Abstract
Somatic cell nuclear transfer (SCNT) has shown tremendous potential for understanding the mechanisms of reprogramming and creating applications in the realms of agriculture, therapeutics, and regenerative medicine, although the efficiency of reprogramming is still low. Somatic nucleus reprogramming is triggered in the short time after transfer into recipient cytoplasm, and therefore, this period is regarded as a key stage for optimizing SCNT. Here we report that CBHA, a histone deacetylase inhibitor, modifies the acetylation status of somatic nuclei and increases the developmental potential of mouse cloned embryos to reach pre- and post-implantation stages. Furthermore, the cloned embryos treated by CBHA displayed higher efficiency in the derivation of nuclear transfer embryonic stem cell lines by promoting outgrowths. More importantly, CBHA increased blastocyst quality compared with trichostatin A, another prevalent histone deacetylase inhibitor reported previously. Use of CBHA should improve the productivity of SCNT for a variety of research and clinical applications, and comparisons of cells with different levels of pluripotency and treated with CBHA versus trichostatin A will facilitate studies of the mechanisms of reprogramming.
Keywords: Development, Histone Modification, Mouse, Reproduction, Somatic Cell Genetics, Histone Acetylation, m-Carboxycinnamic Acid Bishydroxamide (CBHA), Somatic Cell Nuclear Transfer
Introduction
Reprogramming of a terminally differentiated nucleus is successfully achieved in many species through somatic cell nuclear transfer (SCNT)3 technology. However, its efficiency remains very low, limiting its application in agriculture to well bred livestock propagation and in species preservation (1) and regenerative medicine. The reprogramming process remains largely unknown; at the time of its transfer into an enucleated oocyte the somatic donor nucleus is in a configuration very different from a germ cell or an embryonic nucleus. Its own specific program of gene expression will have to be turned on, and the specific epigenetic modifications and chromatin configuration were erased (2). Extensive chromatin remodeling takes place as soon as the somatic nucleus is in contact with the oocyte cytoplasm, and two main types of epigenetic marks are directly involved in gene expression regulation; that is, methylation of histones or DNA and acetylation of histones (3–5). During normal pre-implantation development, the embryonic genome is progressively demethylated up to the early blastocyst stage. This is followed by differential remethylation in the two lineages of the blastocyst, the pluripotent inner cell mass and the trophoblast (6), but this process has been shown to be error-prone in SCNT embryos (7, 8).
Attempts to improve reprogramming during nuclear transfer have, therefore, focused primarily on modifying the epigenetic configuration of the donor nuclei before nuclear transfer. Therefore, treating donor cells with pharmacological agents to remove some epigenetic marks before NT may improve the ability of the donor cells to be fully reprogrammed by the recipient ooplasm. Pretreatment of bovine fibroblast donor cells by DNA demethylation agents such as 5-aza-2′-deoxycytidine or S-adenosyl homocysteine has been shown to decrease the global methylation levels in the donor cells but provide little or no improvement on the blastocyst development of embryos reconstructed from the treated cells (9, 10). Histone-deacetylase inhibitors (HDACi) such as trichostatin A (TSA) enhance the pool of acetylated histones (11). There have been several previous attempts to use TSA and other HDACis to study the role of acetylation in nuclear reprogramming. TSA, applied to the donor cell before NT in the bovine species was shown to be able to improve the blastocyst rate (12). More recently, a major breakthrough has been made by treating the embryos just at the time of reconstruction in the mouse (13, 14). In these studies TSA was used to treat reconstructed embryos over the period of activation, and this treatment markedly improved the rate of blastocysts and live pups at term. Improvement was observed for different kind of somatic cells, including cumulus, fibroblasts, and spleen cells but not embryonic stem (ES) cells (13). A similar protocol has been applied for bovine NT cells, and it was observed to improve the rate and quality of blastocysts, but the long-term benefit remains to be tested (15). After its introduction into the oocyte cytoplasm, histones of the somatic chromatin are rapidly deacetylated and then reacetylated during formation of the pseudo-pronuclei (16). Although the dynamic pattern of histone acetylation mimics that of a fertilized embryo, the global level of acetylation is lower than in fertilized embryos, and this level increases after TSA treatment (16). TSA also improves chromatin remodeling in SCNT embryos (17).
HDACi are widely used as anti-cancer drugs, as they induce apoptosis in cancer cells, whereas normal cells are relatively resistant (18). Depending on their inhibitory activities on different HDACs, they may have different effects on cells and on nuclear reprogramming (19). Investigation of various HDACi is, therefore, important for two purposes. First, a continuous and reliable supply of developmentally competent SCNT embryos is necessary to advance the field, so even incremental improvements in production efficiency can be highly beneficial for laboratory work. Second, identification of HDACi that have different degrees of influence on the SCNT process will provide differential analysis reagents to investigate the mechanism(s) through which reprogramming efficiency is improved. In the present study, we tested the effect of m-carboxycinnamic acid bishydroxamide (CBHA) in SCNT, one member of HDACi family. We found that CBHA can also increase the somatic cell reprogramming efficiency in the mouse and appears even more efficient than TSA. More importantly, its beneficial action appears in full-term development and NT-ES cells derivation efficiency by elevating the quality of blastocysts.
EXPERIMENTAL PROCEDURES
Animals
B6D2F1 female mice (C57BL/6J x DBA/2) (8∼12 weeks old) were used to prepare oocytes and cumulus donor cells. Foster mice were female ICR strains, 8∼12 weeks old. Fertilized control embryos were obtained from superovulated C57Bl/6J females mated with DBA/2 males. Animals were handled according to the Guidelines for the Care and Use of Laboratory Animals established by the Beijing Association for Laboratory Animal Science.
Oocytes and Cumulus Cell Preparation
B6D2F1 females were superovulated with 10 IU pregnant mare's serum gonadotropin followed by 10 IU human chorionic gonadotropin 48 h later. Cumulus-oocyte complexes were collected from oviducts 15 h after human chorionic gonadotropin injection, and cumulus cells were removed using hyaluronidase 300 IU/ml (ICN Pharmaceuticals, Costa Mesa, CA). Before micromanipulation, oocytes were cultured in CZB (Cold Spring Harbor Laboratory) medium supplemented with 3 mg/ml BSA, covered with paraffin oil, and incubated at 37 °C (5% CO2, air). Cumulus cell suspensions were washed with Hepes-CZB medium twice and stored at 4 °C.
Nuclear Transfer
For cumulus nuclear transfer, the “one-step micromanipulation” method was performed as previously described (20). Briefly, a cumulus nucleus was injected into a recipient oocyte, and the meiotic metaphase plate was withdrawn while removing the pipette from cytoplasm after injection. Batches of 20–30 oocytes were placed into a drop of Hepes-CZB medium containing 5 μg/ml of cytochalasin B. The pipettes with an inner diameter of 6–8 μm used to inject cumulus donor nuclei were driven by a Piezo-electric device (Primatech). 1–2 h after nuclear transfer, the reconstructed embryos were activated by 10 mm SrCl2 in calcium-free CZB medium in the presence of 5 μg/ml cytochalasin B for 6 h and cultured in CZB medium at 37 °C, 5% CO2 for 4 days. The one-step micromanipulation method was also used in the R1 ES nuclear transfer process, and the method to prepare the R1 donor cell was performed as previously described (21).
TSA and CBHA Treatment Protocol
TSA (Sigma) was dissolved in DMSO to prepare a 40 mm concentration stock solution. Before use the 40 mm stock solution was diluted to 1 mm using DMSO. This intermediate solution can be stored at −20 °C.
CBHA (Calbiochem, lot D00008432, 5 mg) was also dissolved in DMSO, and the stock solution was 100 mm and stored at −80 °C. The stock solution was also diluted in 10 mm solution in DMSO and stored at −20 °C. Working solutions were freshly prepared just before use, and 1 mm TSA and 10 mm CBHA was added to the activation or culture media according to each experimental procedure, respectively.
Embryo Transfer
Two-cell stage cloned embryos were transferred into oviducts of E0.5 pseudopregnant ICR surrogate mothers. Embryos were dissected at E7 or left until term. Caesarean sectioning was performed at E18.5.
Differential Staining of the Blastocysts for Inner Cell Mass and Trophectoderm Cell Counting
The number of cells in the inner cell mass (ICM) and the trophectoderm of blastocysts was determined as described elsewhere (22). In brief, cloned embryos reaching the blastocyst stage 96 h after activation were denuded with acid PBS, rinsed in HEPES-CZB medium, and exposed to a 1:10 dilution of rabbit anti-mouse whole serum (Sigma) for 30 min. Afterward they were briefly rinsed in HEPES-CZB medium and incubated in a 1:10 dilution of guinea pig complement (Sigma) with propidium iodide (10 μg/ml) and Hoechst 33342 (1 μg/ml) for 30 min at 37 °C. They were then rinsed in HEPES-CZB medium, flattened with coverslips, and immediately examined using an inverted NIKON fluorescent microscope. Cell counts were made directly under the microscope.
Apoptosis Assays
The TUNEL method was used for the detection and quantification of apoptosis at single cell level (in situ Cell Death Detection kit, Fluorescein, Roche Applied Science). According to the instruction manual, NT blastocysts were fixed in 4% paraformaldehyde for 30 min, rinsed in PBS, and permeabilized with 0.5% Triton X-100 for 20 min at room temperature. They were then incubated with fluorescein-labeled dUTP and terminal transferase in the dark for 1 h at 37 °C. Propidium iodide (10 μg/ml) was used for nuclear counterstaining. Embryos were observed with a confocal microscope (Zeiss, LSM 510 META).
Establishment of NT-ES Cell Lines
The NT-ES cell lines were established and cultured as previously described (23, 24). DMEM/F-12 (1:1, Invitrogen no.11320-033) with LIF 2000U (leukemia inhibitory factor, Chemicon, ESG1107) was used for NT-ESC lines establishment, and the concentration of leukemia inhibitory factor decreased by half for ES cells culture.
Immunofluorescence Staining of Embryos and Outgrowths
Immunostaining was performed as previously described with some modifications (6). One-cell-stage-cloned embryos (from 10 min and 1 h after nuclear transfer to 3, 6, and 10 h after activation) or cells grown on coverslips were fixed with 4% paraformaldehyde overnight at 4 °C. All steps were performed at room temperature unless otherwise mentioned. Samples were permeabilized with 0.5% Triton X-100 in PBS for 30 min. After washing three times, they were blocked in 2% BSA in PBS for 1 h. Embryos were incubated with the first antibodies for 1 h at 37 °C. Antibodies against acetylated histones were diluted 1/200 (AcH3K9, Cell Signaling Technology; AcH4K5 and AcH3K18, Upstate Biotechnology), and anti-5-methyl-cytosine antibody was diluted 1:4000 (Eurogentec). Goat anti-Oct3/4 was diluted 1/400 (Santa Cruz). For the detection of 5-methylcytosine, embryos were first treated with 2 n HCl at room temperature for 1 h after permeabilization. After three washings, the embryos were incubated with a fluorescein isothiocyanate-conjugated goat anti-rabbit, donkey anti-goat, or goat anti-mouse secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at 37 °C. Finally, the DNA was stained with propidium iodide (10 μg/ml; Molecular Probes, OR) for 5 min at 37 °C, and samples were mounted in 50% glycerol on holding glass. For each separate sample more than 15 embryos were used, and the experiments were replicated at least three times. Observations were made using a confocal microscope (Zeiss, LSM 510 META).
Outgrowth Proliferation Ability and Oct4-positive Cell Counting
Outgrowth morphologies were recorded every 24 h starting 2 days after seeding of the blastocysts. The outgrowth area was measured using ImageJ software (rsb.info.nih.gov). Outgrowths were fixed after 4 days of culture, and Oct3/4 expression was detected by immunostaining. ImageJ software was used to count the Oct4 positive and the total cell number.
Statistical Analysis
Statistical analysis was performed using SPSS 13.0 statistical software. One-way analysis of variance and Fisher's Exact test were used for statistical analysis. For all statistical analyses, a value of p < 0.05 was considered to be statistically significant.
RESULTS
Improved Preimplantation Development of SCNT Embryos Treated by CBHA
To test whether modification of acetylation could benefit early development, SCNT embryos were treated during pre-implantation development with CBHA. Different concentrations ranging from 0.1 to 300 μm was used, and we found that all SCNT embryos cleaved with a very similar rate, around 90–95%. The effect of the treatment by TSA and CBHA was observed from the morula stage onwards. CBHA and TSA both improved the morula and blastocyst rate. Although a wide range of CBHA concentration could be used, the optimal window was between 1 and 20 μm. In this case, the rate of blastocyst was even higher than in the TSA-treated group (69–71% compared with 54.8%, p < 0.05). A high concentration of CBHA was found to be toxic for development as early as four-cell stage (Table 1).
TABLE 1.
Effect of different concentration of CBHA on the development of SCNT embryos in vitro
The development efficiencies of blastocysts (shown as %) are calculated based on the number of two-cell-stage embryos.
| Treatment, μm/h | No. reconstructed | No. with pronucleus | No. 2 cell | No. 4 cell | No. morulas | No. (% 2-cell) blastocysts |
|---|---|---|---|---|---|---|
| No treatment | 230 | 186 | 185 | 164 | 108 | 60 (32.9 ± 2.9)a |
| TSA, 0.1/10 | 121 | 93 | 90 | 88 | 58 | 49 (54.8 ± 4.1)b |
| CBHA | ||||||
| 0.1/10 | 88 | 76 | 76 | 69 | 58 | 41 (54.1 ± 4.2)b |
| 1/10 | 87 | 70 | 69 | 67 | 58 | 49 (71.2 ± 2.1)d |
| 20/10 | 89 | 72 | 71 | 69 | 63 | 49 (69.5 ± 7.4)d,e |
| 100/10 | 81 | 51 | 48 | 44 | 34 | 28 (56.7 ± 5.1)b,e |
| 300/10 | 85 | 73 | 72 | 46 | 33 | 13 (17.4 ± 3.01)c |
a–e Values with different superscripts are significantly different, p < 0.05.
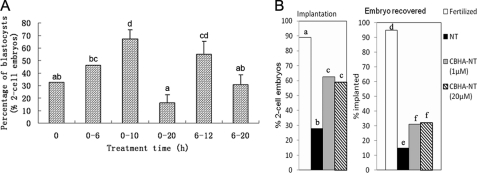
To optimize the treatment of acetylation of histones, we compared the blastocyst rate after various incubation times (Fig. 1A). CBHA was added at the beginning of activation or 6 h later; in both cases, the optimum effect was reached when CBHA was present during the first 4–6 h after activation, its presence during the activation time not being required. Therefore, for the rest of the studies, we chose to add CBHA in the medium after reconstruction and for 10 h, as previously done for TSA in the literature.
FIGURE 1.
CBHA treatment significantly improved the development potential of SCNT embryos in vitro and in vivo. A, shown is the influence of the length and the period of exposure to CBHA on the blastocyst development rate of SCNT embryos. B, shown is the influence of CBHA treatment on implantation and recovery of SCNT embryos. The implantation sites were dissected at E7.5, and the embryos were collected.
To evaluate the quality of blastocysts from SCNT embryos, differential ICM staining and the TUNEL method were performed. The results indicated that CBHA treatment significantly increased the total cell number and ICM cell number in SCNT blastocyst and also reduced the apoptotic cell number (Table 2).
TABLE 2.
Influence of CBHA treatment on cell number and apoptosis of cloned blastocysts
ICM in blastocyst day 0 is relative to the ES derivation, at seeding the blastocyst on the feeder. Total cell data are the total cell number of all 23 blastocysts tested.
| CBHA Concentration | Tested blastocysts | Total no. of cells in blastocyst (day 0) | ICM in blastocyst (day 0) | No. of apoptotic blastocysts | Total cell | Apoptotic cell |
|---|---|---|---|---|---|---|
| μm | % | % | ||||
| 0 | 23 | 46.47 ± 3.60a | 11.73 ± 0.96a | 23 (100)b | 890 | 118 (13.9 ± 1.3)a |
| 20 | 37 | 63.59 ± 2.68b | 18.14 ± 5.82b | 36 (97)b | 1907 | 132 (7.1 ± 0.7)b |
a,b Values with different superscripts are significantly different in one volume with p < 0.05.
CBHA Improves the Post-implantation and Full-term Development of SCNT Embryos
Furthermore, we investigated the post-implantation development of SCNT embryos treated with CBHA in this critical period by dissecting the recipient mice 7 days after transfer of 2-cell embryos.
CBHA treatment greatly improved both the implantation and the survival of embryos. Only 28% of the non-treated 2-cell SCNT embryos were able to implant, whereas more than 60 percent did when treated with CBHA (Fig. 1B, left). When implantation sites were dissected at E7.5, only 15% of them contained an embryo in the case of SCNT without treatment, which is significantly lower than those treated by CBHA (30%, Fig. 1B, right).
We classified the recovered embryos using the benchmark of an apparent primitive streak (Table 3) to compare the development of SCNT embryos with or without CBHA treatment to the fertilized embryos. We found that SCNT embryos were delayed, as at E7.5 only 60% of them were gastrulating, compared with more than 80% in the treated group, a percentage quite similar to fertilized controls.
TABLE 3.
Influence of CBHA treatment on developmental stage of SCNT embryos
The pre-streak embryos percentages (shown as %) are calculated based on the number of recovered embryos. The gastrulating embryo percentages (shown as %) are calculated based on the number of recovered embryos.
| CBHA treatment | No. 2 cell Transferred | No. embryos Recovered | % Pre-streak | % Gastrulating embryos |
|---|---|---|---|---|
| μm | ||||
| Fertilized | 45 | 38 | 5a | 95a |
| 0 | 900 | 38 | 40b | 60b |
| 1 | 128 | 25 | 12.5a | 87.5a |
| 20 | 129 | 24 | 18a | 82a |
a,b Values with different superscripts are significantly different in one volume with p < 0.05 (K2 test).
We compared the development to term of SCNT embryos with or without TSA and CBHA treatment. Two-cell SCNT embryos were transferred to recipient females, and live pups were delivered by caesarian section at E18.5 (Table 4). As described previously in the literature, we verified that the rate of living pups was three times higher after TSA treatment (from 0.8 to 2.6%). CBHA was even more efficient, as it improved this rate by a factor of more than 4 (3.7%). In all cases, SCNT placentas were found to be hypertrophic (Table 4). All SCNT pups survived and grew into healthy adults. As recipient females often had three to five live pups, we decided to let some females deliver naturally at E19. These pups were successfully born, which was usually not the case for control SCNT embryos, as only one fetus survived within the pregnant female.
TABLE 4.
Influence of CBHA and TSA treatment on SCNT embryos full-term development
The percentages of live offspring (shown as %) are calculated based on the number of transferred two-cell-stage embryos. Average placenta weight is the average weight of all the placenta of cloned pups.
| Treatment | No. 2 cell transferred | No. live offspring | Average placenta wt (S.E.) |
|---|---|---|---|
| μm/h | % | g | |
| Control, 0 | 486 | 4 (0.8)a | 0.25 ± 0.02a |
| TSA, 0.1/10 | 117 | 3 (2.6)a,b | 0.33 ± 0.16a |
| CBHA | |||
| 1/10 | 162 | 6 (3.7)b | 0.29 ± 0.03a |
| 5/10 | 240 | 9 (3.8)b | 0.27 ± 0.03a |
| 20/10 | 197 | 7 (3.6)b | 0.21 ± 0.02a |
a Values with different superscripts are significantly different in one volume. p < 0.05.
To test whether CBHA had any adverse effect on the health of the cloned adults and their reproductive performance, we mated 20 cloned females obtained from CBHA-treated embryos with B6D2F1 males. We compared the number, coat color distribution, sex, and weight of the offspring with those of fertilized control females (Table 5). No differences could be detected for any of these parameters in both groups.
TABLE 5.
Reproductive performance of mice obtained from CBHA treated SCNT embryos
Body mass was examined at 3 weeks after birth. Mice were mated with B6D2 F1 males. No differences were found between control and SCNT mice.
| Group | Tested mice | Pregnant mice (%) | Pups (%) | Phenotype |
Sex |
Body mass |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Black | Brown | Dilute brown | % Females | % Males | Female | Male | ||||
| % | g | |||||||||
| Fertilized | 8 | 8 (100.0)a | 77 (10)a | 43 (56)a | 22 (29)a | 12 (16)a | 48 | 52 | 10.0 ± 0.4a | 10.8 ± 0.5a |
| CBHA-NT | 20 | 18 (90)a | 127 (7)a | 80 (63)a | 27 (21)a | 20 (16)a | 51 | 49 | 10.3 ± 0.2a | 10.7 ± 0.2a |
a,a Values in the same column denote that difference is not significant (p < 0.05).
Increased Efficiency of NT-ES Derivation from CBHA-treated SCNT Embryos
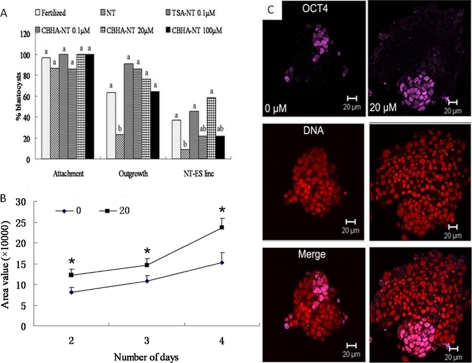
ES cells can be derived from NT blastocysts, but the efficiency is lower than from fertilized embryos (23, 24). To test whether CBHA was beneficial for ES derivation, we compared the ability of NT embryos to give rise to outgrowths and then to ES cell lines when they had been treated with TSA or different doses of CBHA (Fig. 2A).
FIGURE 2.
ES derivation efficiency from SCNT embryos were increased with CBHA treatment. A, improved efficiency of NT-ES cell derivation from CBHA- and TSA-treated SCNT blastocysts is shown. B, expansion of the outgrowths formed by non-treated (0) and CBHA-treated (20 μm) SCNT blastocysts is shown. Area of each outgrowth was measured at days 2, 3, and 4 after plating of the blastocysts. C, shown is expression of Oct4 in day 4 outgrowths derived from NT blastocysts derived from non-treated (0 μm) embryos or CBHA treated ones at 20 μm.
Outgrowth formation is conditioned by the ability of the trophoblast to spread on the substrate and of the ICM to proliferate. It is supposed to mimic in vitro the peri-implantation period of development. Indeed, few SCNT blastocysts (23%) were successful in forming an outgrowth. By contrast, when treated with CBHA or TSA, the rate of outgrowth formation was significantly increased, up to 91% for TSA-treated embryos and 64–86% for CBHA-treated embryos. This is well correlated with the improved survival rate at implantation as shown above.
From these outgrowths we derived ES cell lines. TSA-treated embryos displayed an improved ability to give rise to ES cell lines (45%). The beneficial effect of CBHA treatment was clearly dose-dependent, the best concentration being 20 μm, with the rate of ES derivation being 59% (Fig. 2A). With a lower dose, the rate was 21%, still higher, although not significantly, than without CBHA (10%). A higher dose (100 μm) was probably detrimental as the rate of derivation dropped to 21%.
To gain insight into the origin of the improved outgrowth formation after CBHA treatment, we measured the proliferation of the cells during the outgrowth phase. First, the area covered by each outgrowth was measured at days 2, 3, and 4 (Fig. 2B). The starting point was higher for CBHA-treated embryos, reflecting the fact that CBHA-treated blastocysts had more cells than untreated embryos (see above data). The increase in area was more pronounced for outgrowth of the CBHA-treated batch, evidenced by the different slopes of the growth curves (Fig. 2B). To quantify the increase in cell number, we fixed the outgrowths after 4 days of culture (the time at which the outgrowth is normally dissociated) and counted both the total cell number and Oct4-positive cell number after immunolabeling and DNA staining (Fig. 2C). We compared these numbers with the initial cell number in the SCNT blastocysts (Table 6). CBHA treatment clearly improved the proliferation of the cells, as the net increase in cells was 12 times in outgrowths formed by CBHA-treated embryos and only 5 times for non-treated embryos. Obviously Oct4-positive cells were more abundant in the treated batch (Fig. 2C). However, the Oct4-positive cell population increased by a factor of 4 for both types of cultures when compared with the initial ICM cell number (Table 6).
TABLE 6.
Expansion of the outgrowths formed by non-treated (0) and CBHA-treated (20 μm) SCNT blastocysts
Data were by ImageJ software; outgrowths were scanned under confocal microscopy side by side.
| CBHA concentration | No. of outgrowths tested at day 4 | Total no. of cells at day 4 | No. of OCT4+s cells per outgrowth (day 4) | OCT4 ratio (%) (day 4) |
|---|---|---|---|---|
| μm | ||||
| 0 | 9 | 246.4 ± 29.4a | 44.1 ± 5.9a | 16.2 ± 1.9a |
| 20 | 3 | 744.7 ± 121.6b | 74.7 ± 4.4b | 8.7 ± 0.7a |
a,b Values with different superscripts in the same column are significantly different in one volume with p < 0.05.
Taken together, these data show that CBHA treatment increases the proliferation in SCNT embryos. By doing this, it gives more chance to the Oct4-positive cell population to expand during outgrowth phase up to a level suitable for subsequent ES cell derivation.
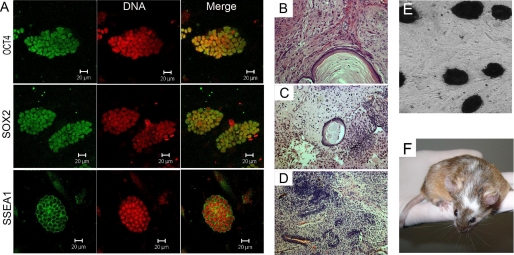
To check whether the ES lines derived from SCNT embryos treated with CBHA were normal, we stained them for the usual pluripotency markers, such as Oct4, Sox2, SSea1, and alkaline phosphatase that were found to be positive in all cell lines (Fig. 3, A and E). Their karyotypes were found to be in the normal range (data not shown). Their differentiation potential was tested in vivo by chimera and teratoma formation. High contribution to the coat color was observed in the chimeric mice (Fig. 3F), and differentiation into the three germ layers was evidenced in histology sections of the teratoma (Fig. 3, B–D).
FIGURE 3.
NT-ES cells derived from CBHA treated embryos exhibit normal features. A, correct expression of Oct4, Sox2, and SSEA1 is shown. B–D, teratomas from mouse NT-ES cells show differentiated derivatives of all three embryonic germ layers: B, minor salivary gland (endoderm); C, cartilage tissue (mesoderm); D, neural tube (ectoderm). E, positive alkaline phosphatase activity is shown. F, shown is a chimera mouse born from CBHA NT-ES.
Increased Acetylation after CBHA Treatment of Reconstructed Embryos
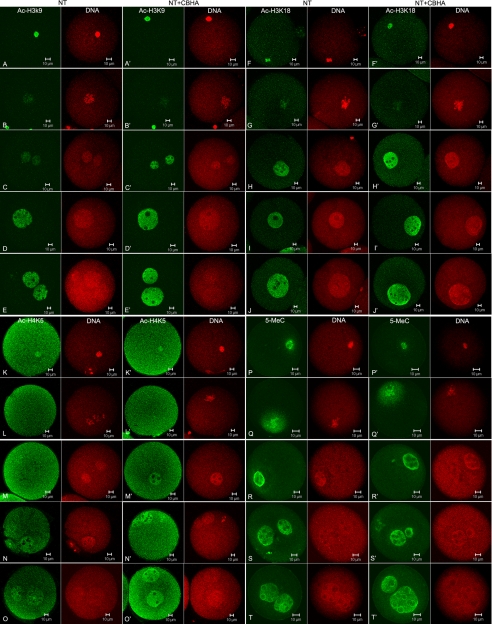
To analyze the cause of CBHA improvement of SCNT embryo developmental potential, the key histone acetylation sites of H3K9, H4K5, and H3K18 were investigated in the primary reprogramming period of SCNT embryos. Oocytes injected with cumulus donor nuclei were treated with CBHA at 20 μm at the beginning of activation and for 10 h. At different time points (10 min and 1 h after injection; 3, 6, and 10 h after activation), the pattern of histone acetylation of SCNT embryos was examined by immunostaining (Fig. 4, A–O). The donor chromatin was initially acetylated, as shown for example for Ac-H3K9 in Fig. 4A and became deacetylated in about 1 h after its injection into the oocyte cytoplasm (Fig. 4B). Re-acetylation occurred at the end of activation when the pseudo-pronucleus was newly formed (Fig. 4D). In the presence of CBHA this re-acetylation seemed to occur earlier; as it was already evidenced at 3 h post-activation and at the end of activation, the nucleus was more strongly stained than without CBHA. A very similar result was obtained with H3K18 and H4K5 (Fig. 4, F–O). By contrast, the level of methylation assessed by 5 methylcytosine staining remained unchanged with or without CBHA treatment, and the chromatin remained strongly methylated during the 1-cell stage (Fig. 4, P–T).
FIGURE 4.
Identification of acetylation sites and DNA methylation in SCNT embryos treated with or without 20 μm at 1-cell stage at different times, including 10 min after SCNT, 1 h after SCNT, 3 h after the beginning of activation, and 6 h after the beginning of activation and 10 h after activation. A–E, reprogramming of H3K9 acetylation (green) in SCNT 1-cell embryos and DNA counterstaining (red) is shown. F–J, reprogramming of H3K18 acetylation (green) in SCNT 1-cell embryos and DNA counterstaining (red) is shown. K–O, reprogramming of H4K5 acetylation (green) in SCNT 1-cell embryos and DNA counterstaining (red) is shown. P–T, reprogramming of DNA methylation (green) in SCNT 1-cell embryos is shown.
DISCUSSION
In this study we used CBHA, a new HDAC inhibitor different from those used previously, to improve SCNT. We have once more proved that elevating the level of histone acetylation is really beneficial for the rate of development to blastocyst to term and even for the efficiency of ES cell derivation from SCNT blastocysts. In addition, our study has brought insight into key features of the development of the treated embryos; CBHA treatment improves the quality of the blastocysts by markedly increasing the full-term developmental potential of implanted embryos and derivation of NT-ES cell lines. By comparison, CBHA seems more effective than TSA, the first HDAC inhibitor proved to be useful for SCNT embryo treatment. The use of CBHA to boost SCNT blastocyst production efficiency can provide immediate benefits by increasing the number of viable embryos available to laboratories that need a continuous and reliable supply to advance their research. Characterization of the differences in molecular targets or activities of TSA versus CBHA may be very useful for identifying what processes are responsible for improving reprogramming efficiencies and will perhaps provide clues for how HDACi could be further optimized for this purpose.
After nuclear transfer, the donor chromatin is rapidly deacetylated in the oocyte cytoplasm, concomitant with premature chromosome condensation, confirming previous results obtained with TSA (14, 16). In MII oocytes, a similar deacetylation happens, mainly driven by HDAC1, which associates with the metaphase chromosomes (25). A high deacetylase activity probably remains in the ooplasm, as deacetylation of the injected chromatin occurs even in the absence of HDACi. Then the forming pseudo-pronuclei start to be reacetylated, and the global acetylation level is increased when CBHA is added during this time. With TSA it has been shown that the level of acetylation in the one-cell NT embryo becomes similar to that of a one-cell fertilized embryo (16). As Lys-5 is the last lysine to be acetylated, acetylated H4K5 reflects hyperacetylated histone H4, and this is correlated with a transcriptionally permissive state of the chromatin (26) that may be a necessary step for the embryonic genome activation occurring at the two-cell stage. Indeed, a recent study by Van Thuan et al. (27) has shown that Scriptaid, another inhibitor of HDAC, is able to increase the rate of de novo RNA synthesis in SCNT embryos at the two-cell stage. Comparison of the transcriptome profile at one- and two-cell stages in mouse SCNT and fertilized embryos revealed that a large subset of genes is indeed abnormally expressed during the embryonic genome activation (27). Among them, many transcription factors were found to be either up-regulated or repressed, therefore, affecting the reprogramming of the somatic genome into a totipotent one.
Apart from being involved in the control of gene expression, the level of acetylation of the chromatin also plays a role in the spatial configuration of the chromatin (28, 29). During normal development, a dynamic reorganization of the heterochromatin occurs that seems to be required for subsequent development to occur (5). After nuclear transfer, such reorganization occurs very inefficiently. However, it is markedly improved by treating the embryos with TSA (17); this means that the drugs including TSA and CBHA probably improved reprogramming by prolonging the acetylation duration of cloned embryos, which inhibits the deacetylation process using HDACi. Interestingly, CBHA is useless to cloned embryos reconstructed from ES cells (data not shown), which is a result similar to that reported before in TSA. Moreover, this shows that manipulating the level of epigenetic marks such as acetylated histones can have very a different impact depending on the initial configuration of the chromatin. This is well illustrated by the opposite effects of the treatment of ES cells and EpiSCs with HDACi. These two types of pluripotent cells are derived from different development stages of the embryo, the ICM and the epiblast of post-implanted embryos, respectively. Although both are pluripotent in vitro and in vivo, only ES cells are able to colonize the germ cell lineage in chimeras (30, 31) and, thus, can be considered as more pluripotent. When treated with sodium butyrate or TSA, EpiSCs regress to a more pluripotent state, whereas ES cells convert to a less pluripotent state (32). This could explain why treating ES cells with either TSA (17, 33) or CBHA (this study) have no beneficial effect on development or chromatin remodeling.
The implantation efficiency is not different between treated and untreated embryos, which means that most NT blastocysts are already able to implant in absence of any treatment. This is in good correlation with the fact that Cdx2, a marker essential for trophoblast maintenance, is correctly expressed in most SCNT embryos (34). Despite their ability to implant, the survival of SCNT embryos is rapidly compromised, most of them having resorbed 2 days after their implantation. Our data show that CBHA treatment is able to increase by a factor of more than two the survival of implanted SCNT embryos.
Our data also show that CBHA treatment reduces the delay in development affecting SCNT embryos. The onset of cavitation occurs earlier than in non-treated SCNT morula, and later, the proportion of gastrulating embryos at E7 is increased. Such an effect is correlated with the increase in cell number observed at blastocyst stage. Indeed, in normal development, the onset of gastrulation indicated by the appearance of the primitive streak is dependent on the cell number in the epiblast (35). The increase in cell number at blastocyst is correlated with the reduced rate of apoptosis we have evidenced in the blastocysts obtained from CBHA-treated SCNT embryos. It can probably be also attributed to a faster cleavage rate. We indeed observed an increased proliferation during the outgrowth phase of ES derivation from CBHA-treated SCNT blastocysts.
Reprogramming a somatic cell into a pluripotent one is now feasible using another method, the induction of pluripotency by exogenous factors leading to iPS cells. It has been shown that treating somatic cells with HDAC inhibitors concomitant with the induction of the four factors greatly improves the efficiency of reprogramming. HDAC inhibitors can replace c-Myc, the main action of which is to initiate chromatin remodeling and to shut down the somatic cell gene expression program (36, 37).
In recent comparisons of iPS cell lines, microRNA and gene expression levels from the Dlk1-Dio3 region of mouse chromosome 12 were shown to be correlated with the degree of iPS cell pluripotency (38, 39). This region is subject to imprinting (40), the maternal- or paternal-specific silencing of gene expression through histone deacetylation and DNA methylation. Considering the potential regulatory networks formed by histone H3 deacetylation and DNA methylation (41), permanent gene silencing by HDAC activity during differentiation (42), and microRNA feedback control of the Dlk1-Dio3 region genes (38), further investigation of HDACi influence on pluripotency will certainly include work to identify the regulatory mechanisms operating at this chromosomal region. Indeed, an iPS cell line silenced at Dlk1-Dio3 showed reactivation of the region's genes upon HDACi treatment, which then conferred full pluripotency and development to all-iPS mice (39).
In summary, the present study using CBHA indicates that this drug can significantly improve the developmental potential of SCNT embryos in vitro and in vivo. More importantly, this drug increases the ES derivation efficiency by accelerating outgrowth propagation. Therefore, this study of CBHA is not only helpful for the study of reprogramming but is also a key step toward using NT-ES cells in therapeutic cloning as well as cell replacement therapy.
Acknowledgment
We acknowledge Dr. Nathalie Beaujean for helpful criticism of the manuscript.
This study was supported by 973 Program Grants 2006CB944003 and 2007CB947702, 863 Program Grant 2006AA02A101, and National Science Foundation of China Grant 30525040.
- SCNT
- somatic cell nuclear transfer
- CBHA
- m-carboxycinnamic acid bishydroxamide
- HDACi
- histone-deacetylase inhibitor
- TSA
- trichostatin A
- ICM
- inner cell mass
- ES
- embryonic stem
- NT
- nuclear transfer.
REFERENCES
- 1.Campbell K. H., Fisher P., Chen W. C., Choi I., Kelly R. D., Lee J. H., Xhu J. (2007) Theriogenology 68, S214–S231 [DOI] [PubMed] [Google Scholar]
- 2.Dean W., Santos F., Reik W. (2003) Semin. Cell Dev. Biol. 14, 93–100 [DOI] [PubMed] [Google Scholar]
- 3.Gao S., Czirr E., Chung Y. G., Han Z., Latham K. E. (2004) Biol. Reprod. 70, 1162–1170 [DOI] [PubMed] [Google Scholar]
- 4.Teranishi T., Tanaka M., Kimoto S., Ono Y., Miyakoshi K., Kono T., Yoshimura Y. (2004) Dev. Biol. 266, 76–86 [DOI] [PubMed] [Google Scholar]
- 5.Martin C., Beaujean N., Brochard V., Audouard C., Zink D., Debey P. (2006) Dev. Biol. 292, 317–332 [DOI] [PubMed] [Google Scholar]
- 6.Santos F., Hendrich B., Reik W., Dean W. (2002) Dev. Biol. 241, 172–182 [DOI] [PubMed] [Google Scholar]
- 7.Dean W., Santos F., Stojkovic M., Zakhartchenko V., Walter J., Wolf E., Reik W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13734–13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang Y. K., Park J. S., Koo D. B., Choi Y. H., Kim S. U., Lee K. K., Han Y. M. (2002) EMBO J. 21, 1092–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright B. P., Sung L. Y., Chang C. C., Yang X., Tian X. C. (2005) Biol. Reprod. 72, 944–948 [DOI] [PubMed] [Google Scholar]
- 10.Jeon B. G., Coppola G., Perrault S. D., Rho G. J., Betts D. H., King W. A. (2008) Reproduction 135, 815–828 [DOI] [PubMed] [Google Scholar]
- 11.Yoshida M., Kijima M., Akita M., Beppu T. (1990) J. Biol. Chem. 265, 17174–17179 [PubMed] [Google Scholar]
- 12.Enright B. P., Kubota C., Yang X., Tian X. C. (2003) Biol. Reprod. 69, 896–901 [DOI] [PubMed] [Google Scholar]
- 13.Kishigami S., Bui H. T., Wakayama S., Tokunaga K., Van Thuan N., Hikichi T., Mizutani E., Ohta H., Suetsugu R., Sata T., Wakayama T. (2007) J. Reprod. Dev. 53, 165–170 [DOI] [PubMed] [Google Scholar]
- 14.Rybouchkin A., Kato Y., Tsunoda Y. (2006) Biol. Reprod. 74, 1083–1089 [DOI] [PubMed] [Google Scholar]
- 15.Iager A. E., Ragina N. P., Ross P. J., Beyhan Z., Cunniff K., Rodriguez R. M., Cibelli J. B. (2008) Cloning Stem Cells 10, 371–379 [DOI] [PubMed] [Google Scholar]
- 16.Wang F., Kou Z., Zhang Y., Gao S. (2007) Biol. Reprod. 77, 1007–1016 [DOI] [PubMed] [Google Scholar]
- 17.Maalouf W. E., Liu Z., Brochard V., Renard J. P., Debey P., Beaujean N., Zink D. (2009) BMC Dev. Biol. 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi G., Jiang G. (2006) Cell. Mol. Immunol. 3, 285–290 [PubMed] [Google Scholar]
- 19.Dokmanovic M., Marks P. A. (2005) J. Cell Biochem. 96, 293–304 [DOI] [PubMed] [Google Scholar]
- 20.Zhou Q., Renard J. P., Le Friec G., Brochard V., Beaujean N., Cherifi Y., Fraichard A., Cozzi J. (2003) Science 302, 1179. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Q., Jouneau A., Brochard V., Adenot P., Renard J. P. (2001) Biol. Reprod. 65, 412–419 [DOI] [PubMed] [Google Scholar]
- 22.Papaioannou V. E., Ebert K. M. (1988) Development 102, 793–803 [DOI] [PubMed] [Google Scholar]
- 23.Wakayama S., Ohta H., Kishigami S., Thuan N. V., Hikichi T., Mizutani E., Miyake M., Wakayama T. (2005) Biol. Reprod. 72, 932–936 [DOI] [PubMed] [Google Scholar]
- 24.Zhao C., Yao R., Hao J., Ding C., Fan Y., Dai X., Li W., Hai T., Liu Z., Yu Y., Wang Y., Hou X., Ji W., Zhou Q., Jouneau A., Zeng F., Wang L. (2007) Cell Res. 17, 80–87 [DOI] [PubMed] [Google Scholar]
- 25.Ma P., Schultz R. M. (2008) Dev. Biol. 319, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urnov F. D., Wolffe A. P. (2001) Oncogene 20, 2991–3006 [DOI] [PubMed] [Google Scholar]
- 27.Van Thuan N., Bui H. T., Kim J. H., Hikichi T., Wakayama S., Kishigami S., Mizutani E., Wakayama T. (2009) Reproduction 138, 309–317 [DOI] [PubMed] [Google Scholar]
- 28.Taddei A., Maison C., Roche D., Almouzni G. (2001) Nat. Cell Biol. 3, 114–120 [DOI] [PubMed] [Google Scholar]
- 29.Bártová E., Pacherník J., Harnicarová A., Kovarík A., Kovaríková M., Hofmanová J., Skalníková M., Kozubek M., Kozubek S. (2005) J. Cell Sci. 118, 5035–5046 [DOI] [PubMed] [Google Scholar]
- 30.Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., Vallier L. (2007) Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- 31.Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. (2007) Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- 32.Ware C. B., Wang L., Mecham B. H., Shen L., Nelson A. M., Bar M., Lamba D. A., Dauphin D. S., Buckingham B., Askari B., Lim R., Tewari M., Gartler S. M., Issa J. P., Pavlidis P., Duan Z., Blau C. A. (2009) Cell Stem Cell 4, 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishigami S., Mizutani E., Ohta H., Hikichi T., Thuan N. V., Wakayama S., Bui H. T., Wakayama T. (2006) Biochem. Biophys. Res. Commun. 340, 183–189 [DOI] [PubMed] [Google Scholar]
- 34.Kishigami S., Hikichi T., Van Thuan N., Ohta H., Wakayama S., Bui H. T., Mizutani E., Wakayama T. (2006) FEBS Lett. 580, 1801–1806 [DOI] [PubMed] [Google Scholar]
- 35.Power M. A., Tam P. P. (1993) Anat. Embryol. 187, 493–504 [DOI] [PubMed] [Google Scholar]
- 36.Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008) Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. (2009) Cell 136, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L., Luo G. Z., Yang W., Zhao X., Zheng Q., Lv Z., Li W., Wu H. J., Wang L., Wang X. J., Zhou Q. (2010) J. Biol. Chem. 285, 19483–19490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. (2010) Nature, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagan J. P., O'Neill B. L., Stewart C. L., Kozlov S. V., Croce C. M. (2009) PLoS One 4, e4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory R. I., Randall T. E., Johnson C. A., Khosla S., Hatada I., O'Neill L. P., Turner B. M., Feil R. (2001) Mol. Cell Biol. 21, 5426–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Oevelen C., Wang J., Asp P., Yan Q., Kaelin W. G., Jr., Kluger Y., Dynlacht B. D. (2008) Mol. Cell 32, 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]