lys63-linked ubiquitination modification is often a signaling event and has been shown to participate in diverse cellular functions, including endocytosis, autophagy, NF-κB activation, and DNA damage repair (4–7).
Opposing the functions of E3 ligases that promote protein ubiquitination, deubiquitinating enzymes (DUBs) are proteases that specifically remove ubiquitin moieties from substrates. Although there are around 600 E3 ligases in humans, there are only ∼80 DUBs, implying that DUB activities may be regulated by their associated proteins. These DUBs can be divided into five subfamilies: UCH (ubiquitin C-terminal hydrolase), USP (ubiquitin-specific protease), OUT (ovarian tumor protease), Josephin, and JAMM/MPN+ (Joesphin and JAB1/MPN/MOV34 metallloenzyme) families (8–10). Except for the JAMM/MPN+ family of DUBs that are zinc metalloproteases, all of the other DUBs are cysteine proteases.
The JAMM domain is found in all three major kingdoms of life, bacteria, archaea, and eukarya, although bacteria do not have deubiquitinating activity. Therefore, it is suggested that JAMM domain may have adopted a new function as a protease during evolution. At least five JAMM/MPN+ domain-containing DUBs have been reported. These include the 26 S proteasome-associated POH1 (a human PAD1 homolog, also known as Rpn11 in yeast) (11, 12), CSN5 (COP9 signalosome subunit 5) (13), the ESCRT machinery-associated DUBs AMSH and AMSH-LP (14), and BRCC36 (BRCA1-BRCA2-containing complex subunit 36) (15). These five DUBs have distinct cellular functions. POH1 cleaves at or near the proximal end of the polyubiquitin chain and is required for proteasome integrity (11, 12), whereas CSN5 removes Ub-like protein Nedd8 from Cullin1 (13). Incorporation into large protein complexes is required for the activation of POH1 and CSN5 enzymatic activities, but this is not the case for AMSH and AMSH-LP. These two have intrinsic Lys63-specific DUB activity, which is determined by their abilities to specifically recognize Lys63-linked ubiquitin chains over other linkages (14, 16, 17).
The fifth member of this family is BRCC36. BRCC36 is a component of the BRCA1-A complex, which also contains a ubiquitin-binding motif (UIM) domain-containing protein (RAP80), a coiled-coil domain-containing protein (CCDC98/ABRAXAS), BRCC45/BRE, MERIT40/NBA1, and BRCA1. This complex is responsible for the stable accumulation of BRCA1 at sites of DNA breaks and thus plays a role in DNA damage response (15, 18–21). However, exactly how this complex works in vivo remains elusive. Especially, it is not yet clear how BRCC36 activity is controlled in the cell. To gain further insight into BRCC36 function in vivo, we generated cell line with stable BRCC36 knockdown and observed accumulation of chromatin-associated Lys63-linked Ub chains in these cells, indicating that BRCC36 is normally involved in the regulation of Lys63-linked ubiquitin chain formation in the nucleus. Interestingly, besides the BRCC36-containing BRCA1-A complex, we also found that BRCC36 associates with KIAA0157, a protein that shares extensive sequence homology with CCDC98/ABRAXAS. Our further analyses of these two BRCC36-containing complexes suggest that BRCC36 activity and its localization are highly regulated in the cell.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
RAP80, CCDC98, and BRCC36 polyclonal antibodies were generated as described previously (18, 22, 23). KIAA0157 polyclonal antibody was raised by immunizing rabbits with GST-KIAA0157 fusion protein (residue 1–196 and 296–419) and was subsequently affinity-purified. Additional antibodies used in this study are as follows: anti-β-actin antibody (Sigma), anti-FLAG antibody M2 (Sigma), anti-Myc antibody 9E10 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-ubiquitin antibody P4D1-A11 (Upstate Biotechnology Inc.), anti-ubiquitinated-H2A antibody E6C5 (Millipore), anti-BRCA1 antibody (D-9, Santa Cruz Biotechnology, Inc.), anti-H3 antibody (Upstate Cell Signaling), and anti-BRCC36 antibody (ProSci Inc.). RAP80 UIM-agarose beads, poly-Ub Lys63-linked chains (Ub2 to -7), and poly-Ub Lys48-linked chains (Ub2 to -7) (Boston Biochem). BRCC36 shRNA-resistant constructs were generated by introducing silent mutations (5′-CCAACAGCATTTACAAGAGCT-3′) in the shBRCC36 targeting sequence.
The BRCC36 inactive mutant (S132A/D135N) was generated using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by sequencing. Plasmid encoding SFB-Lys63 only was a gift from Michael S. Y. Huen (Hong Kong University of Science and Technology), and plasmids encoding His-Ub was a gift from Richard Baer (Columbia University).
Cell Culture, Transfection, and Establishment of Stable Cell Lines
HeLa cells were maintained in RPMI/1640 medium containing 10% bovine serum and penicillin/streptomycin. Transient transfection was performed with the polyethylenimine (25 kDa) method. Stable knockdown cell lines were established by transfecting HeLa with pLKO.1 empty vector or shRNAs (Open Biosystems) that specifically target BRCC36 (5′-CCAACAGCATTTGCAGGAATT-3′), CCDC98 (5′-GCATGTCTGAACAACTGGGTT-3′), or KIAA0157 (5′-CAGAGCCTTCTAATAGTGAAT-3′). Puromycin (2 μg/ml)-resistant clones were selected, and down-regulation of targeted genes was verified by Western blotting. Puromycin was withdrawn during subsequent culture.
Chromatin Fractionation and Pull-down Assay
HeLa cells were harvested, resuspended in high salt NETN buffer (20 mm Tris, pH 8.0, 500 mm NaCl, 0.5% Nonidet P-40, 1 mm EDTA) supplemented with protease inhibitor and 5 mm NEM, and then incubated on ice for 30 min. Pellets were washed twice using the same buffer and extracted with 5 volumes of 0.2 n HCl on ice for 30 min. The extracted chromatin fractions were neutralized with 1 volume of 1 m Tris (pH 8.8). 10 volumes of normal NETN buffer supplemented with protease inhibitor and 5 mm NEM were added for the pull-down assay.
Cytoplasmic and Nuclear Fractionation
The cell pellet was resuspended in 10 volumes of cold buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 1 mm DTT) containing protease inhibitor and allowed to swell on ice for 30 min. Nonidet P-40 was then added to a final concentration of 0.2%. After votexing continuously for 5 s, the homogenate was spun for 5 min at 3000 rpm. The supernatant containing the cytoplasmic fraction was transferred to a new tube, and the concentration of NaCl was adjusted to 200 mm with an equal volume of buffer C (20 mm HEPES, pH 7.9, 0.4 m NaCl, 0.4% Triton X-100, 1 mm DTT). The crude nuclear pellet was washed once in buffer A and then suspended in buffer C with protease inhibitor, vigorously vortexed, and put on ice for 30 min. The homogenate was centrifuged again at high speed. The clarified supernatant containing the nuclear fraction was transferred to a new tube, and the concentration of NaCl was adjusted to 200 mm by adding an equal volume of buffer A.
In Vitro Deubiquitination Assay
Purified proteins were incubated with 250 ng of Lys63-linked ubiquitin chains in DUB reaction buffer (20 mm HEPES, pH 7.4, 100 mm NaCl, 1 mm DTT) at 37 °C. Reactions were stopped at the indicated time by the addition of SDS sample buffer. Samples were resolved on 12% SDS-polyacrylamide gels and blotted with anti-ubiquitin antibody.
Detection of H2A Ubiquitination in Vivo
HeLa cells were transfected with various constructs as indicated and harvested 48 h later. Cells were lysed in phosphate/guanidinium buffer (6 m guanidinium HCl, 0.1 m phosphate, pH 8.0, 1 mm DTT, and 5 mm imidazole) with sonication. The ubiquitinated proteins were affinity-purified on nickel affinity gel (Sigma), eluted with SDS sample buffer, and immunoblotted with anti-Myc antibody.
Baculoviral Expression and Purification
We generated baculoviruses encoding the following proteins: GST-RAP80, SFB-BRCC36, SFB-KIAA0157, SFB-CCDC98, His6-BRCC45, and untagged MERIT40. Sf9 cells were co-infected with various baculovirus stocks and harvested 48 h later. Cells were lysed in NETN buffer supplemented with protease inhibitor and centrifuged to remove insoluble material. Supernatants were subjected to sequential affinity chromatography using streptavidin-Sepharose and S-protein-agarose beads (18).
Tandom Affinity Purification (TAP), Irradiation, Immunostaining, and Immunoprecipitation
All of these procedures were performed as described previously (18).
RESULTS
BRCC36 Regulates Lys63-linked Ubiquitin Conjugates in Chromatin Fractions
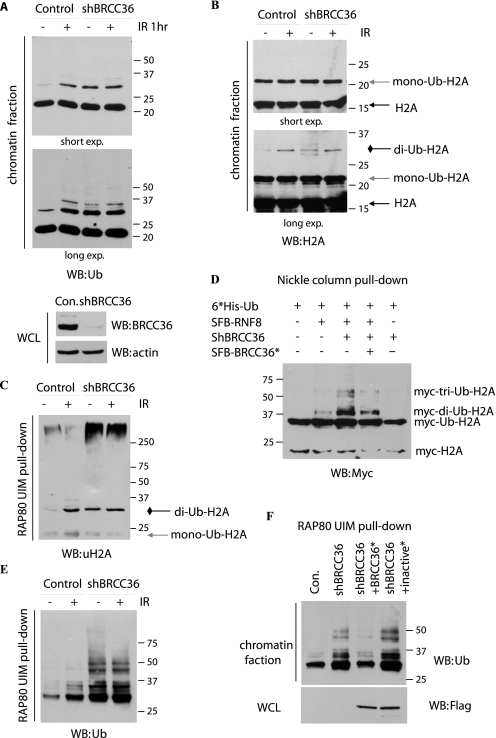
Earlier studies have already established that BRCC36 only cleaves Lys63-linked polyubiquitin chains (2, 24, 25), and it localizes at sites of DNA double strand breaks (DSBs) (20). These observations suggest that this highly specified DUB may modulate Lys63-linked polyubiquitin chains that are known to be involved in DNA damage response. Thus, we tested whether the down-regulation of BRCC36 would enhance polyubiquitin chain formation in the chromatin fraction. We used high salt condition to remove all soluble fractions and proteins that only loosely associate with chromatin. Western blotting with anti-Ub antibody indicated that the most abundant ubiquitinated proteins in chromatin fraction are proteins of ∼22 kDa, which correspond to monoubiquitinated histones (see below). The abundance of these monoubiquitinated histone species did not change after DNA damage. However, another ubiquitinated band of ∼30 kDa quickly appeared following IR treatment (Fig. 1A, lanes 1 and 2), which is likely to be the diubiquitinated H2A/H2AX, as reported previously (26, 27) (see below). Consistent with the report that BRCC36 could not cleave monoubiquitinated substrates (24), the intensity of the 22-kDa ubiquitinated species did not change in BRCC36-deficient cells. However, the ∼30-kDa ubiquitinated proteins were up-regulated in BRCC36-deficient cells even in the absence of IR (Fig. 1A, lanes 3 and 4), indicating that this ubiquitinated protein(s) is probably a chromatin substrate(s) of BRCC36 in vivo.
FIGURE 1.
BRCC36 deubiquitinates chromatin-associated Lys63-linked ubiquitin chains. A, HeLa cells stably transfected with vector alone or BRCC36-specific shRNA were mocked-treated or treated with ionizing radiation. Cells were collected 1 h later. Chromatin fractions were isolated and analyzed by immunoblotting using anti-ubiquitin antibody. Two different exposures of the same blot were shown (top). Whole-cell lysates (WCL) were immunoblotted to indicate the expression of BRCC36 in control or knockdown cells (bottom). B, experiments were carried out as described in A, except that immunoblotting was conducted using anti-H2A antibody. C, control or BRCC36 knockdown cells were irradiated or left untreated. Chromatin fractions were prepared and subjected to a pull-down assay using RAP80-UIM agarose beads. Bound proteins were separated by SDS-PAGE and subjected to anti-uH2A immunoblotting. D, ubiquitination of H2A in the presence of RNF8 and BRCC36. Cells were lysed in denaturing buffer, and the His-tagged ubiquitinated proteins were purified using a nickel column and blotted with anti-Myc antibody. See “Experimental Procedures” for details. E, experiments were conducted similarly to that described in C and immunoblotted with anti-ubiquitin antibody. F, BRCC36-deficient HeLa cells were transfected with plasmids encoding shRNA-resistant SFB-tagged wild type or catalytic inactive mutant (S132A/D135N) of BRCC36. Experiments were conducted similarly to that described in E.
Next we wanted to determine whether or not H2A/H2AX are substrates of BRCC36. Histone H2A and H2AX are targets of E3 ligases RNF8 and RNF168. For example, RNF8 is known to be responsible for increased H2AX diubiquitination upon IR (26). RNF168 interacts with diubiquitinated H2A through its MIU domains (28). Moreover, a Lys63-specific E2 enzyme UBC13 functions together with RNF8 and RNF168 (26–29). These data suggest that histones H2A and H2AX are substrates of RNF8/RNF168, which probably promote Lys63-dependent ubiquitination events. Based on these findings, we reasoned that the same Lys63-linked H2A might also be the substrates of BRCC36. Indeed, BRCC36 knockdown was accompanied by an increase in the level of diubiquitinated H2A, which is ∼30 kDa (Fig. 1B). To further confirm that the 30-kDa H2A is Lys63-polyubiquitinated H2A, we took advantage of the fact that the two UIM domains of RAP80 have preferential binding to Lys63-linked Ub chains (30, 31). Thus, we used RAP80 UIM agarose beads as affinity matrix to pull down Lys63-linked ubiquitinated chains from chromatin fractions. As expected, the ∼30-kDa band was recognized by anti-uH2A antibody (Fig. 1C). Moreover, higher molecular weight ubiquitinated H2A species were also enriched in BRCC36-deficient cells (Fig. 1D). These results suggest that diubiquitinated and poly-Lys63-ubiquitinated H2A are BRCC36 substrates in vivo. To further confirm these results, HeLa cells were transfected with plasmids encoding Myc-tagged H2A together with plasmids encoding His6-tagged Ub, RNF8, or BRCC36. Nickel column affinity chromatography was performed, followed by Western blotting using anti-Myc antibody. As reported previously (26, 27), RNF8 induced di- and polyubiquitination of H2A (Fig. 1E, lane 2), which was further enhanced with the knockdown of BRCC36 (Fig. 1D, lane 3). In addition, reconstitution using shRNA-resistant BRCC36 restored normal levels of H2A ubiquitination (Fig. 1E, lane 4), indicating that BRCC36 antagonizes RNF8-mediated H2A ubiquitination. These results suggest that at least one of the substrates of BRCC36 is ubiquitinated H2A.
We also observed higher molecular weight bands following ionizing radiation or in BRCC36-deficient cells (Fig. 1A, long exp.). The pattern of these ubiquitinated bands is very similar in control cells following ionizing radiation with that observed in BRCC36-deficient cells, suggesting that BRCC36 is the major DUB involved in the removal of ubiquitin conjugates in chromatin fractions. Using a RAP80 UIM-agarose bead pull-down assay similar to that shown in Fig. 1C, Lys63-specific ubiquitin conjugates increased quickly upon IR in control cells (Fig. 1E, lanes 1 and 2); however, such an increase was not observed in BRCC36-depleted cells (Fig. 1E, lanes 3 and 4). The major difference between control and BRCC36-depleted cells is at the basal levels of Lys63-specific ubiquitin conjugates in non-irradiated cells (Fig. 1E, lanes 1 and 3). These data suggest that BRCC36 negatively regulates the chromatin-associated, Lys63-linked ubiquitin chain formation in vivo. To confirm that this involvement of BRCC36 in Lys63-linked ubiquitin chain formation requires the enzymatic activity of BRCC36, we introduced the shRNA-resistant wild-type BRCC36 or its inactive mutant S132A/D135N (32) into BRCC36-deficient cells. Only wild-type BRCC36 and not its catalytic inactive mutant decreased the ubiquitin conjugates that associated with RAP80 UIM-agarose beads (Fig. 1F), validating that BRCC36 DUB activity is needed for the regulation of chromatin-associated Lys63-linked ubiquitin chains.
Only BRCC36-containing Complexes and Not BRCC36 Alone Have DUB Activity
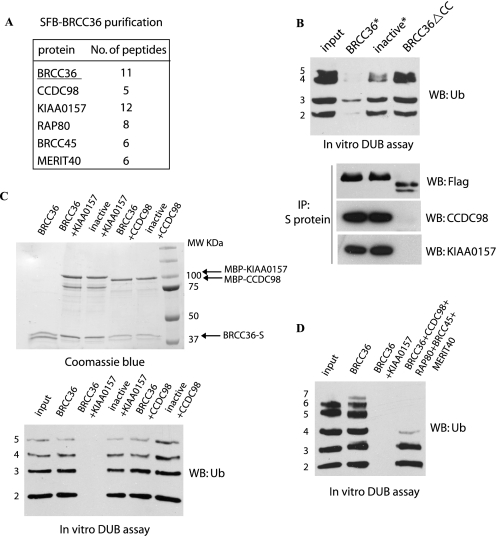
Besides the BRCA1-A complex including RAP80, CCDC98/Abraxas, BRCC45/BRE, and MERIT40/NBA1, we also identified another protein, KIAA0157, as a BRCC36-associated protein (Fig. 2A). KIAA0157 is 39% identical to CCDC98 at its N-terminal region, which contains the JAMM/MPN+ domain and a coiled-coil domain (33, 34). Previous studies suggest that the JAMM/MPN+ domain binds to BRCC45/BRE, whereas the coiled-coil domain is responsible for its interaction with BRCC36 (18, 19, 21, 33). The major difference between CCDC98 and KIAA0157 is that KIAA0157 lacks the pSXXF motif at its very C terminus, which is the motif that mediates the interaction between CCDC98 and BRCA1 (23, 34).
FIGURE 2.
Characterization of BRCC36 deubiquitination activity in vitro. A, 293T cells stably expressing SFB-tagged BRCC36 were used for Tandom Affinity Purification (TAP). The table is a summary of proteins identified by mass spectrometry analysis. B, BRCC36-deficient HeLa cells were transfected with constructs encoding shRNA-resistant SFB-tagged wild-type, inactive mutant (S132A/D135N) or coiled-coil domain deletion mutant (ΔCC) of BRCC36. Cell lysates were subjected to precipitation (IP) using S-protein beads. 250 ng of Lys63-linked ubiquitin chains (Ub2 to -7) were incubated with the indicated immunocomplexes for 2 h at 37 °C. Products were analyzed by anti-ubiquitin immunoblotting (WB). C, in vitro DUB assay using bacterially expressed S-tagged BRCC36 alone (BRCC36-S) or its inactive mutant (S132A/D135N) coexpressed and co-purified with MBP-tagged KIAA0157 or MBP-tagged CCDC98. DUB reactions were performed similarly to that described in B. BRCC36 and its associated proteins were visualized by Coomassie Blue staining. D, in vitro DUB assay using insect cell-expressed BRCC36, BRCC36-KIAA0157 complex, or the five-subunit complex containing BRCC36-CCDC98-BRCC45-MERIT40-RAP80. Experiments were conducted similarly to that described in B.
A previous study (15) indicated that BRCC36 expressed and purified from insect cells was catalytically inactive. However, the BRCC36 complexes isolated from HeLa cells were proficient in cleaving Lys63-linked ubiquitin chains (Fig. 2B, top), suggesting that BRCC36 DUB activity is likely to be regulated by its associated proteins. Indeed, the coiled-coil domain deletion mutant of BRCC36, which still contains the intact JAMM/MPN+ domain, failed to associate with either CCDC98 or KIAA0157 and did not display any DUB activity in vitro (Fig. 2B). To determine how BRCC36 activity is regulated by its binding partners, we purified BRCC36 alone, BRCC36-KIAA0157 complex, or BRCC36-CCDC98 complex from bacteria. In agreement with a recent report (15, 24), although BRCC36 alone was catalytically inactive, the BRCC36-KIAA0157 complex showed robust DUB activity (Fig. 2C). Surprisingly, a similar CCDC98-BRCC36 complex was catalytically inactive (Fig. 2C). In addition, we failed to detect DUB activity in either the RAP80-CCDC98-BRCC36 subcomplex or the CCDC98-BRCC36-BRCC45-MERIT subcomplex (data not shown). Only the five-subunit complex containing RAP80, CCDC98, BRCC45, MERIT40, and BRCC36 displayed in vitro DUB activity (Fig. 2C). This scenario is very similar to POH1 and CSN5, which also need to be assembled into multisubunit protein complexes like proteasome (11, 12) or COP9 signalosome (13) to exhibit their DUB activities.
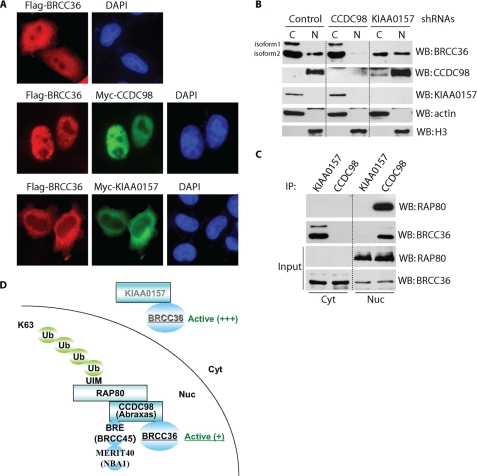
The Two Scaffold Proteins KIAA0157 and CCDC98 Determine the Subcellular Localization of BRCC36
Although both RAP80 and CCDC98 are nuclear proteins (20, 22, 23, 34, 35), the KIAA0157-BRCC36 complex was recently isolated from S100 fraction (24), indicating that KIAA0157 may exist in cytoplasm. Indeed, epitope-tagged KIAA0157 mainly localized in cytosol (Fig. 3A). Interestingly, ectopically expressed SFB-tagged BRCC36 showed both cytoplasmic and nuclear localization; however, co-transfection of BRCC36 with CCDC98 resulted in predominant nuclear localization of BRCC36. In contrast, co-transfection with KIAA0157 promoted cytoplasmic translocation of BRCC36 (Fig. 3A). Similarly, Western blot analysis indicated that endogenous BRCC36 existed in both cytosol and nucleus, and knockdown of endogenous CCDC98 resulted in a dramatic reduction of nuclear BRCC36 (Fig. 3B). Although the reduction of KIAA0157 expression decreased the cytoplasmic pool of BRCC36, it did not affect the abundance of BRCC36 in nucleus (Fig. 3B). Co-immunoprecipitation experiments further confirmed that KIAA0157 only interacted with BRCC36 in cytosol, whereas CCDC98 (and RAP80) associated with BRCC36 in nuclear fractions (Fig. 3C). Together, these data indicate that there are two cellular pools of BRCC36. The KIAA0157-BRCC36 complex mainly exists in cytosol and may regulate cytoplasmic function of BRCC36, whereas CCDC98 determines the nuclear localization of BRCC36, and they form a nuclear complex with three additional components, RAP80, BRCC45, and MERIT40, which is important for nuclear function of BRCC36, especially in response to DNA damage (Fig. 3D).
FIGURE 3.
KIAA0157 and CCDC98 determine the subcellular localization of BRCC36. A, HeLa cells were transfected with constructs encoding SFB-tagged BRCC36 alone or together with constructs encoding Myc-tagged KIAA0157 or Myc-tagged CCDC98. Immunostaining was conducted using antibodies as indicated. B, HeLa cells stably transfected with empty vector, KIAA0157, or CCDC98-specific shRNAs were used for cellular fractionation experiments. Equal amounts of cytoplasmic (C) or nuclear (N) proteins were loaded and blotted (WB) with antibodies as indicated. Please note that BRCC36 has two splicing isoforms. C, cytoplasmic and nuclear fractions were subjected to immunoprecipitation (IP) using anti-KIAA0157 or CCDC98 antibodies and immunoblotted with antibodies as indicated. D, a model of BRCC36 in two distinct protein complexes.
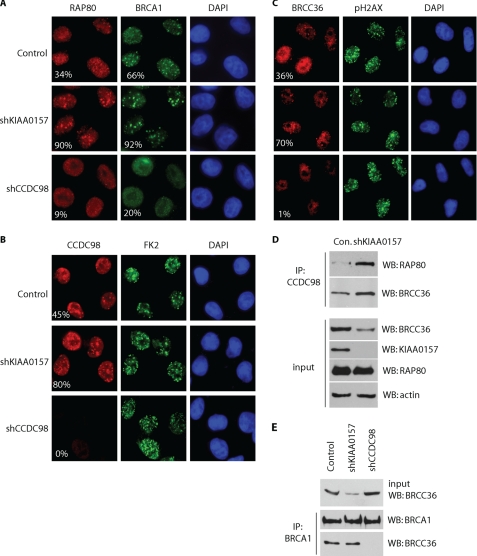
Loss of KIAA0157 Expression Enhances the Assembly of Nuclear BRCC36-containing Complex
Next we asked whether KIAA0157-BRCC36 and CCDC98-BRCC36 are two independent complexes or if they can communicate with each other. We did not observe any clear cytoplasmic to nuclear trans-localization of BRCC36 following ionizing radiation (data not shown). However, we noticed that IRIF of the BRCA1-A complex was enhanced in KIAA0157-depleted cells, as suggested by immunostaining using antibodies recognizing RAP80, BRCA1, CCDC98, or BRCC36 (Fig. 4, A–C). On the other hand, loss of CCDC98 greatly reduced the foci formation of RAP80, BRCA1, and BRCC36, as previously reported (18, 20, 21, 23, 34). These results indicate that the loss of cytoplasmic BRCC36 complex could promote the assembly of nuclear BRCC36 complex.
FIGURE 4.
Loss of cytoplasmic scaffold protein KIAA0157 enhances the formation and focus localization of BRCA1-A complex. A and B, HeLa cells stably transfected with empty vector, KIAA0157, or CCDC98-specific shRNAs were irradiated (10 grays). 2 h later, cells were fixed and immunostained with anti-RAP80 and BRCA1 antibodies (A), anti-CCDC98 and anti-conjugated ubiquitin FK2 antibodies (B), or anti-BRCC36 antibody (C). For the visualization of BRCC36 in nuclear foci (C), cells were pre-extracted with 0.5% Triton before fixation. The staining of BRCC36 in CCDC98 knockdown cells was dim because CCDC98 is responsible for nuclear localization of BRCC36. More than 150 cells in each sample were counted to evaluate the percentage of IRIF-positive nuclei (cells with more than five foci). D, HeLa cells stably transfected with empty vector, KIAA0157, or CCDC98-specific shRNAs were harvested. Cell lysates were immunoprecipitated (IP) with anti-CCDC98 antibody and immunoblotted (WB) with indicated antibodies. E, CCDC98, but not KIAA0157, is required for BRCC36/BRCA1 interaction. Lysates prepared from HeLa cells stably transfected with empty vector, KIAA0157, or CCDC98-specific shRNAs were immunoprecipitated with anti-BRCA1 antibody and immunoblotted with antibodies as indicated.
As shown in Fig. 4D, although depletion of KIAA0157 destabilized BRCC36 and led to an overall reduction of BRCC36 expression in the cell, the loss of KIAA0157 actually enhanced the interaction between endogenous CCDC98 with RAP80 and BRCC36 (Fig. 4D). We also examined BRCC36/BRCA1 interaction in the absence of either KIAA0157 or CCDC98. Consistent with previous reports (18, 21), knockdown of CCDC98 abolished the interaction between BRCC36 and BRCA1. In contrast, although KIAA0157 depletion greatly decreased total level of BRCC36, the BRCC36/BRCA1 interaction was not affected (Fig. 4D).
DISCUSSION
In this study, we provide evidence indicating that BRCC36 is a Lys63 chain-specific DUB that acts to modulate chromatin-associated ubiquitin chain formation. Besides BRCC36, another DUB USP3 has also been implicated in DNA damage response (36). USP3 belongs to the USP family, and its depletion enhances monoubiquitination of H2A and H2B (36). USP3 does not seem to localize to DNA break sites, which may be explained by the transient nature of its interaction with its chromatin substrates (36). On the contrary, BRCC36 is readily detected at DSB sites and its foci formation depends on RAP80, which contains tandem UIMs that specifically bind to Lys63-linked polyubiquitinated chains (20, 22, 34). The paradox is that although BRCC36 depletion leads to the accumulation of chromatin-associated Lys63-linked ubiquitin chains (Fig. 1), it compromises RAP80 IRIF formation (18, 19, 21). Our in vitro DUB assay showed that the RAP80-CCDC98-BRCC36-BRCC45-MERIT40 complex is not a very active DUB, and the disassembly of the Lys63 chain is incomplete even after long term incubation (Fig. 2D). Thus, we speculate that the main function of this five-subunit BRCC36-containing nuclear complex is not just to remove all Lys63-linked ubiquitin chains at DSB sites. Instead, it may act with BRCA1 and promote BRCA1-dependent non-canonical K6-linked protein ubiquitination (37, 38). Further studies are needed to address whether the DUB activity of BRCC36 in the RAP80-CCDC98-containing complex would facilitate BRCA1 E3 ligase activity and more importantly whether a ubiquitin chain editing event occurs at DSB sites. One can image that with the help of a nuclear BRCC36-containing complex, the initial Lys63-linked ubiquitin chains formed at DSB sites may be gradually switched to BRCA1-dependent Lys6-linked ubiquitin chains for certain yet-to-be-identified functions in DNA damage response.
Another unexpected observation is that depletion of BRCC36 mainly affects the formation of ubiquitin conjugates in non-irradiated cells (Fig. 1), implying that a key aspect of BRCC36 function is to diminish the basal level of chromatin-associated ubiquitin chains. It is likely that this function of BRCC36 is to prevent premature activation of DNA damage response. This negative role of BRCC36 in ubiquitin chain formation can be overcome following DNA damage by the specific recruitment of E3 ligases RNF8 and RNF168 to sites of DNA damage and thus allow the proper activation of ubiquitin-dependent DNA damage signaling pathways. As we discussed above, the exact task of BRCC36 at DNA damage foci remains to be determined.
Besides nuclear BRCC36, there is also a fraction of BRCC36 existing in the cytoplasm, which is activated by a CCDC98-like protein KIAA0157. Although the binding of KIAA0157 to BRCC36 is sufficient to activate BRCC36, the association of CCDC98 with BRCC36 is not. These observations clearly indicate that these two scaffold proteins differentially regulate BRCC36 catalytic activities. In addition, within the JAMM/MPN+ family, only AMSH and AMSH-LP have intrinsic Lys63-specific DUB activity because they have two unique insertions at their JAMM domain, which are absent in BRCC36, POH1, or CSN5 (17). These unique insertions allow AMSH and AMSH-LP to bind specifically to Lys63-linked ubiquitin chains, which may be responsible for its specificity toward Lys63-linked ubiquitin chains (17). However, KIAA0157 binds to both Lys48 and Lys63 chains (24, 33), and thus the chain specificity of KIAA0157-BRCC36 complex is not due to the selective binding of this complex to Lys63-linked ubiquitin chain. Further structural studies are needed to explore the molecular mechanism of linkage selectivity of these DUB complexes.
Acknowledgments
We thank all colleagues in the Chen laboratory for insightful discussion and technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA089239 (to J. C.) and CA016672 (to M. D. Anderson Cancer Center).
- Ub
- ubiquitin
- DUB
- deubiquitinating enzyme
- UIM
- ubiquitin-binding motif
- DSB
- double strand break
- SFB
- S-tag, FLAG tag, and streptavidin-binding peptide tag.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeda F., Dikic I. (2008) EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 5.Kirkin V., Dikic I. (2007) Curr. Opin. Cell Biol. 19, 199–205 [DOI] [PubMed] [Google Scholar]
- 6.Welchman R. L., Gordon C., Mayer R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 7.Clague M. J., Urbé S. (2006) Trends Cell Biol. 16, 551–559 [DOI] [PubMed] [Google Scholar]
- 8.Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 9.Komander D., Clague M. J., Urbé S. (2009) Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 10.Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao T., Cohen R. E. (2002) Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 12.Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 13.Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., Deshaies R. J. (2002) Science 298, 608–611 [DOI] [PubMed] [Google Scholar]
- 14.McCullough J., Clague M. J., Urbé S. (2004) J. Cell Biol. 166, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong Y., Hakimi M. A., Chen X., Kumaraswamy E., Cooch N. S., Godwin A. K., Shiekhattar R. (2003) Mol. Cell 12, 1087–1099 [DOI] [PubMed] [Google Scholar]
- 16.McCullough J., Row P. E., Lorenzo O., Doherty M., Beynon R., Clague M. J., Urbé S. (2006) Curr. Biol. 16, 160–165 [DOI] [PubMed] [Google Scholar]
- 17.Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., Fukai S. (2008) Nature 455, 358–362 [DOI] [PubMed] [Google Scholar]
- 18.Feng L., Huang J., Chen J. (2009) Genes Dev. 23, 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao G., Patterson-Fortin J., Messick T. E., Feng D., Shanbhag N., Wang Y., Greenberg R. A. (2009) Genes Dev. 23, 740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., Greenberg R. A. (2007) Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B., Elledge S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H., Chen J., Yu X. (2007) Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 23.Kim H., Huang J., Chen J. (2007) Nat. Struct. Mol. Biol. 14, 710–715 [DOI] [PubMed] [Google Scholar]
- 24.Cooper E. M., Boeke J. D., Cohen R. E. (2010) J. Biol. Chem. 285, 10344–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper E. M., Cutcliffe C., Kristiansen T. Z., Pandey A., Pickart C. M., Cohen R. E. (2009) EMBO J. 28, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 28.Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 29.Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 30.Sato Y., Yoshikawa A., Mimura H., Yamashita M., Yamagata A., Fukai S. (2009) EMBO J. 28, 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sims J. J., Cohen R. E. (2009) Mol. Cell 33, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambroggio X. I., Rees D. C., Deshaies R. J. (2004) PLoS Biol. 2, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B., Hurov K., Hofmann K., Elledge S. J. (2009) Genes Dev. 23, 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., Elledge S. J. (2007) Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan Z., Kim Y. S., Jetten A. M. (2002) J. Biol. Chem. 277, 32379–32388 [DOI] [PubMed] [Google Scholar]
- 36.Nicassio F., Corrado N., Vissers J. H., Areces L. B., Bergink S., Marteijn J. A., Geverts B., Houtsmuller A. B., Vermeulen W., Di Fiore P. P., Citterio E. (2007) Curr. Biol. 17, 1972–1977 [DOI] [PubMed] [Google Scholar]
- 37.Wu-Baer F., Lagrazon K., Yuan W., Baer R. (2003) J. Biol. Chem. 278, 34743–34746 [DOI] [PubMed] [Google Scholar]
- 38.Morris J. R., Solomon E. (2004) Hum. Mol. Genet. 13, 807–817 [DOI] [PubMed] [Google Scholar]