Abstract
The steroid hormone ecdysone triggers the rapid destruction of larval tissues through transcriptional cascades that culminate in rpr and hid expression and caspase activation. Here we show that mutations in Mdh2 and Med24 block caspase cleavage and larval salivary gland cell death. Mdh2 encodes a predicted malate dehydrogenase that localizes to mitochondria. Consistent with this proposed function, Mdh2 mutants have significantly lower levels of ATP and accumulate late-stage citric acid cycle intermediates, suggesting that the cell death defects arise from a deficit in energy production. Med24 encodes a component of the Mediator transcriptional coactivator complex. Unexpectedly, however, expression of the key death regulator genes is normal in Med24 mutant salivary glands. This study identifies novel mechanisms for controlling the destruction of larval tissues during Drosophila metamorphosis and provides new directions for our understanding of steroid-triggered programmed cell death.
Keywords: ecdysone signaling, programmed cell death, mitochondria, metabolism
INTRODUCTION
Programmed cell death plays a central role in the development of multicellular organisms by controlling cell numbers, removing abnormal cells, sculpting complex structures, and eliminating obsolete tissues (Jacobson et al., 1997). Pioneering genetic studies in C. elegans provided critical insights into the molecular mechanisms that regulate cell death, with subsequent efforts showing that the central regulators in this pathway are conserved through evolution (Danial and Korsmeyer, 2004). In Drosophila, these cell death regulators include the caspases Dronc and Drice, the CED-4/Apaf-1 homolog Dark, and two Bcl-2 family members Debcl and Buffy (Hay and Guo, 2006). In addition, genetic studies in Drosophila identified three critical cell death activator genes: rpr, hid and grim. Deletion of all three genes blocks most programmed cell death during embryogenesis, while ectopic expression of any of these three genes is sufficient to induce caspase-dependent cell death (White et al., 1994; Grether et al., 1995; Chen et al., 1996; White et al., 1996). Rpr, Hid and Grim trigger cell death by interacting with the essential death inhibitor DIAP1 and directing its degradation, thereby overcoming its inhibitory effects on caspase activation and cell death (Danial and Korsmeyer, 2004).
Upstream signaling pathways dictate the appropriate temporal and spatial patterns of programmed cell death during development, ensuring that this response is restricted to cells that are fated to die. One of these signals is steroid hormones that act through members of the nuclear receptor family of transcription factors, exemplified by the glucocorticoid-induced apoptosis of immature thymocytes and mature T cells (Winoto and Littman, 2002). Although relatively little is known about the mechanisms of steroid-regulated programmed cell death in vertebrates, significant insights into this pathway have been gained in Drosophila, where the steroid ecdysone directs the massive and rapid destruction of larval tissues during metamorphosis. At the end of the third larval instar, a pulse of ecdysone triggers puparium formation and adult tissue morphogenesis, marking the onset of metamorphosis and initiating the prepupal stage of development. Several larval tissues are destroyed in response to this ecdysone pulse, including the midgut. A subsequent ecdysone pulse, about ten hours after puparium formation (APF), triggers adult head eversion, marking the prepupal-to-pupal developmental transition and inducing the rapid destruction of the larval salivary glands. Destruction of the larval midguts and salivary glands is accompanied by classic hallmarks of apoptosis, including acridine orange staining, caspase activation, and the breakdown of nuclear lamins (Jiang et al., 1997; Martin and Baehrecke, 2004). Ecdysone triggers cell death through a transcriptional cascade that converges on rpr and hid induction, overcoming the inhibitory effect of DIAP1 and inducing the apical caspase Dronc and caspase adaptor Dark (Jiang et al., 2000; Lee et al., 2002; Daish et al., 2004; Mills et al., 2006). The larval salivary glands also display hallmarks of autophagy, characterized by the formation of intracellular autophagic vesicles (Lee and Baehrecke, 2001). Autophagy is induced just prior to salivary gland cell death and appears to act in parallel with caspases to drive tissue destruction (Berry and Baehrecke, 2007; McPhee and Baehrecke, 2009).
In an effort to further our understanding of ecdysone-triggered programmed cell death, we have undertaken an open-ended genetic screen for defects in larval salivary gland destruction (Wang et al., 2008). This screen resulted in the identification of seven multiallelic complementation groups, at least five of which represent novel regulators of cell death. We mapped three of these complementation groups to specific genes: CG5146, which encodes a protein of unknown function, Med24, which encodes a component of the Mediator complex, and Mdh2 (CG7998), which encodes a predicted mitochondrial malate dehydrogenase (Wang et al., 2008). Here, we show that mutations in Mdh2 and Med24 effectively block cell death, caspase activation, and the breakdown of nuclear lamins. The ecdysone-triggered transcriptional cascade that directs rpr and hid induction, however, occurs normally in Mdh2 and Med24 mutant salivary glands, indicating that these factors control cell death through distinct pathways. We show that the malate dehydrogenase encoded by Mdh2 localizes to mitochondria, and that disruption of Mdh2 function results in a severe energy deficit in prepupae, providing a possible mechanism for the defects in cell death. These studies provide new directions for understanding the regulation of steroid-triggered programmed cell death during development.
RESULTS AND DISCUSSION
Med24 and Mdh2 Mutations Block Salivary Gland Cell Death
Alleles of Med24 and Mdh2 were identified in a large scale EMS screen on the third chromosome, selecting for mutations that disrupt larval salivary gland destruction (Wang et al., 2008). Homozygous mutants for each of these alleles arrest their development during the pupal stage, with more than 40% of the mutant pupae containing persistent larval salivary glands (Wang et al., 2008). The Med241 mutation is due to a premature stop codon, while Mdh21 carries a 236 bp deletion within the protein coding region (Wang et al., 2008). Hemizygous mutants of each allele [Med241/Df(3L)Exel6112 and Mdh21/Df(3R)Exel6178] display effects on lethality and salivary gland cell death that are similar to those of homozygous mutants, indicating that they behave as null alleles (data not shown). These defects can be attributed to a loss of Med24 and Mdh2 function because they can be rescued by expressing the corresponding wild-type gene, using UAS-Med24 or UAS-Mdh2, under the control of an Act5c-GAL4 driver (Table 1).
Table 1.
Genetic rescue of Med24 and Mdh2 mutants
| PSG | Eclosion | |||||
|---|---|---|---|---|---|---|
| Studies | Genotype | n | % | n | % | |
| a | control | w1118 | 70 | 0 | 82 | 100 |
| b | Mdh2− | w; +; Mdh21/Mdh22 | 99 | 46 | 98 | 0 |
| c | Mdh2−, geno. rescue | w; Mdh2-myc/+; Mdh21/Mdh22 | 87 | 1 | 97 | 83 |
| d | Mdh2− , Act5c-GAL4 | w; Act5c-GAL4 /+; Mdh21 /Df(3R)Exel6178 | 72 | 40 | 90 | 0 |
| e | Mdh2− , UAS-Mdh2 | w; +; Mdh21, UAS-Mdh2/Df(3R)Exel6178 | 33 | 42 | 71 | 0 |
| w; Act5c-GAL4/+; Mdh21, | ||||||
| f | Mdh2− , Act5c>Mdh2 | UAS-Mdh2/Df(3R)Exel6178 | 49 | 2 | 90 | 99 |
| g | Med24− , Act5c-GAL4 | w; Act5c-GAL4 /+; Med241/Med241 | 46 | 46 | 97 | 2 |
| h | Med24− , UAS-Med24 | w; +; Med241, UAS-Med24/Med241 | 48 | 42 | 98 | 0 |
| w; Act5c-GAL4/+; Med241, | ||||||
| i | Med24− , Act5c>Med24 | UAS-Med24/Med241 | 60 | 0 | 71 | 99 |
| j | Med24− , UAS-Mdh2 | w; +; Med241, UAS-Mdh2/Med241 | 45 | 42 | 91 | 0 |
| w; Act5c-GAL4/+; Med241, | ||||||
| k | Med24− , Act5c>Mdh2 | UAS-Med24/Med241 | 51 | 41 | 89 | 3 |
From top to bottom are studies for (a) control, (b) Mdh2 trans-heterozygous mutant, (c) Mdh2 trans-heterozygous mutant carrying the genomic Mdh2-myc transgene, (d) Mdh2 hemizygous mutant carrying the Act5c-GAL4 driver alone, (e) Mdh2 hemizygous mutant carrying the UAS-Mdh2 transgene alone, (f) rescue of the Mdh2 hemizygous mutant with Act5c>Mdh2, (g) Med24 homozygous mutant carrying the Act5c-GAL4 driver alone, (h) Med24 homozygous mutant carrying the UAS-Med24 transgene alone, (i) rescue of the Med24 homozygous mutant with Act5c>Med24, (j) Med24 homozygous mutant carrying the UAS-Mdh2 transgene alone and (k) failure of Act5c>Mdh2 to rescue Med24 homozygous mutants. Persistent salivary glands (PSG) were scored by dissecting pupae staged at 24 hr APF. Eclosion was scored by counting empty pupal cases. “n” indicates the total number of pupae scored. Number of pupae that possessed PSG or successfully eclosed is shown as a percentage (%) relative to “n”.
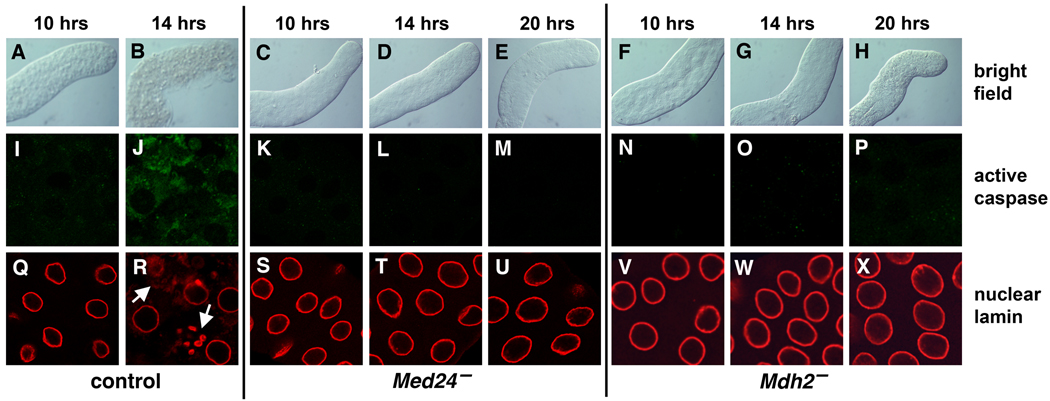
Med24 and Mdh2 mutant pupae appear virtually identical to control pupae at 12 hrs after puparium formation (APF), with no apparent defects in ecdysone-triggered adult head eversion and leg elongation. In addition, there is no delay between puparium formation and head eversion in the mutant pupae, indicating appropriate developmental progression in response to the late larval and prepupal ecdysone pulses. In contrast to control salivary glands, however, which become ruptured and show clear signs of degradation soon after head eversion (Fig. 1A,B), Med24 and Mdh2 mutant salivary glands remain intact and maintain their morphological integrity. This is seen at 14 hrs APF (Fig. 1D,G) as well as at 20 hrs APF (Fig. 1E,H), when control salivary glands have been completely eliminated.
Fig. 1.
Med24 and Mdh2 mutant salivary glands fail to undergo cell death. Salivary glands were dissected from w1118 control, Med241/Df(3L)Exel6112, and Mdh21/Df(3R)Exel6178 mutant prepupae or pupae at either 10, 14, or 20 hrs after puparium formation (APF) and imaged using bright field microscopy (A–H) or after staining with antibodies directed against active Caspase-3 (I–P) or the nuclear lamin protein DmO (Q–X). Control salivary glands lose their morphological integrity soon after head eversion (B), and display caspase activation (J) and breakdown of nuclear lamins (R), indicative of the onset of cell death. In contrast, Med24 and Mdh2 mutant salivary glands remain intact and fail to display either caspase activation or changes in nuclear lamin. Arrows in panel R mark nuclear lamin breakdown.
As a first step toward understanding why Med24 and Mdh2 mutant salivary glands fail to be destroyed, we examined caspase integrity in salivary glands from staged prepupae and early pupae. Salivary glands were stained with an antibody that is directed against the cleaved active form of caspase-3, which detects the Drosophila DrICE effector caspase and accurately reflects the onset of cell death (Yu et al., 2002). We interpret a positive result from this experiment as evidence of caspase activation. As expected, caspases become activated in control salivary glands soon after head eversion, at 14 hrs APF (Fig. 1I,J). In contrast, caspases fail to be activated in Med24 and Mdh2 mutant salivary glands, at 14 hrs APF (Fig. 1L,O) or at 20 hrs APF (Fig. 1M,P), when control glands have been completely degraded. Consistent with this observation, nuclear lamins begin to fragment in control salivary glands at 14 hrs APF (Fig. 1R, arrows), in parallel with caspase activation. Med24 and Mdh2 mutant salivary glands, however, display fully intact nuclear lamins at 14 hrs APF (Fig. 1T,W) or at 20 hrs APF (Fig. 1U,X). These results indicate that both Med24 and Mdh2 mutant salivary glands display an effective block in caspase-driven programmed cell death.
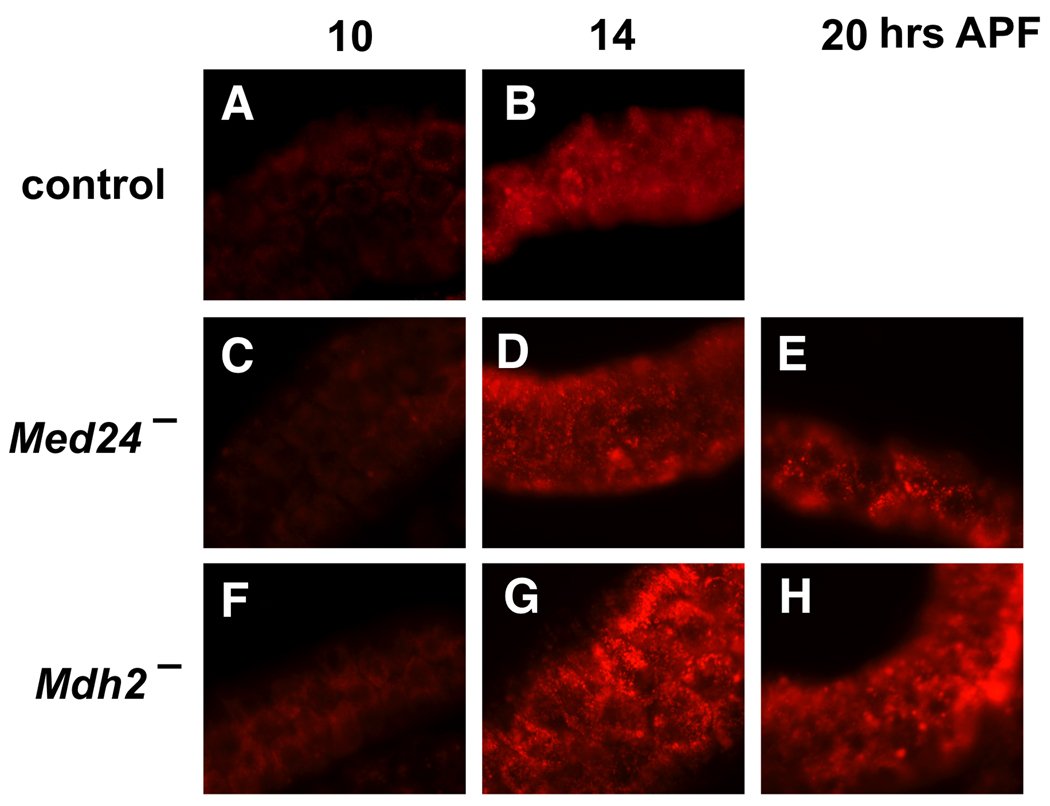
Both apoptosis and autophagy contribute to the complete destruction of larval salivary glands in response to ecdysone (McPhee and Baehrecke, 2009). Accordingly, we examined the effect of the Med24 and Mdh2 mutations on the induction of salivary gland autophagy using the dye Lysotracker, which labels acidic autophagic vacuoles (Scott et al., 2004). As expected, control salivary glands display a significant increase in punctate Lysotracker staining soon after head eversion, at 14 hrs APF (Fig. 2A,B). A similar increase in Lysotracker staining, however, was seen in both Med24 (Fig. 2C–E) and Mdh2 (Fig. 2F–H) mutant salivary glands, with strong Lysotracker staining evident at 20 hrs APF, when control glands have been completely degraded. These results suggest that autophagy is induced normally in Med24 and Mdh2 mutant salivary glands. Similar results have been seen in studies of other mutants that block salivary gland cell death. For example, mutations in the Broad-Complex (BR-C) selectively block caspase activation but do not affect autophagy in persistent mutant salivary glands (Lee and Baehrecke, 2001; Martin and Baehrecke, 2004). Similarly, autophagic vacuoles are present in persistent salivary glands from dark mutants (Akdemir et al., 2006). Taken together, our results indicate that Med24 and Mdh2 mutant salivary glands display a specific block in caspase-mediated programmed cell death.
Fig. 2.
Autophagy is induced normally in Med24 and Mdh2 mutant salivary glands. Salivary glands were dissected from w1118 control (A,B), Med241/Df(3L)Exel6112 (C–E), and Mdh21/Df(3R)Exel6178 (F–H) mutant prepupae or pupae, at either 10 hrs (A,C,F), 14 hrs (B,D,G), or 20 hrs (E,H) APF, and stained with Lysotracker to detect acidic autophagic vacuoles. Autophagy is activated soon after head eversion in both control and mutant salivary glands.
Cell Death Regulators are Expressed Normally in Med24 and Mdh2 Mutant Glands
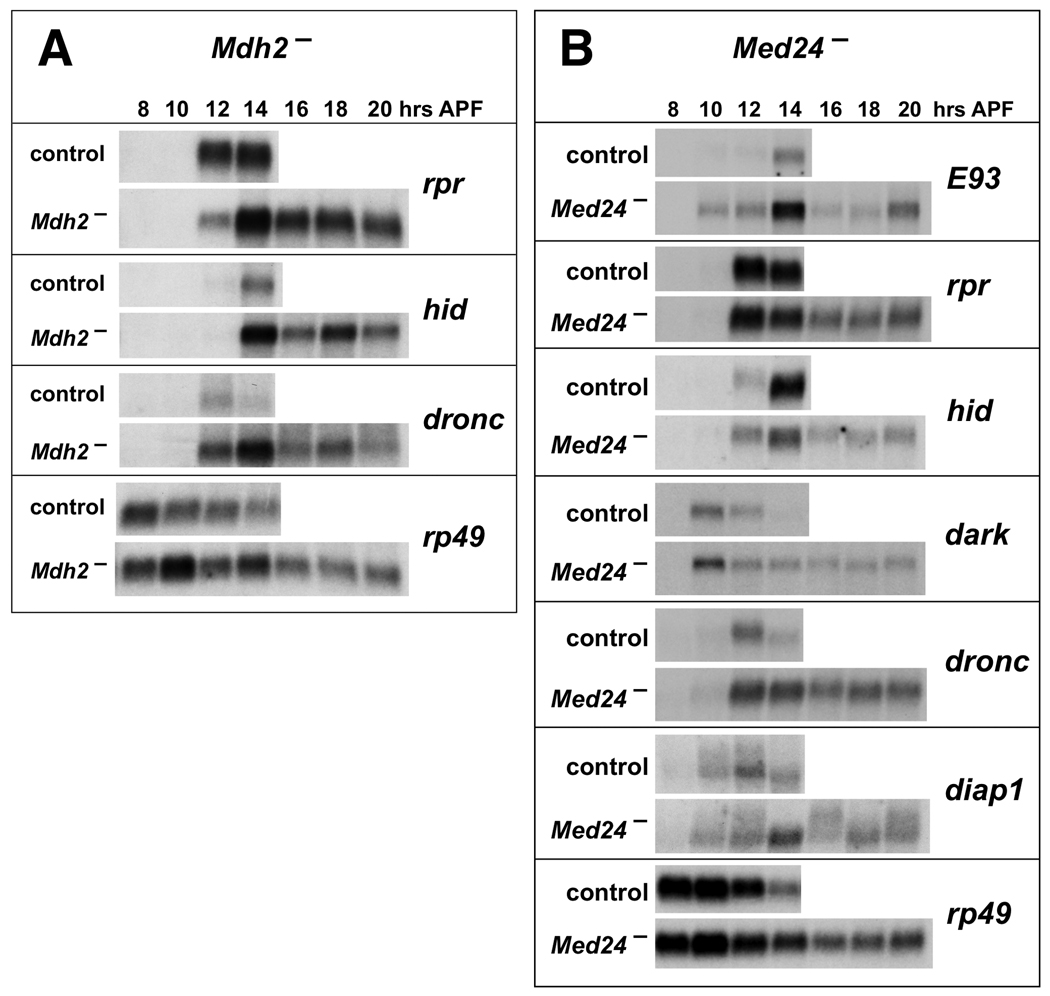
Ecdysone triggers caspase activation in doomed salivary glands through a transcriptional cascade that results in the induction of key death activator genes, including rpr and hid (Yin and Thummel, 2005). To determine if defects in this regulatory response might account for the failure of Med24 and Mdh2 mutant salivary glands to undergo cell death, we examined the expression of key ecdysone-inducible cell death regulators. RNA was extracted from salivary glands dissected from staged control animals and Mdh2 mutants and analyzed by northern blot hybridization to detect rpr, hid, and dronc expression (Fig. 3A). Consistent with the identification of Mdh2 as encoding a putative mitochondrial-localized malate dehydrogenase, we observed no defects in rpr, hid and dronc induction. Levels of rpr, hid and dronc mRNA in mutant salivary glands were similar or higher than those in control glands and were maintained in persistent mutant salivary glands at 20 hrs APF. These results indicate that the ecdysone-triggered transcriptional cascade remains intact in Mdh2 mutant salivary glands.
Fig. 3.
Death regulators are expressed normally in Mdh2 and Med24 mutant salivary glands. Total RNA was extracted from salivary glands dissected from w1118 control animals, Mdh21/Df(3R)Exel6178 mutants (A), or Med241/Df(3L)Exel6112 mutants (B) staged at 8 hrs APF and at 2 hour intervals thereafter. RNA was analyzed by northern blot hybridization to detect the expression of indicated death regulators. Hybridization to detect rp49 was used as a control for loading and transfer. Mutant and control blots were treated together to allow direct comparison.
In contrast, Med24 encodes a component of the Drosophila Mediator complex, which regulates gene expression by bridging specific transcription factors with the RNA polymerase II basal transcriptional machinery (Boube et al., 2000). Based on this function, we predicted that a mutation in Med24 would affect the ecdysone-triggered transcriptional induction of death activator genes. Surprisingly, however, we found that expression of rpr, hid, dronc, dark, and the ecdysone-inducible transcription factor E93, is essentially normal in Med24 mutant salivary glands (Fig. 3B). The slight reduction in hid expression seen in Med24 mutant salivary glands at 14 hrs APF cannot account for the persistent salivary glands found in more than 40% of Med24 mutant pupae because efficient inactivation of hid expression by RNAi results in only about 25% of treated animals having persistent glands (Yin and Thummel, 2004). These effects on death gene expression are inconsistent with the block in cell death seen in Med24 mutant salivary glands because expression of any one of these death activators is sufficient to drive an efficient death response (Grether et al., 1995; White et al., 1996; Dorstyn et al., 1999; Lee et al., 2000). This apparent paradox, however, might be explained by up-regulation of diap1, which is the only known death regulator that acts in parallel with, or downstream from, these death activators (Hay and Guo, 2006). We found, however, that diap1 expression is essentially normal in Med24 mutant salivary glands (Fig. 3B). Similar results were seen when DIAP1 protein was detected by antibody staining of staged salivary glands from control, Mdh2, and Med24 mutants, indicating that changes in DIAP1 expression cannot account for the failure of these salivary glands to undergo cell death (data not shown).
Myc-Tagged Mdh2 is Localized to Mitochondria and Rescues Mdh2 Mutants
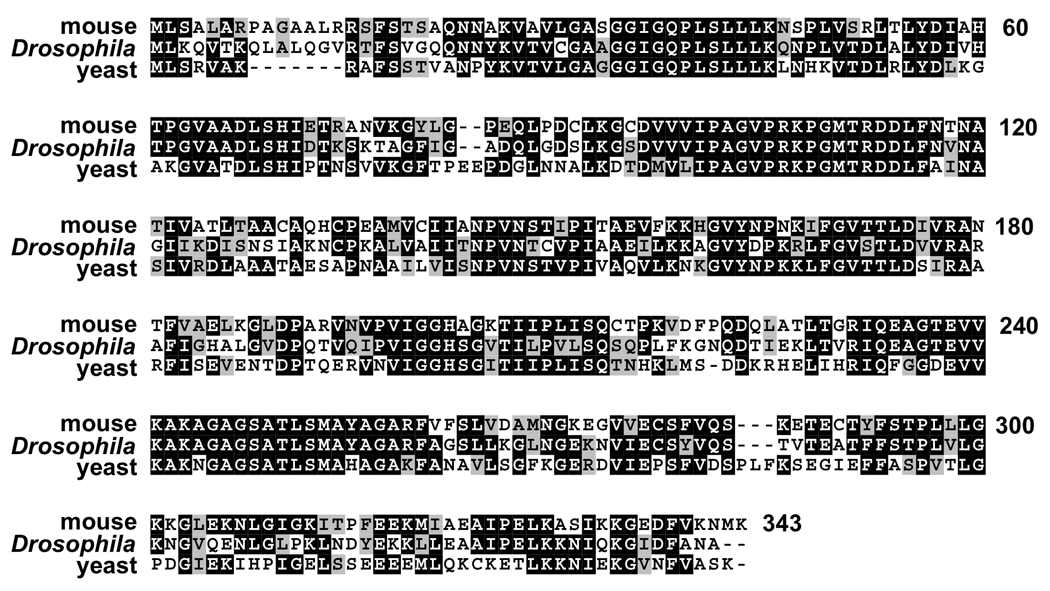
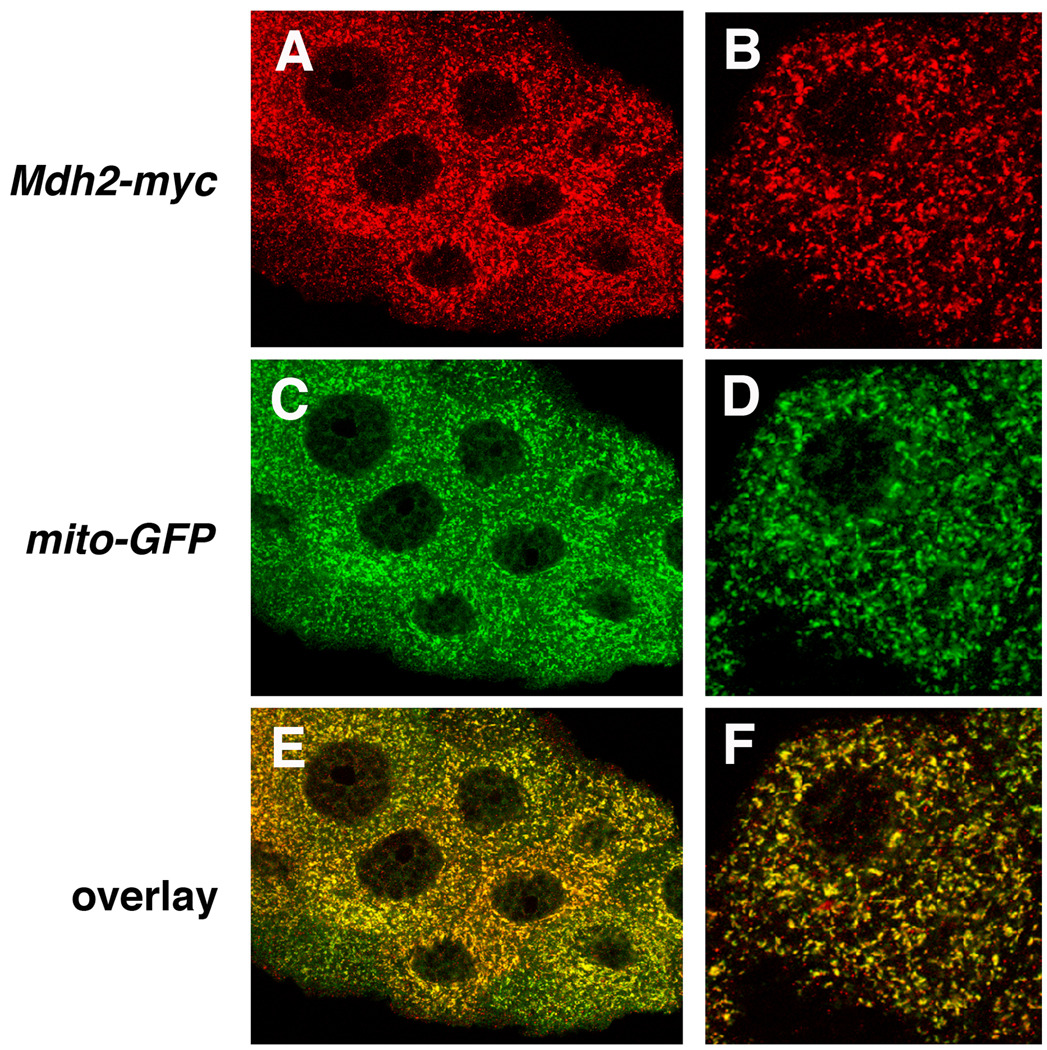
The protein encoded by Mdh2 is closely related to mitochondrial malate dehydrogenases from multiple animal species, including mouse Mdh2 and yeast Mdh1 (Fig. 4), consistent with its original proposed function (Voelker et al., 1979). As a first step toward characterizing this activity, we determined if Mdh2 localizes to mitochondria in vivo. Transformants were established using a transgene that carries the entire Mdh2 genomic locus with the coding region for a tandem Myc epitope tag fused in frame at the 3’ end of the Mdh2 protein coding region. Including one copy of this transgene in a Mdh2 mutant background restores normal salivary gland cell death and efficiently rescues pupal lethality (Table 1 a–c). In addition, detection of the Myc epitope tag in salivary glands dissected from Mdh2-myc transformants revealed a pattern of punctate staining that is indistinguishable from the expression of a mitochondrial-targeted GFP reporter, mito-GFP (Fig. 5) (LaJeunesse et al., 2004). This is consistent with the presence of a predicted mitochondrial localization signal at the N-terminus of Mdh2 (0.97 probability, Claros and Vincens, 1996). These results indicate that the protein encoded by Mdh2 localizes to mitochondria in vivo.
Fig. 4.
Mitochondrial malate dehydrogenases are highly conserved across species. Amino acid sequences of mitochondrial malate dehydrogenases from mouse (Mdh2, Swiss-Prot P08249) and yeast (Mdh1, Swiss-Prot P17505) were aligned with Drosophila Mdh2 (CG7998) using ClustalW and Boxshade programs. Black shading indicates identical amino acid residues, and gray shading indicates similar amino acid residues. A total of 215 residues are either identical (145 residues, 42%) or similar (70 residues, 20%) between all three proteins.
Fig. 5.
Myc-tagged Mdh2 localizes to mitochondria. Salivary glands dissected from w; Mdh2-myc; mito-GFP transgenic animals were stained with antibodies directed against Myc (A,B) and GFP (C,D). Mdh2-myc is expressed in a punctate subcellular pattern that is identical to that of the mitochondrial-localized GFP marker. Panels on the right depict a single cell viewed at high magnification.
Mdh2 Mutants Have Reduced ATP levels and Accumulate Citric Acid Cycle Intermediates
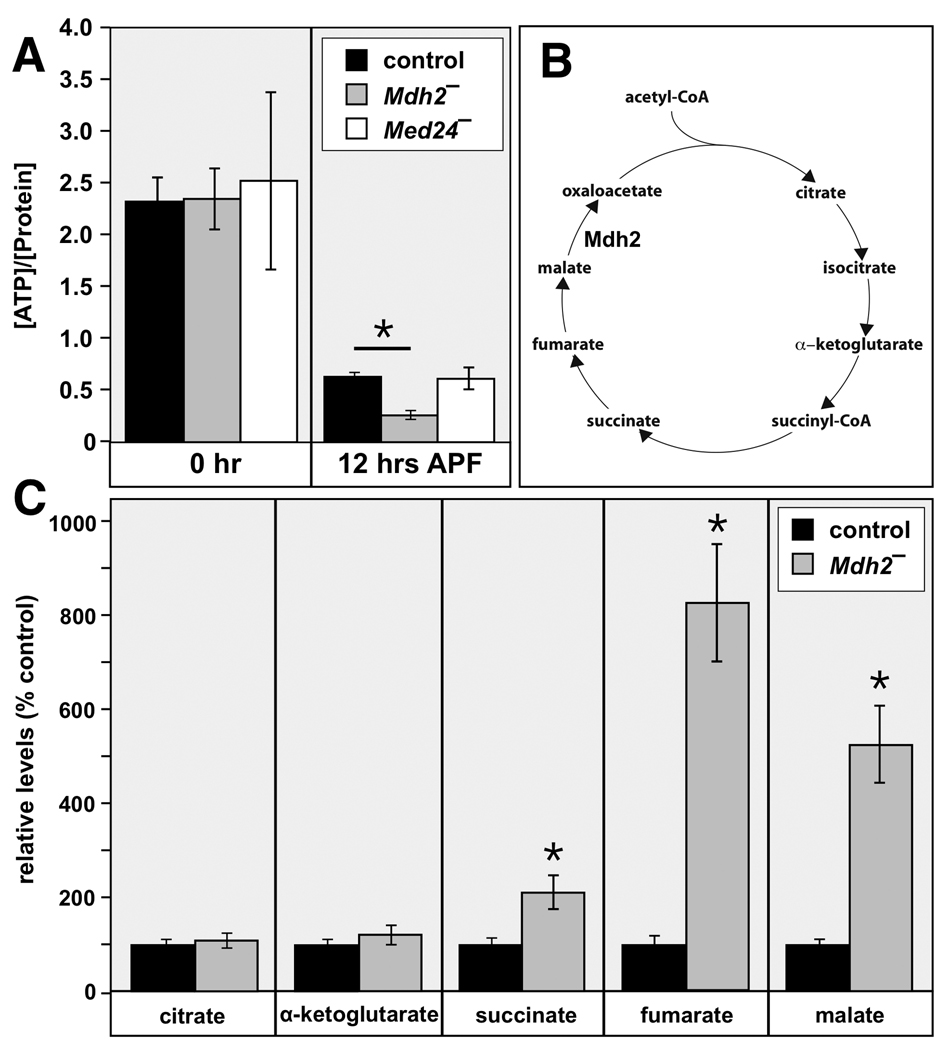
Mitochondrial-localized malate dehydrogenase controls a key step in the citric acid cycle and is critical for cellular energy production (Fig. 6B). To determine if Mdh2 exerts this function in Drosophila, we measured whole animal ATP levels in control and Mdh2 mutants at puparium formation (0 hr APF) and immediately after pupation (12 hr APF). Interestingly, ATP levels drop significantly in control prepupae between these two time points, indicating that metabolic activity is down-regulated after puparium formation (Fig. 6A). This is consistent with earlier gene profiling studies, which have demonstrated that genes involved in oxidative metabolism are repressed by ecdysone at this stage in development (Li and White, 2003). Although there is no significant difference in ATP levels between control and Mdh2 mutants at 0 hrs APF, ATP levels are significantly lower in Mdh2 mutants at 12 hrs APF (Fig. 6A). These results indicate that Mdh2 mutants display a significant energy deficiency at the stage that immediately precedes salivary gland destruction.
Fig. 6.
Mdh2 mutants display metabolic defects. (A) ATP levels were measured in prepupae or pupae staged at puparium formation (0 hr APF) or immediately after head eversion (12 hr APF) from w1118 controls, Med2 mutants (w; Mdh21/Df(3R)Exel6178), and Med24 mutants (w; Med241/Df(3L)Exel6112). The y-axis depicts ATP concentration (nmol) normalized to total protein content (µg). Mdh2 mutants display significantly reduced ATP levels at 12 hrs APF. (B) A schematic representation of the citric acid cycle is depicted with the proposed step catalyzed by Mdh2. (C) Pupae staged at 12 hrs APF from w1118 controls or Mdh2 mutants (w; Mdh21/Df(3R)Exel6178) were analyzed by GC/MS to detect citric acid cycle intermediates. The relative levels of each metabolite were determined for both control and Mdh2 mutant genotypes and are presented as normalized to a level of 100% for the control. Compared to the control, mutant pupae accumulate significantly higher levels of the late-step citric acid cycle intermediates succinate, fumarate, and malate. *p < 0.001, using an unpaired t-test. Error bars are 2× SEM.
To confirm and extend this observation, we measured citric acid cycle intermediates in control and Mdh2 mutant pupae at 12 hrs APF using gas chromatography coupled with mass spectrometry (GC/MS) (Fig. 6C). Normal levels of citrate and α-ketoglutarate are seen in Mdh2 mutant pupae. In contrast, however, late-step citric acid cycle intermediates succinate, fumarate, and malate accumulate to higher levels in Mdh2 mutants (Fig. 6C). The selective effect on these intermediates is consistent with the identification of Mdh2 as encoding a critical mitochondrial malate dehydrogenase, and biochemically identifies the point at which energy metabolism is blocked in Mdh2 mutants (Fig. 6B). These results suggest that there is a specific energy requirement from the citric acid cycle for proper progression through Drosophila metamorphosis, and that an energy deficiency may lead to a selective block in salivary gland cell death.
Mdh2 Expression is Reduced in Med24 Mutants
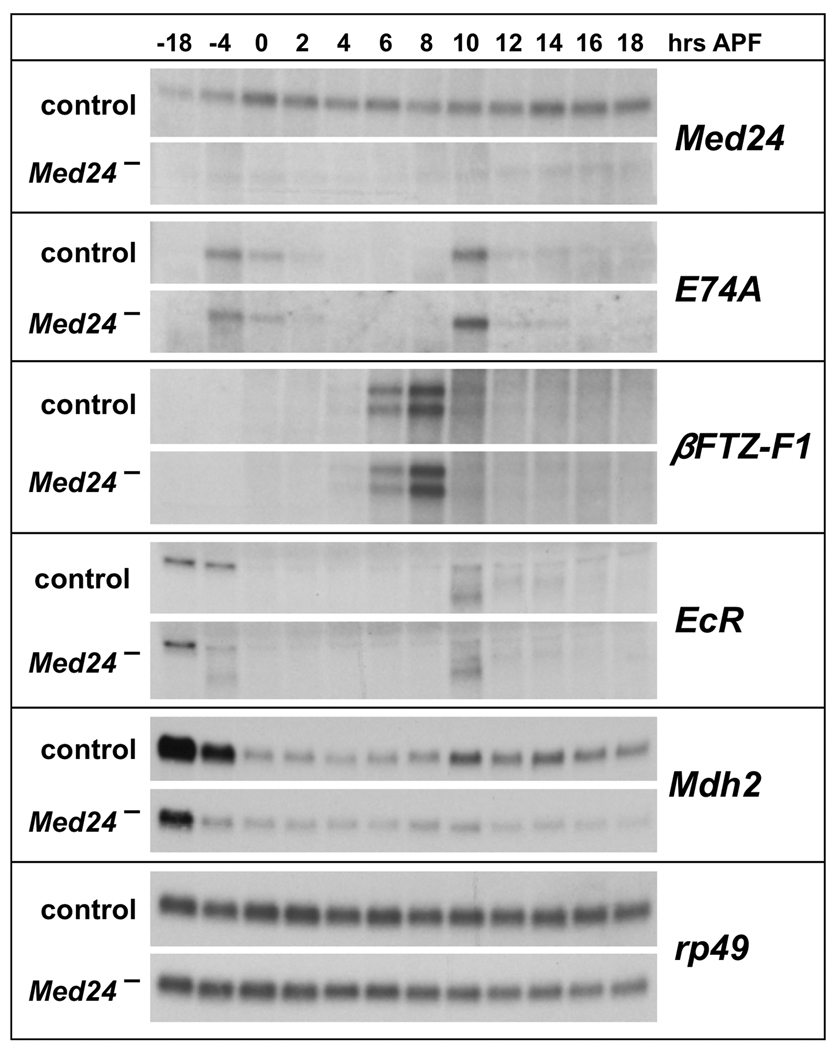
The Mediator complex was originally identified through its interactions with thyroid hormone receptor, which regulates maturation in vertebrate organisms, analogous to the role of ecdysone in directing progression through the Drosophila life cycle. We thus considered the possibility that the pupal lethality of Med24 mutants might arise from defects in the ecdysone-regulated responses that drive Drosophila maturation. To test this possibility, total RNA was isolated from staged control and Med24 mutant late third instar larvae, prepupae and early pupae, spanning the time when the two sequential ecdysone pulses trigger puparium formation and the prepupal-pupal transition. This RNA was analyzed by northern blot hybridization to detect the expression of key ecdysone-regulated transcription factors that control developmental progression during early metamorphosis. Levels of Med24 mRNA are significantly reduced in Med24 mutants, likely due to nonsense-mediated mRNA decay (Fig. 7). The other key ecdysone-regulated transcription-factor encoding genes examined, however, appear to be expressed normally in Med24 mutants, including E74A, βFTZ-F1, BR-C, and E75A (Fig. 7 and data not shown). A slight reduction in EcR transcription is seen in Med24 mutants shortly before puparium formation, at 4 hours before puparium formation (Fig. 7). The absence of any effects on puparium formation or the induction of ecdysone primary-response genes such as E74A and BR-C, however, indicates that this reduction in EcR expression is not of functional significance. In contrast, expression of Mdh2 is markedly reduced in Med24 mutants (Fig. 7). This result raises the interesting possibility that reduced levels of Mdh2 expression in Med24 mutants might lead to energy deficits that could contribute to the developmental defects seen in Med24 mutants. In addition, it is interesting to note that Mdh2 mRNA levels are reduced at puparium formation in control animals, in parallel with the reduction in ATP levels that occurs after this stage (Fig. 6A). This observation suggests that down-regulation of Mdh2 expression may contribute to the normally reduced metabolic activity seen in prepupae.
Fig. 7.
Mdh2 is expressed at reduced levels in Med24 mutants. Total RNA isolated from staged w1118 control and Med241/Df(3L)Exel6112 late third instar larvae, prepupae, and early pupae was analyzed by northern blot hybridization to detect Med24, E74A, βFTZ-F1, EcR and Mdh2 transcripts. Hybridization to detect rp49 was used as a control for loading and transfer. Numbers at the top indicate hours relative to puparium formation. Mutant and control blots were treated together to allow direct comparison.
To test the possibility that Med24 acts, at least in part, through its effects on Mdh2 expression, we examined the effect of expressing wild-type Mdh2 in Med24 mutants. While combining the Act5c-GAL4 and UAS-Mdh2 transgenes fully rescues larval salivary gland destruction and pupal lethality in Mdh2 mutants (Table 1, f), it fails to have an effect in Med24 mutants (Table 1, g, j and k). We also measured ATP levels in Med24 mutant prepupae, to determine if the reduced levels of Mdh2 expression might have an effect on overall energy levels in these animals. No significant difference in ATP levels, however, was detected when comparing control and Med24 mutants at either 0 hr or 12 hr APF (Fig. 6A). These results indicate that, although the reduced expression of Mdh2 in Med24 mutants may contribute to their block in salivary gland cell death, it is not the primary cause of this phenotype.
Future Directions
A number of transcription-factor encoding genes are required for ecdysone-triggered salivary gland cell death, including E74A, βFTZ-F1, E93, and BR-C (Yin and Thummel, 2005). Interestingly, Med24 is distinct from these regulators because it has no detectable effect on the transcriptional cascade that culminates in rpr and hid induction (Fig. 3B). This disconnect between death activator expression and salivary gland cell death is reminiscent of our earlier studies of dCBP mutants, which display a penetrant block in salivary gland cell death in spite of normal death activator gene expression (Yin et al., 2007). This effect is due to an earlier role for dCBP in down-regulating diap1 expression in the middle of the third instar, reducing the levels of this death inhibitor to a critical threshold that is susceptible to subsequent ecdysone-induced rpr- and hid-mediated cell death. The high levels of DIAP1 protein present in dCBP mutant salivary glands effectively block ecdysone-triggered cell death in spite of normal rpr and hid expression. Levels of diap1 expression in Med24 mutant salivary glands, however, are similar to those in control glands (Fig. 3B). In addition, antibody stains show that DIAP1 protein levels are normal during prepupal stages in both Med24 and Mdh2 mutant salivary glands (data not shown). These results indicate that Med24 and Mdh2 mutant salivary glands are competent to respond to death activators such as rpr and hid. Other unidentified factors must therefore be responsible for the cell death defects in Med24 mutants. Further studies are required to identify Med24-regulated target genes in the salivary glands as a step toward defining its essential role in cell death.
In contrast, our studies suggest that the block in salivary gland cell death in Mdh2 mutants can be attributed to defects in mitochondrial function. Mitochondrial factors play critical roles in regulating caspase activation in mammalian cells (Wallace and Fan, 2009). The most prominent example of this is the release of cytochrome c from mitochondria into the cytosol, which triggers the Apaf-1-mediated formation of the apoptosome and downstream caspase activation in mammalian cells (Schafer and Kornbluth, 2006). In Drosophila, however, this function of cytochrome c does not appear to be conserved, and mitochondrial regulation of caspase activation has remained controversial (Hay and Guo, 2006). Several recent studies have shown that Rpr and Hid are localized to mitochondria where they induce mitochondrial remodeling and regulate the dynamics of DIAP1 degradation (Abdelwahid et al., 2007; Goyal et al., 2007; Freel et al., 2008). It is thus possible that, in larval salivary glands, mutations in Mdh2 might impair Rpr- and Hid-mediated caspase activation by disrupting mitochondrial remodeling. Although our preliminary results indicate that mitochondrial morphology is normal in Mdh2 mutant salivary glands, additional studies are required to determine if these glands undergo the normal mitochondrial changes associated with cell death. Another possibility is that Mdh2 interacts with the Apaf-1 homolog Dark. This proposal is supported by our observation of melanotic masses on the developing wings of Mdh2 mutant pupae at a low penetrance (10–15%), a phenotype previously reported in dark mutants (Rodriguez et al., 1999) (data not shown). Further studies are required to test this hypothesis by determining if Mdh2 mutations display genetic interactions with dark alleles. In conclusion, our studies of Med24 and Mdh2 mutants have identified novel mechanisms for caspase activation in larval salivary glands and provide a new context to define the molecular basis by which steroids control programmed cell death responses during development.
EXPERIMENTAL PROCEDURES
Stocks and Developmental Staging
The Med241 (originally referred to as Med249293), Mdh21 (CG799814144), and Mdh22 (CG79986753) mutant alleles are described in Wang et al. (2008). Deficiency stocks used to generate hemizygous Med24 and Mdh2 mutants were obtained from the Bloomington stock center, Df(3L)Exel6112 (stock #7591) and Df(3R)Exel6178 (stock #7657), as was the mito-GFP transformant (stock #7194). Embryos were collected at 2- to 6-hr intervals on molasses/agar plates supplemented with yeast paste, and allowed to age as necessary. Third instar larvae were grown on standard cornmeal agar food supplemented with 0.05% bromophenol blue, and staged using dye clearance in the gut as described (Andres and Thummel, 1994). For analysis during early metamorphosis, animals were staged relative to the white prepupal stage as 0 hrs APF, and aged on moist black filter paper in a petri dish at 25°C, as necessary.
Tissue Stains
Larval salivary glands were dissected from animals at the appropriate stage, fixed and stained with antibodies essentially as described (Boyd et al., 1991). Glands were stained using antibodies directed against the active form of Caspase-3 (Cell Signaling Technologies), Lamin Dm0 (ADL84, Developmental Studies Hybridoma Bank), Myc (9E10, Developmental Studies Hybridoma Bank), GFP (MBL International), or DIAP1 (provided by Bruce A. Hay, Caltech), all at 1:200 dilution. Cy3- or Cy5-labeled secondary antibodies were obtained from Jackson Laboratories and used at 1:200 dilutions. Samples were mounted in Vectashield (Vector Laboratories) and imaged on a Leica TCS SP2 confocal microscope. Lysotracker staining was performed by exposing salivary glands to 25 nM Lysotracker Red (Molecular Probes) in PBS for 5 min at room temperature. Glands were rinsed twice in PBS and imaged immediately using a Zeiss Axioskop 2 microscope equipped with fluorescence.
RNA Isolation and Northern Blot Hybridizations
Total RNA was isolated from dissected larval salivary glands or staged whole animals using Tripure (Roche). Equal amounts of RNA were fractionated on 1% formaldehyde gels and transferred to nylon membranes for northern blot hybridization. Probes were prepared as previously described (Andres et al., 1993; Jiang et al., 1997). The probes to detect dronc, dark and diap1 mRNA were generated by PCR using the following primers: dronc- 5’-CAATCACGTTTCAACCGAGGCGTT, 5’-GCACG-CGAGCACAGAACACATAAT, dark-5’-AGTGACATTCTGCGTATGGCGGTA, 5’-TGTTGGGACTGACCTGTTTGAGGT, and diap1- 5’-ATTTGAGGACTT-GGGTGCGCATTG, 5’-TTAAACCGCGAGGAGACGCGATTA. Med24 and Mdh2 mRNAs were detected by synthesizing probes from cloned Med24 and Mdh2 cDNAs.
Rescue Transgenes
A 5.7 kb fragment encompassing the entire Mdh2 genomic locus, including about 1.1 kb upstream of the transcription start site and about 2.3 kb downstream of the transcription stop site, was amplified by PCR from the genomic clone BACR27G04 (CHORI). To insert a tandem Myc tag at the end of the Mdh2 coding region, the fragment was cloned in two parts. The first 3.2 kb part starts from the 5’end of the 5.7 kb fragment and ends before the stop codon of Mdh2, with the coding region for a tandem Myc tag added to the 3’end. A NotI site was introduced at the 5’ end, along with a stop codon to block potential upstream translation from the neighboring gene. This 3.2 kb fragment was amplified by PCR using the following primers: 5’-GTGCGGCCGCTCAGGCACGTATGTTGGCCGACTTGATT and 5 ’-CAGATCCTCTTCGGAGATCAGCTTCTGCTCCAGATCCTCTTCGGAGATCAGCTTCTGCTCGGCGTTGGCAAAGTCAATGCCCTTCTG. The second 2.5 kb fragment has a ScaI site before the stop codon of Mdh2 and ends at the 3’ end of the 5.7 kb fragment with an Asp718 site. This fragment was amplified by PCR using the following primers: 5’-AGTACTTAAGCGATCCAACAAATCCCATCCGCTAC and 5’-GTGGTACCAGGGAAAGTGAGAGATGACGCCAA. PCR reactions were conducted using the proof-reading Herculase II (Stratagene) that produces blunt ends. The 2.5 kb fragment was digested by Asp718 and inserted into an Asp718/EcoRV digested pCR-Blunt-II-Topo vector (Invitrogen). The resulting plasmid was digested by NotI and ScaI and ligated to the NotI-digested 3.2 kb fragment, generating the 5.7 kb Myc-tagged Mdh2 rescue fragment. The integrity of the 5.7 kb fragment was confirmed by restriction enzyme mapping. The 5.7 kb fragment was excised using NotI and Asp718 digestion and inserted into the pCasPeR4 vector for standard P-element mediated transformation. A homozygous viable transgenic line that carries an insertion on the second chromosome was used in this study.
The 3.2 kb full-length Med24 cDNA was amplified by PCR from a reverse transcription reaction using total RNA isolated from control (w1118) white prepupae and oligo(dT) primers. The following primers that include a NotI and XbaI site were used for PCR: 5’-ATAAGAATGCGGCCGCATTACTTCCACGGACCGAGAGAT and 5’-GCTCTAGATTTCGCCCACCCGTCTCTTAATCA. PCR-amplified Med24 cDNA was inserted into the pCR-Blunt-II-Topo vector (Invitrogen) and checked for accuracy by sequencing. The Med24 cDNA was excised by digestion with NotI and XbaI and cloned into the pUAST vector for standard P-element mediated transformation. Several homozygous viable transgenic lines were established. One line was selected, which carries a UAS-Med24 insertion on the third chromosome, and recombined with the Med241 mutation on the same chromosome.
The full length Mdh2 cDNA (DGC gold BDGP clone RE60471) was sequenced, excised by digestion with EcoRI and BglII, and cloned into the pUAST vector. Several homozygous viable transgenic lines were established using standard P-element mediated transformation. One line that carries a UAS-Mdh2 insertion on the third chromosome was selected and recombined with either the Mdh21 mutation or the Med241 mutation on the same chromosome. The Gal4/UAS system was used to drive the expression of UAS-Mdh2 and UAS-Med24 transgenes. An Act5c-GAL4 was recombined with UAS-GFP on the second chromosome to positively mark the animals that carry the driver.
Metabolic Assays
Total ATP content was measured using the luciferin-luciferase-based ATP determination kit (Molecular Probes). Staged pupae were homogenized in cold PBS and the homogenates were cleared by centrifugation. Supernatants were diluted with cold PBS, added to the reaction buffer, and assayed using a microplate reader (Bio-Tek) for luminescence. Each sample was assayed in triplicate and normalized to protein concentration as determined by a Bradford Assay (BioRad).
Control (w1118) and Mdh2 (w; Mdh21/Exel6178) mutant pupae were collected at 12 hrs APF and snap frozen in liquid nitrogen to preserve their metabolic status for GC/MS analysis. Vials containing three pupae each were transferred into a bead mill tube containing 1.4 mm ceramic beads (MoBio Laboratories, Carlsbad, CA) and 400 µl of 80% methanol that had been chilled to −20°C. Using a FastPrep 24 bead mill (MP Biomedicals, Solon, OH), the pupae were extracted for 30 seconds at 6.5 m/sec. Cell debris was removed by centrifugation at 14000g. The supernatant was dried in a vacuum and resuspended in 40 µl pyridine containing O-methyl hydroxylamine hydrochloride (40 mg/ml), sonicated for 5 minutes to fully dissolve the dried sample, followed by an hour of incubation at 34°C. Samples were clarified by quick centrifugation and transferred to a MPS 2 autosampler (Gerstel, Linthicum, MD) for a second derivatization step. One microliter of derivatized sample was injected to a CIS4 PTV injector (Gerstel) that was programmed to an initial hold temperature at 75°C for 30 seconds, followed by a 10°C/sec ramp to 275°C and a final hold time of 35 minutes. An Agilent 6890 (Agilent, Santa Clara, CA) gas chromatograph was employed during the initial temperature held at 75°C for two minutes, followed by a 40°C/min ramp to 110°C. A second 5°C/min ramp was employed to a temperature of 250°C, followed by a final 25°C/min ramp to 330°C. This final temperature was held for 3 minutes. A Restek (Bellefonte, PA) 30 m RTX-5MS column fitted with a 10 m guard column was employed for metabolite separation. A Waters (Beverly, MA) GCT Premier TOF mass spectrometer was used for metabolite detection. Normal EI conditions were used for detecting in the positive mode. Data was collected using MassLynx 4.1 software (Waters) with deconvolution and peak analysis performed in MarkerLynx (Waters). Principle component analysis (PCA) and partial least squares-discriminate analysis (PLS-DA) analysis was performed using SIMCA-P 12.0 (Umetrics, Kinellon, NJ). Further statistics was performed using Statistica Data Miner (StatSoft, Tulsa, OK).
ACKNOWLEDGEMENTS
We thank James Cox at the Health Sciences Center Core Research Facilities of the University of Utah for assistance with the GC/MS metabolomic assays, Eric Baehrecke and the Bloomington Stock Center for providing fly stocks, Bruce Hay and the Developmental Studies Hybridoma Bank for providing antibodies, and the DGRC for providing cDNA clones. We also thank members of the Thummel lab for their assistance and suggestions during the course of this study. L.W. was partially supported by a University of Utah Graduate Research Fellowship and Atherton Scholarship. This work was supported by a grant from the National Institute of Health (5R01GM073670) to C.S.T.
REFERENCES
- Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved'ova L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006;133:1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Fletcher JC, Karim FD, Thummel CS. Molecular analysis of the initiation of insect metamorphosis: A comparative study of Drosophila ecdysteroid-regulated transcription. Dev. Biol. 1993;160:388–404. doi: 10.1006/dbio.1993.1315. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical Uses in Cell and Molecular Biology. New York: Academic Press; 1994. pp. 565–573. [DOI] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boube M, Faucher C, Joulia L, Cribbs DL, Bourbon HM. Drosophila homologs of transcriptional mediator complex subunits are required for adult cell and segment identity specification. Genes Dev. 2000;14:2906–2917. doi: 10.1101/gad.17900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd L, O'Toole E, Thummel CS. Patterns of E74A RNA and protein expression at the onset of metamorphosis in Drosophila. Development. 1991;112:981–995. doi: 10.1242/dev.112.4.981. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Daish TJ, Mills K, Kumar S. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev Cell. 2004;7:909–915. doi: 10.1016/j.devcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Dorstyn L, Colussi PA, Quinn LM, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc Natl Acad Sci U S A. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel CD, Richardson DA, Thomenius MJ, Gan EC, Horn SR, Olson MR, Kornbluth S. Mitochondrial localization of Reaper to promote inhibitors of apoptosis protein degradation conferred by GH3 domain-lipid interactions. J Biol Chem. 2008;283:367–379. doi: 10.1074/jbc.M708931200. [DOI] [PubMed] [Google Scholar]
- Goyal G, Fell B, Sarin A, Youle RJ, Sriram V. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev Cell. 2007;12:807–816. doi: 10.1016/j.devcel.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes & Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Jiang C, Baehrecke EH, Thummel CS. Steroid regulated programmed cell death during Drosophila metamorphosis. Development. 1997;124:4673–4683. doi: 10.1242/dev.124.22.4673. [DOI] [PubMed] [Google Scholar]
- Jiang C, Lamblin AF, Steller H, Thummel CS. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol Cell. 2000;5:445–455. doi: 10.1016/s1097-2765(00)80439-6. [DOI] [PubMed] [Google Scholar]
- LaJeunesse DR, Buckner SM, Lake J, Na C, Pirt A, Fromson K. Three new Drosophila markers of intracellular membranes. Biotechniques. 2004;36:784–788. 790. doi: 10.2144/04365ST01. [DOI] [PubMed] [Google Scholar]
- Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- Lee CY, Simon CR, Woodard CT, Baehrecke EH. Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev Biol. 2002;252:138–148. doi: 10.1006/dbio.2002.0838. [DOI] [PubMed] [Google Scholar]
- Lee CY, Wendel DP, Reid P, Lam G, Thummel CS, Baehrecke EH. E93 directs steroid-triggered programmed cell death in Drosophila. Mol Cell. 2000;6:433–443. doi: 10.1016/s1097-2765(00)00042-3. [DOI] [PubMed] [Google Scholar]
- Li TR, White KP. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev Cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Martin DN, Baehrecke EH. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- McPhee CK, Baehrecke EH. Autophagy in Drosophila melanogaster. Biochim Biophys Acta. 2009;1793:1452–1460. doi: 10.1016/j.bbamcr.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K, Daish T, Harvey KF, Pfleger CM, Hariharan IK, Kumar S. The Drosophila melanogaster Apaf-1 homologue ARK is required for most, but not all, programmed cell death. J Cell Biol. 2006;172:809–815. doi: 10.1083/jcb.200512126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Kornbluth S. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev Cell. 2006;10:549–561. doi: 10.1016/j.devcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Voelker RA, Ohnishi S, Langley CH. Genetic and cytogenetic studies of the malate dehydrogenases of Drosophila melanogaster. Biochem Genet. 1979;17:947–956. doi: 10.1007/BF00504314. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Evans J, Andrews HK, Beckstead RB, Thummel CS, Bashirullah A. A genetic screen identifies new regulators of steroid-triggered programmed cell death in Drosophila. Genetics. 2008;180:269–281. doi: 10.1534/genetics.108.092478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Winoto A, Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109 Suppl:S57–S66. doi: 10.1016/s0092-8674(02)00710-9. [DOI] [PubMed] [Google Scholar]
- Yin VP, Thummel CS. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. Proc Natl Acad Sci U S A. 2004;101:8022–8027. doi: 10.1073/pnas.0402647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin VP, Thummel CS. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin Cell Dev Biol. 2005;16:237–243. doi: 10.1016/j.semcdb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Yin VP, Thummel CS, Bashirullah A. Down-regulation of inhibitor of apoptosis levels provides competence for steroid-triggered cell death. J Cell Biol. 2007;178:85–92. doi: 10.1083/jcb.200703206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]