SUMMARY
During an immune response, B cells undergo rapid proliferation and AID-dependent remodeling of immunoglobulin (IG) genes within germinal centers (GCs) to generate memory B and plasma cells. Unfortunately, the genotoxic stress associated with the GC reaction also promotes most B cell malignancies. Here we report that exogenous- and intrinsic AID-induced DNA strand breaks activate ATM, which signals through an LKB1 intermediate to inactivate CRTC2, a transcriptional coactivator of CREB. Using genome-wide location analysis, we determined that CRTC2 inactivation unexpectedly represses a genetic program that controls GC B cell proliferation, self-renewal, and differentiation while opposing lymphomagenesis. Inhibition of this pathway results in increased GC B cell proliferation, reduced antibody secretion, and impaired terminal differentiation. Multiple distinct pathway disruptions were also identified in human GC B cell lymphoma patient samples. Combined, our data show that CRTC2 inactivation, via physiologic DNA damage response signaling, promotes B cell differentiation in response to genotoxic stress.
INTRODUCTION
DNA double-strand breaks (DSBs) are generated during the assembly and diversification of antigen receptor genes in developing lymphocytes. During early B cell maturation in the bone marrow, the recombinase activating gene (RAG) endonuclease generates complete antigen receptor genes by the process of V(D)J recombination (Fugmann et al., 2000; Tonegawa, 1983). The generation of a diverse repertoire of high-affinity antibodies (Abs) requires further modifications of the IG genes (Rajewsky, 1996; Revy et al., 2000) in secondary lymphoid follicles within compartments known as germinal centers (GCs). GCs are sites within lymphoid tissues where mature B cells rapidly proliferate, modify IG gene sequences, and differentiate in response to a stimulating antigen. A key feature of IG remodeling is class switch recombination (CSR), a process that modifies the effector function of an Ab by replacing one constant region of the IG gene with another. CSR requires activation-induced cytidine deaminase (AID)-generated DSB intermediates (Chaudhuri et al., 2003; Muramatsu et al., 2000) and subsequent repair of distal severed ends. This genomic remodeling is critical for a robust Ab response, but genotoxic stress associated with the GC reaction also promotes most human lymphomas (Kuppers and Dalla-Favera, 2001).
In order to preserve genomic integrity, mammalian cells undergoing genotoxic stress usually respond by activating a complex DNA damage response (DDR). This response, which is required to prevent tumor formation, includes inhibition of cellular proliferation and/or induction of apoptosis (Khanna and Jackson, 2001). In GC B cells, the DDR is coordinated by the ATM serine/threonine kinase, which senses DSBs in concert with the MRN (MRE11-RAD50-NBS1) complex (Kastan and Bartek, 2004). This response is critical for humoral immunity and evasion of tumorigenesis, as defects in CSR and increased chromosomal lesions occur in activated mature B cells from mice lacking ATM (Lumsden et al., 2004; Reina-San-Martin et al., 2004) or its target proteins 53BP1 (Manis et al., 2004; Ward et al., 2004), H2AX (Franco et al., 2006), NBS1 (Kracker et al., 2005; Reina-San-Martin et al., 2005), or MDC1 (Lou et al., 2006).
During the GC reaction, B cells express the BCL6 oncoprotein, which functions as a transcriptional repressor of the PRDM1 gene encoding BLIMP-1 (Shaffer et al., 2000), the master regulator of plasma cell differentiation (Turner et al., 1994). Importantly, BCL6 also suppresses key components of the DDR in the GC by repressing the expression of ATR (Ranuncolo et al., 2007), TP53 (Phan and Dalla-Favera, 2004), and CDKN1A (P21) (Phan et al., 2005). This suppression may enable GC B cells to proliferate rapidly without triggering cellular senescence or apoptosis programs, although the resulting modified DDR increases the susceptibility of GC B cells to malignant transformation. Accordingly, BCL6 downregulation is required for post-GC B cell differentiation and evasion of tumorigenesis (Cattoretti et al., 2005).
ATM promotes and BCL6 represses the DDR, representing antagonistic forces in the life of a GC B cell. To terminate the GC reaction, rapidly proliferating B cells must tip this balance toward exiting the cell cycle to allow for terminal differentiation, although a mechanism initiating this shift has not been identified. Previously, we showed that B cell antigen receptor (BCR) engagement led to cytoplasmic sequestration and inactivation of the CREB transcriptional coactivator CRTC2 (TORC2), causing downregulation of the TCL1 oncogene in GC B cells (Kuraishy et al., 2007). Studies of glucose metabolism regulation have shown that CRTC2 inactivation results from phosphorylation at S-171 (Screaton et al., 2004) and/or S-275 (Jansson et al., 2008) by members of the AMPK family, promoting a physical association between CRTC2 and the cytoplasmic chaperone 14-3-3. However, the physiologic event(s) that inactivate CRTC2 in GC B cells are unknown. As GC B cells experience both DNA damage and CRTC2 inactivation-dependent TCL1 repression, we hypothesized that CRTC2 is inhibited by the DDR and that CRTC2 controls an extended gene program beyond TCL1. Testing of this hypothesis led to the discovery of a novel DDR pathway in GC B cells, with exogenous or intrinsic AID-induced DSBs activating ATM signaling to LKB1, a master kinase for AMPK family member proteins (Lizcano et al., 2004), that then resulted in the inactivation of CRTC2. Suggesting a role as a key homeostatic regulator, changes in gene expression resulting from CRTC2 inactivation were essential for cessation of the GC reaction, plasma cell differentiation, and suppression of tumorigenesis.
RESULTS
DNA Double-Strand Breaks Inactivate CRTC2
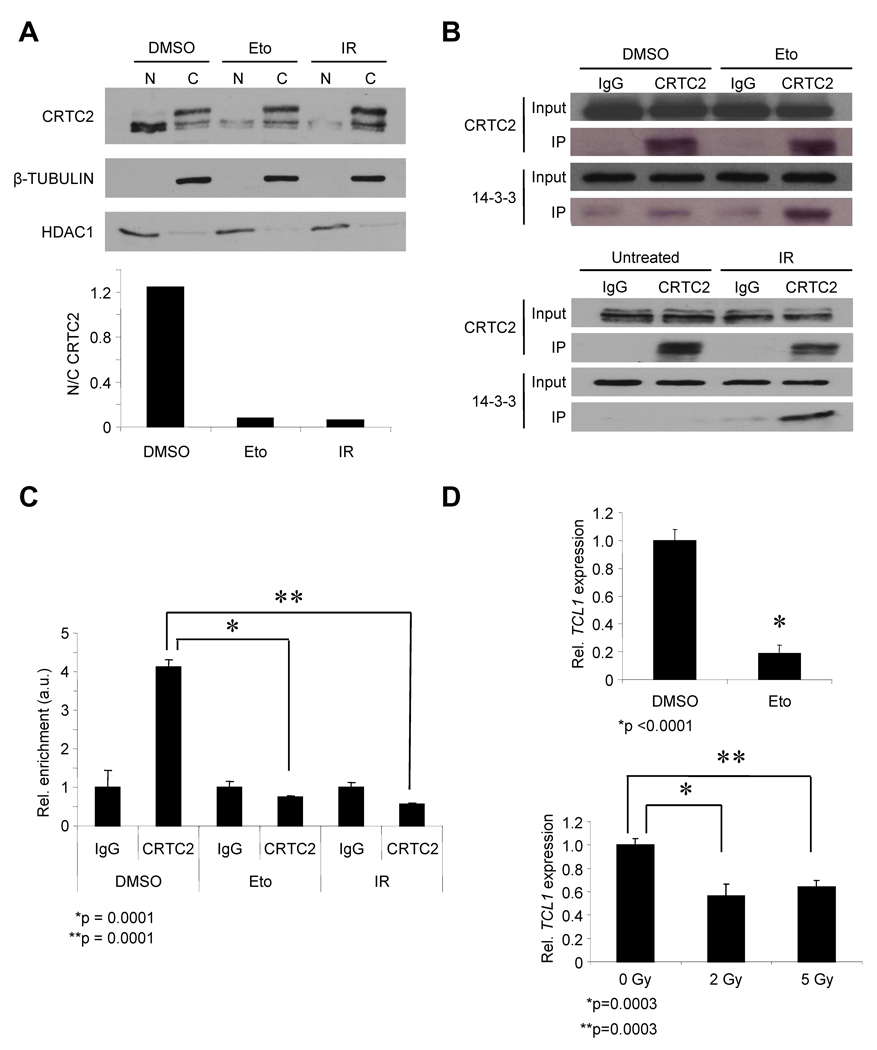
To determine whether DNA damage inactivates CRTC2, DSBs were induced in the Ramos human GC B cell line using etoposide (Eto) or γ-irradiation (IR), which are known to generate γ-H2AX foci (Phan et al., 2007). Subcellular fractionation showed that DSBs caused a shift in CRTC2 localization from the nucleus to the cytoplasm (Figure 1A). This change in CRTC2 location was accompanied by an increased association between CRTC2 and the cytoplasmic chaperone 14-3-3 (Figure 1B) (Jansson et al., 2008; Screaton et al., 2004). Chromatin immunoprecipitation (ChIP) showed a >4-fold reduction in the association between CRTC2 and the CRTC2-responsive TCL1 promoter with DSBs (Figure 1C). DSBs also repressed expression of the TCL1 promoter (Figures 1D and S1A–C). Combined, these data show that DSBs inactivate CRTC2, leading to repression of CRTC2-dependent gene expression.
Figure 1. DNA Double-Strand Breaks Inactivate CRTC2.
(A) (Top) Immunoblot showing CRTC2 protein expression in the nucleus (N) and cytoplasm (C) of Ramos B cells exposed to DMSO (control), Eto (20 µM, 1h), or IR (5 Gy). Upper bands in the cytoplasm lanes indicate phosphorylated CRTC2. β-TUBULIN (cytoplasm) and HDAC1 (nucleus) are loading controls. (Bottom) Nuclear/cytoplasmic ratios for CRTC2 are plotted from densitometry.
(B) (Top) Immunoprecipitation of CRTC2 or rabbit IgG from lysates of Ramos cells exposed to DMSO or Eto (20 µM, 1h). (Bottom) Untreated cells were compared to cells exposed to IR (5 Gy). Immunoblots for lysates (Input) and CRTC2 or 14-3-3 immunoprecipitates (IP) are shown.
(C) ChIP for CRTC2 or rabbit IgG using chromatin from Ramos cells after DMSO, Eto (20 µM, 1h), or IR (5Gy). Immunoprecipitates were analyzed by QPCR for the TCL1 promoter. Values were normalized to the ACTB promoter and shown as arbitrary units (a.u.). Values are expressed as the mean ± SEM for three independent experiments.
(D) (Top) QPCR for TCL1 in Ramos cells after DMSO or Eto (20 µM, 6h). (Bottom) Untreated cells were compared to cells exposed to the indicated doses of IR. Values were normalized to 36B4. Values are expressed as the mean ± SEM for three independent experiments.
DSB-Induced CRTC2 Inactivation Requires Activation of ATM and LKB1
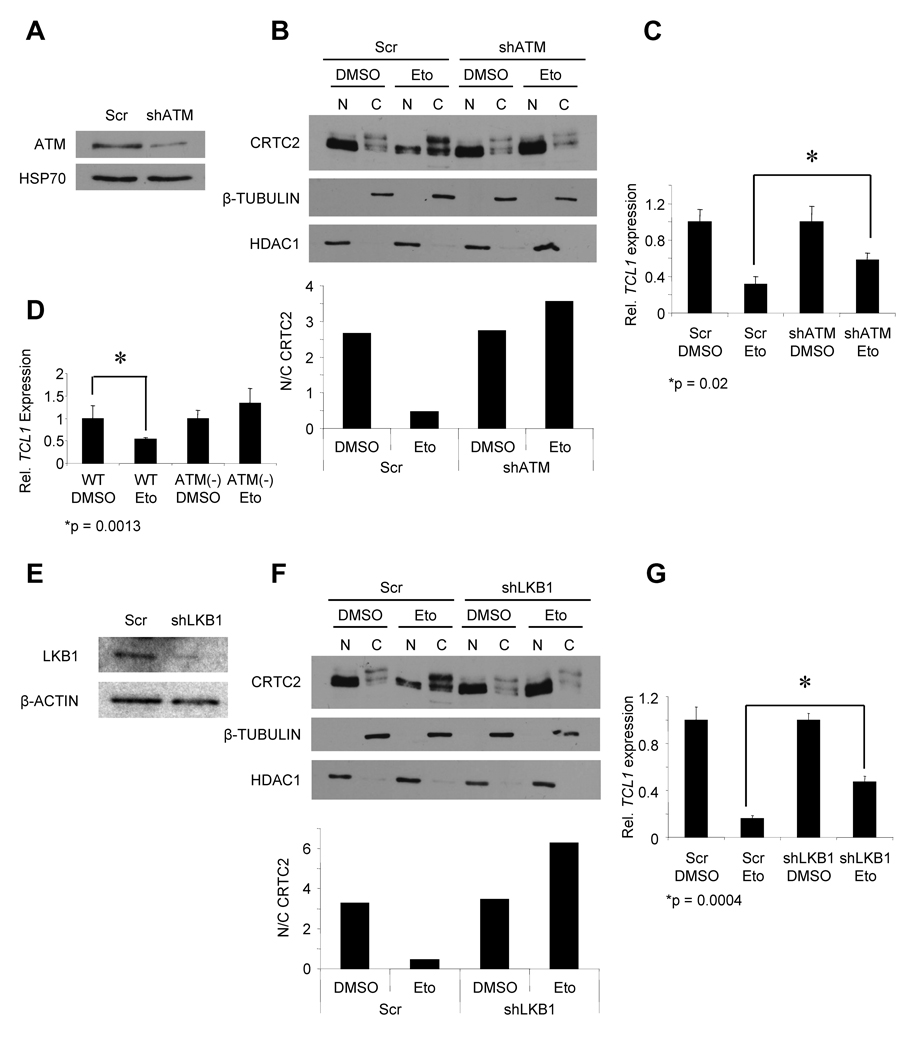
We next tried to identify a link between DSBs and CRTC2 inactivation. Since the DNA damage-sensing kinase ATM is required for CSR (Lumsden et al., 2004; Reina-San-Martin et al., 2004), we evaluated ATM for a role in CRTC2 inactivation. Induced DSBs in Ramos activated ATM (Figure S2A). ATM loss-of-function, using 2 different shRNA sequences targeting ATM, pharmacological inhibition with the ATM inhibitor Kudos, and the use of B cell lines from ATM-deficient ataxia-telangiectasia (A-T) patients showed a requirement for ATM in DSB-induced CRTC2 inactivation (Figures 2A–D, S2B–D). ATM phosphorylates multiple substrates during the DDR (Matsuoka et al., 2007), potentially including T366 of the tumor suppressor LKB1 (Fernandes et al., 2005; Sapkota et al., 2002). In turn, LKB1 phosphorylates and inactivates CRTC2 through AMPK family members (Fu and Screaton, 2008; Katoh et al., 2006; Shaw et al., 2005), suggesting a pathway from DSBs to CRTC2 inactivation. DSBs caused ATM-dependent phosphorylation of LKB1 T366 (Figure S2E,F). Similarly, DSBs induced LKB1 phosphorylation in primary B cells (Figure S2G). Metformin is an anti-diabetic drug that promotes LKB1-dependent activation of AMPK (Shackelford and Shaw, 2009; Shaw et al., 2005). Ramos cells exposed to metformin showed reduced nuclear localization of CRTC2 and TCL1 repression (Figure S2H–J). shRNA knockdown of LKB1 with 2 different sequences lessened CRTC2 inactivation in response to DSBs in Nalm-6 pre B cells (Figure S2K,L) and Ramos cells (Figures 2E–G, S2M,N). These data demonstrate that DSBs inactivate CRTC2 via ATM and LKB1 signaling, providing a novel gene regulation mechanism during the DDR.
Figure 2. DSB-induced CRTC2 Inactivation Requires Activation of ATM and LKB1.
(A) Immunoblot showing ATM knockdown in Ramos cells 4d post-transduction. HSP70 is a loading control.
(B) (Top) Immunoblot showing CRTC2 protein expression in the nucleus (N) and cytoplasm (C) of Ramos cells transduced with control (Scr) shRNA or shATM after exposure to DMSO or Eto (20 µM, 1h). β-TUBULIN (cytoplasm) and HDAC1 (nucleus) are loading controls. (Bottom) Nuclear/cytoplasmic ratios for CRTC2 are plotted from densitometry.
(C) QPCR for TCL1 in shATM or control (Scr) Ramos cells after DMSO or Eto (20 µM, 6h) exposure. Values were normalized to 36B4. Values are expressed as the mean ± SEM for three independent experiments.
(D) QPCR for TCL1 in WT or ATM-deficient lymphoblastoid cells exposed to DMSO or Eto (20 µM, 6h). Values were normalized to 36B4. Values are expressed as the mean ± SEM for three independent experiments.
(E) Immunoblot showing LKB1 knockdown in Ramos cells 4d post-transduction. β-ACTIN iss a loading control.
(F) (Top) Immunoblot showing CRTC2 protein expression in the nucleus (N) and cytoplasm (C) of Ramos cells transduced with control (Scr) shRNA or shLKB1 after exposure to DMSO or Eto (20 µM, 1h). β-TUBULIN (cytoplasm) and HDAC1 (nucleus) are loading controls. (Bottom) Nuclear/cytoplasmic ratios for CRTC2 are plotted from densitometry.
(G) QPCR for TCL1 in shLKB1 or control (Scr) Ramos cells after DMSO or Eto (20 µM, 6h) exposure. Values were normalized to 36B4. Values are expressed as the mean ± SEM for three independent experiments.
CRTC2 Inactivation Occurs During CSR in GC B cells
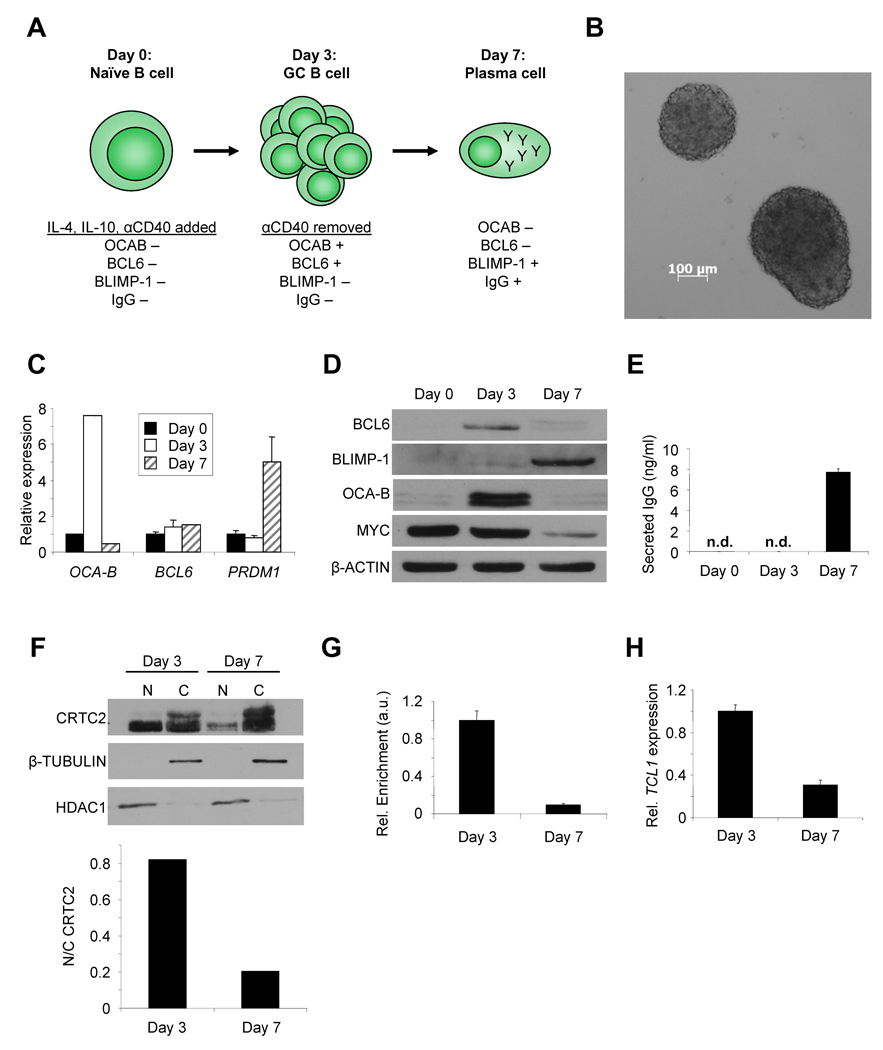
To determine the role of CRTC2 in GC B cells, changes in CRTC2 activity and direct target gene expression were evaluated over the course of a GC reaction. For this, we modified an in vitro B cell differentiation system starting with naïve human tonsil B cells (Figure 3A) (Arpin et al., 1995; Fluckiger et al., 1998). Rapid B cell expansion and correct modulation of established GC B and plasma cell markers (BCL6, MYC, OCA-B, BLIMP-1) occurred over 7 days, as expected for a GC-like reaction (Figure 3B–D) (Allman et al., 1996; Greiner et al., 2000; Shaffer et al., 2008). Though undetectable on day 3, soluble and membrane-bound IgG (32% of cells) was detected by day 7 (Figures 3E, S3A), preceded by γ-H2AX focus formation by day 5 (Figure S3B)(Petersen et al., 2001). These results indicate that CSR followed by plasma cell differentiation was induced during a GC-like reaction between days 3 and 7 of culture.
Figure 3. CRTC2 Inactivation Occurs During CSR in GC B Cells .
(A) Schematic of primary human B cell differentiation system.
(B) Phase contrast image of expanding human B cell clusters 24h post-stimulation (5X).
(C) QPCR for OCA-B, BCL6, and PRDM1 in human B cells at 0, 3, or 7 days of differentiation. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments.
(D) Immunoblot for BCL6, BLIMP-1, OCA-B, and MYC in primary human B cells after 0, 3, or 7 days of differentiation. β-ACTIN is a loading control.
(E) ELISA for total IgG in human B cell culture media harvested after 0, 3, or 7 days of differentiation. n.d.= none detected. Values are expressed as the mean ± SEM for three independent experiments.
(F) (Top) Immunoblot showing CRTC2 protein expression in the nucleus (N) and cytoplasm (C) of primary human B cells on days 3 or 7 of differentiation. β-TUBULIN (cytoplasm) and HDAC1 (nucleus) are loading controls. (Bottom) Nuclear/cytoplasmic ratios for CRTC2 are plotted from densitometry.
(G) ChIP for CRTC2 using chromatin from human B cells after 3 or 7 days of differentiation. Immunoprecipitates were analyzed by QPCR for the TCL1 promoter. Values are normalized to an intergenic region and shown as arbitrary units (a.u.). Values are expressed as the mean ± SEM for three independent experiments.
(H) QPCR for TCL1 in human B cells after 3 or 7 days of differentiation. Values are normalized to 36B4. Values are expressed as the mean ± SEM for three independent experiments.
CRTC2 activity was evaluated during the interval in which CSR occurred. Nuclear CRTC2 decreased between days 3 and 7 (Figure 3F) with a coinciding decrease in the association between CRTC2 and the TCL1 promoter and decreased TCL1 expression, as observed in vivo (Figure 3G,H)(Said et al., 2001; Teitell et al., 1999). Importantly, similar CRTC2 modulation was observed during GC B cell development in vivo (Figure S3C)(Klein et al., 2003; Said et al., 2001), with ~70% of plasma cells containing entirely cytoplasmic CRTC2 and ~30% negative for CRTC2 protein expression. Combined, these results strongly suggest that CRTC2 becomes phosphorylated and sequestered in the cytoplasm and inactivated during CSR in vivo, resulting in reduced expression of CRTC2-dependent target genes, such as TCL1 (Figure S3D).
AID-Dependent, ATM to LKB1 Signaling in GC B Cells Inactivates CRTC2
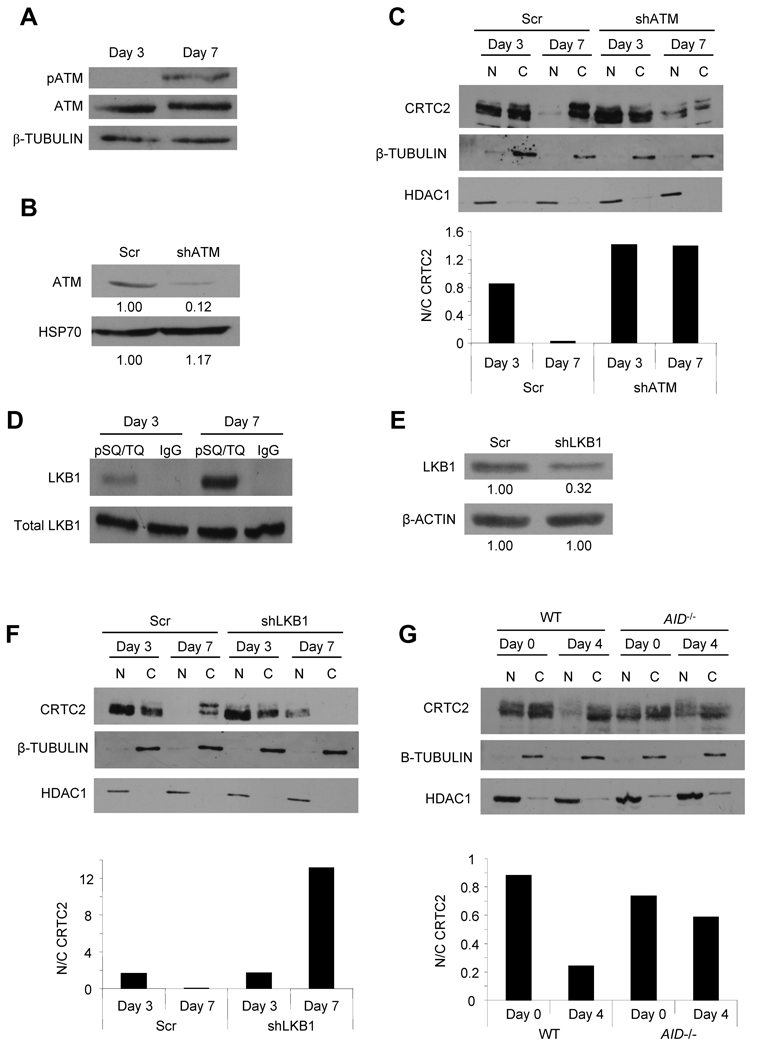
The requirement for ATM and LKB1 as upstream regulators of CRTC2 inactivation was assessed in the modeled GC-like reaction. An increase in phospho-ATM S1981 was detected by day 7 (Figure 4A), coinciding with DSB generation during CSR. ATM knockdown (Figure 4B) resulted in increased CRTC2 nuclear localization during B cell differentiation compared to control cells (Figure 4C). Similar results were obtained for LKB1 (Figure 4D–F). These data strongly suggest that GC-like B cells responding to physiologic DSBs activate ATM to LKB1 signaling to inactivate CRTC2. This pathway is engaged during the period of CSR, further suggesting a link to Ab production and B cell maturation.
Figure 4. AID-Dependent ATM to LKB1 Signaling in GC B Cells Inactivates CRTC2.
(A) Immunoblot of pATM (S1981) and ATM in human B cells after 3 or 7 days of differentiation. β-TUBULIN is a loading control.
(B) Immunoblot showing ATM knockdown in human B cells 3 days post-shATM transduction. HSP70 is a loading control. Densitometric values are indicated below each band.
(C) (Top) Immunoblot showing CRTC2 protein expression in the nucleus (N) and cytoplasm (C) of primary human B cells, transduced with control (Scr) or shATM expressing lentiviruses, on days 3 or 7 of differentiation. β-TUBULIN (cytoplasm) and HDAC1 (nucleus) are loading controls. (Bottom) Nuclear/cytoplasmic ratios for CRTC2 are plotted from densitometry.
(D) Immunoprecipitation of phosphorylated ATM/ATR substrates (pSQ/TQ) or a rabbit IgG control from lysates of human B cells after 3 or 7 days of differentiation. Lysates (Total LKB1) or immunoprecipitates (LKB1) are analyzed by immunoblot for LKB1.
(E) Immunoblot showing LKB1 knockdown in human B cells 3 days post-shLKB1 transduction. β-ACTIN is a loading control. Densitometric values are indicated below each band.
(F) (Top) Immunoblot showing CRTC2 protein expression in the nucleus (N) and cytoplasm (C) of primary human B cells, transduced with control (Scr) or shLKB1 expressing lentiviruses, on days 3 or 7 of differentiation. β-TUBULIN (cytoplasm) and HDAC1 (nucleus) are loading controls. (Bottom) Nuclear/cytoplasmic ratios for CRTC2 are plotted from densitometry.
(G) (Top) Immunoblot showing CRTC2 protein expression in the nucleus (N) and cytoplasm (C) of spleen B cells from wild-type (WT) or AID knockout (AID−/−) mice on days 0 and 4 of differentiation. β-TUBULIN (cytoplasm) and HDAC1 (nucleus) are loading controls. (Bottom) Nuclear/cytoplasmic ratios for CRTC2 are plotted from densitometry.
To directly determine whether CSR drives CRTC2 inactivation, the subcellular localization of CRTC2 was compared in B cells from WT and AID knockout mice. AID-deficient B cells cannot produce DSBs at IG loci and therefore CSR does not occur (Muramatsu et al., 2000). αCD40 and IL-4 induced CSR in ~43% of WT but not in AID knockout B cells (Figure S4). Most importantly, CRTC2 was retained in the nucleus of AID knockout B cells compared to its nuclear depletion in WT B cells (Figure 4G). This result strongly supports the requirement for physiologic DSBs induced by AID during CSR for ATM- and LKB1-dependent CRTC2 inactivation.
CRTC2 Regulates Genes that Control B Cell Development
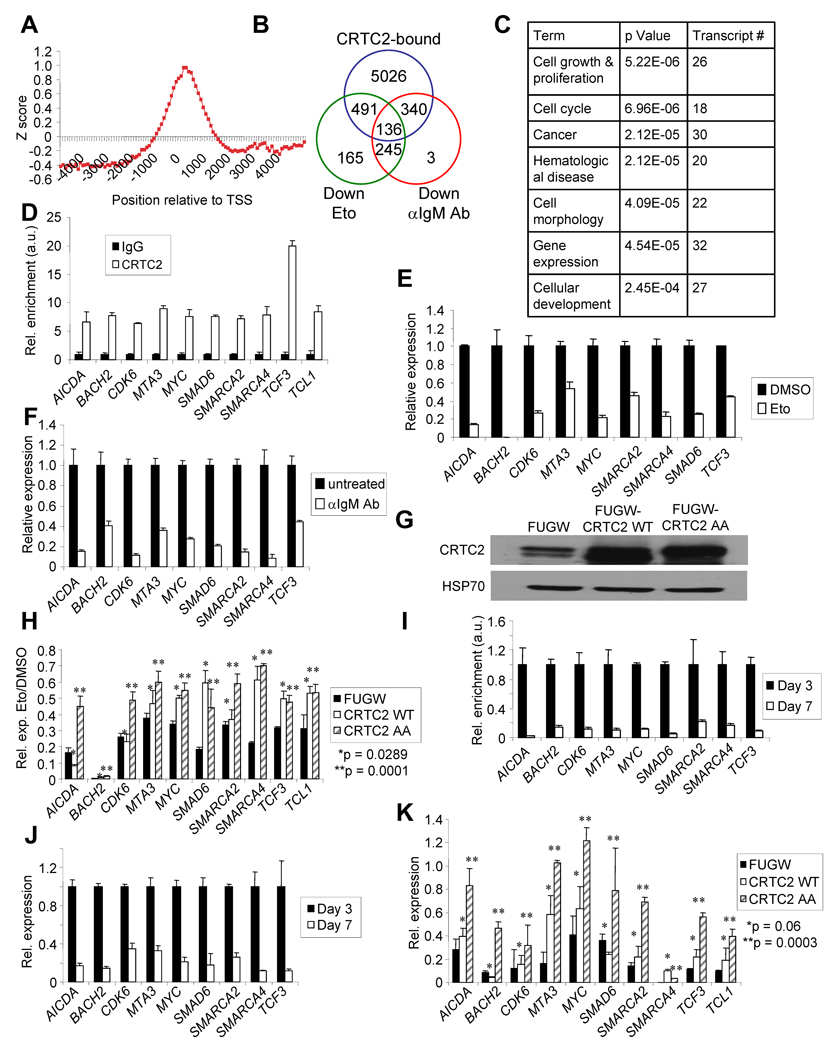
The effect of CRTC2 inactivation on global gene expression in GC B cells was evaluated. ChIP-on-chip for CRTC2 target genes in Ramos cells was performed using two Abs that recognize distinct CRTC2 epitopes (Figure S5A). The genomic DNA bound to CRTC2 and total input DNA were distinctly labeled and co-hybridized to Agilent 244K human promoter microarrays that contained 60-mer oligonucleotide probes covering the region from −5.5 kb to +2.5 kb relative to the transcriptional start sites for ~17,000 annotated human genes. The vast majority of bound sequences were located within 1kb of transcriptional start sites (Figure 5A), consistent with a previous global analysis of CREB promoter occupancy (Zhang et al., 2005). This approach revealed that CRTC2 occupied the promoters of 5993 genes (Figure S5B,C). Motif analysis among the bound sequences identified conventional CRE half-sites as the two most highly enriched of all possible 6-mers (p = 10−22.5, Figure S5D), as anticipated for a coactivator of CREB. Gene expression profiling was also performed using Ramos cells treated with Eto or anti-IgM Ab, which was also shown to inactivate CRTC2 (Kuraishy et al., 2007). This screen identified 136 putative CRTC2-regulated genes (Figure 5B, Table S1) implicated in cellular growth and proliferation, cell cycle, and cancer (Figure 5C).
Figure 5. CRTC2 Regulates a Gene Program that Controls B Cell Development.
(A) Plotted Z-scores for CRTC2 binding to human promoter regions relative to the predicted transcriptional start sites (TSS).
(B) Venn diagram displaying putative CRTC2 direct target genes as defined by downregulation with exposure to α-IgM Ab and Eto, and CRTC2 binding to the promoter region.
(C) Gene ontology (GO) analysis of candidate CRTC2 target genes. p-values are based on a hypergeometric distribution.
(D) ChIP for CRTC2 or rabbit IgG in Ramos cells. Immunoprecipitates were analyzed by QPCR for the promoters of CRTC2 target genes. Values are normalized to intergenic regions and shown as arbitrary units (a.u.). Values are expressed as the mean ± SEM for three independent experiments.
(E) QPCR for CRTC2 target genes in Ramos cells after DMSO or Eto (20 µM, 6h) exposure. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments.
(F) QPCR for CRTC2 target genes in Ramos cells with or without α-IgM (10µg/ml, 6h) exposure. Values are normalized to 36B4. Values are expressed as the mean ± SEM for three independent experiments.
(G) Immunoblot for CRTC2 protein expression in Ramos cells 48h post-infection with the indicated lentivirus. HSP70 is a loading control.
(H) QPCR for CRTC2 target genes in Ramos cells transduced with the indicated lentivirus. DMSO or Eto (20 µM, 6h) exposures were initiated 48h post-infection. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments.
(I) ChIP for CRTC2 using chromatin from human B cells after 3 or 7 days of differentiation. Immunoprecipitates were analyzed by QPCR for the promoters of CRTC2 target genes. Values are normalized to intergenic regions, are shown as arbitrary units (a.u.), and are expressed as the mean ± SEM for three independent experiments.
(J) QPCR for CRTC2 target genes in human B cells after 3 or 7 days of differentiation. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments.
(K) QPCR for CRTC2 target genes in human B cells transduced with the indicated lentivirus after 3 or 7 days of differentiation. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments.
To validate these findings, ten candidate CRTC2 target genes were selected which have functional relevance for GC B cell development and/or lymphomagenesis. Gene-specific ChIP demonstrated enrichment for promoter regions of these ten candidate CRTC2 target genes with a CRTC2 Ab compared to an isotype control (Figure 5D). Gene expression changes were measured with Eto or anti-IgM treatment, with QPCR validating the expression array results (Figure 5E, F). To determine a causal relationship between CRTC2 inactivation and downregulation of candidate CRTC2 target genes, a lentiviral expression system was used to transduce Ramos cells with WT or mutant CRTC2 in which serines 171 and 275 were mutated to alanines (CRTC2-AA; Figures 5G and S5E). CRTC2-AA should remain in the nucleus because both serines require phosphorylation by AMPK to exclude CRTC2 from the nucleus (Jansson et al., 2008; Screaton et al., 2004). CRTC2-WT overexpression or continued activation by CRTC2-AA caused a derepression of the 10 target genes with DSBs or with anti-IgM exposure (Figures 5H, S5F), indicating that expression of these genes is dependent upon CRTC2 activity. Similar results were obtained from primary human B cells (Figures 5I–K). Supporting the connection between genotoxic stress and CRTC2 target gene regulation, pre-treatment of Ramos cells with ATM inhibitor Kudos caused a significant depression of CRTC2 target genes with DSBs (Figure S5I). In addition, a significant derepression of CRTC2 target genes occurred when DSBs were induced in immortalized B cells from A-T patients, which lack functional ATM, compared to WT controls (Figure S5G). A-T cells also exhibited a striking retention of CRTC2 in the nucleus after Eto exposure compared to WT controls (Figure S5H).
CRTC2 Inactivation is Required for Plasma Cell Differentiation
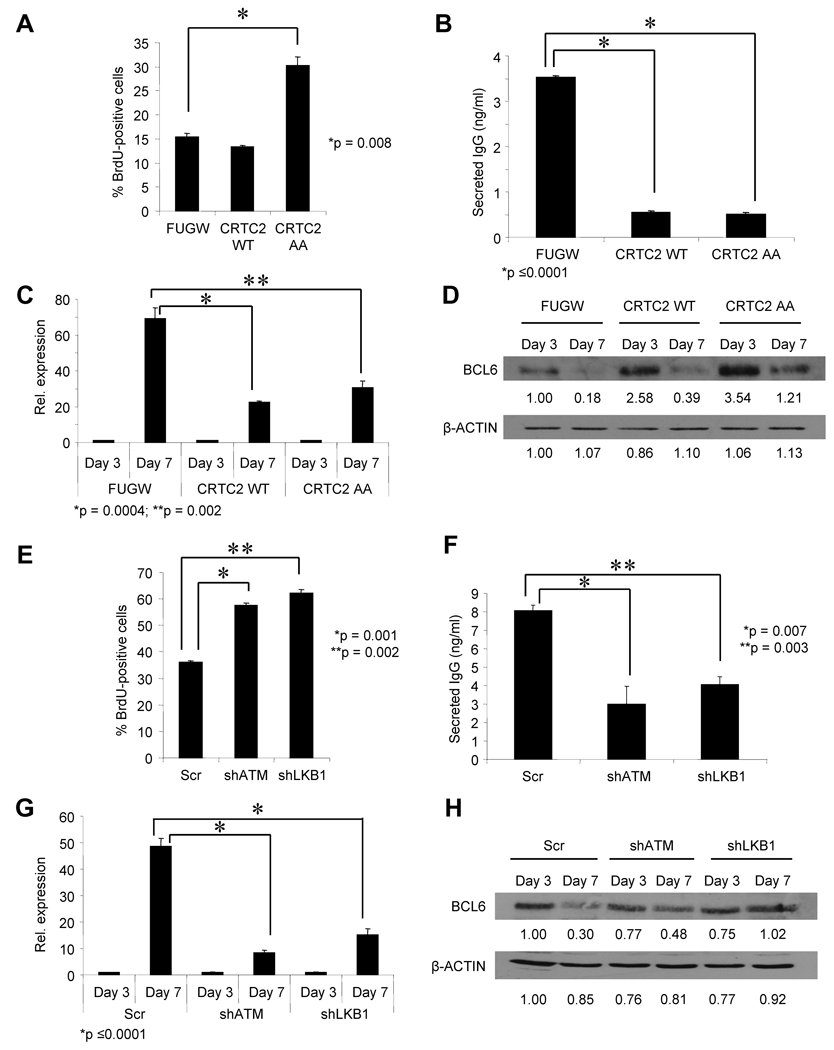
To determine the effect of signaling that inactivates CRTC2 on B cell development, naïve tonsil B cells efficiently transduced with CRTC2-WT or CRTC2-AA were stimulated to generate a GC-like reaction. Proliferating GC B cells, or centroblasts, are among the fastest proliferating cells in the body (Klein and Dalla-Favera, 2008), and plasma cell differentiation is characterized in part by repression of pro-proliferative gene expression (Shaffer et al., 2002). Hyperactive or overexpressed CRTC2 caused a marked increase in proliferation (Figure 6A), a decrease in soluble IgG production (Figure 6B), impaired induction of the plasma cell master regulator BLIMP-1 (Figure 6C), and sustained expression of the GC B cell master regulator BCL6 (Figure 6D) on day 7 of culture. Similar results were obtained when ATM or LKB1 expression was decreased by shRNAs (Figure 6E–H). Interestingly, transduced CRTC2 did not impair CSR, as equivalent levels of productive IGG transcripts were generated in CRTC2-overexpressing and control cells (Figure S6A,B). These data strongly suggest that the DDR pathway leading to CRTC2 inactivation is required for efficient termination of the GC reaction and Ab secretion.
Figure 6. CRTC2 Inactivation is Required for Plasma Cell Differentiation.
(A) BrdU incorporation in human B cells, transduced with the indicated lentivirus, on day 7 of differentiation. BrdU-FITC-positive cells were detected by flow cytometry. Values are expressed as the mean ± SEM for three independent experiments.
(B) ELISA for total secreted IgG in human B cell culture media harvested after day 7 of differentiation. Cells were transduced with the indicated lentivirus prior to the initiation of differentiation. Values are expressed as the mean ± SEM for three independent experiments.
(C) QPCR for PRDM1 expression in human B cells transduced with the indicated lentivirus after 3 or 7 days of differentiation. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments.
(D) Immunoblot for BCL6 in human B cells transduced with the indicated lentivirus after 3 or 7 days of differentiation. β-ACTIN is a loading control. Densitometric values are indicated below each band.
(E) BrdU incorporation in human B cells, transduced with the indicated lentivirus, on day 7 of differentiation. BrdU-FITC-positive cells were detected by flow cytometry. Values are expressed as the mean ± SEM for three independent experiments.
(F) ELISA for total secreted IgG in human B cell culture media harvested after day 7 of differentiation. Cells were transduced with the indicated lentivirus prior to the initiation of differentiation. Values are expressed as the mean ± SEM for three independent experiments.
(G) QPCR for PRDM1 in human B cells transduced with the indicated lentivirus after 3 or 7 days of differentiation. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments.
(H) Immunoblot for BCL6 in human B cells, transduced with the indicated lentivirus, after 3 or 7 days of differentiation. β-ACTIN is a loading control. Densitometric values are indicated below each band.
CRTC2 Inactivation is Disrupted in GC-Derived B Cell Lymphomas
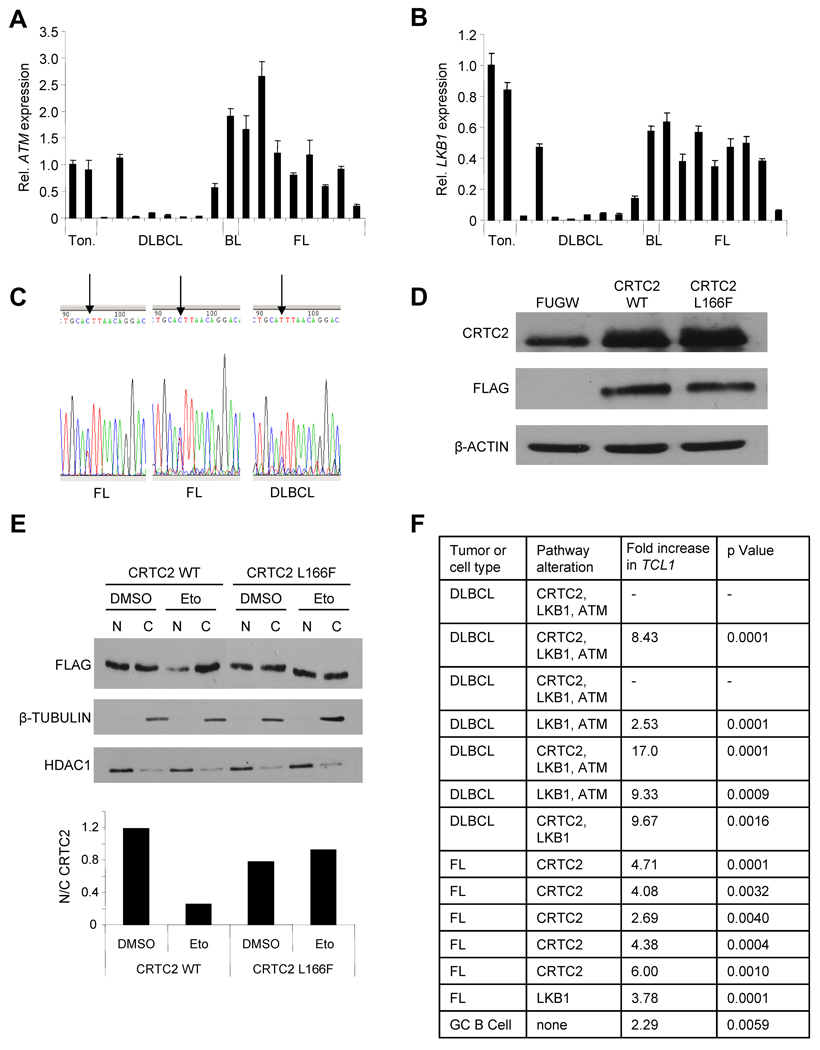
Since TCL1 is often overexpressed in GC B cell lymphomas (Klein et al., 2001; Narducci et al., 2000; Said et al., 2001), we assessed this signaling pathway in human lymphoma samples. QPCR analysis revealed a 10-fold or greater loss of ATM expression in 6/17 (35%), or LKB1 expression in 7/17 (41%), clinical samples (Figure 7A,B). CRTC2 expression was not altered in these tumors (data not shown), so the CRTC2 coding sequence was evaluated for alterations. A C→T missense mutation was identified in 10/17 tumor samples, compared with 0/14 normal tonsil samples (p < 0.0005, one-sided Fisher’s exact test)(Figures 7C and S7). This change results in a L→F amino acid substitution in the AMPK recognition sequence of the CRTC2 protein (Screaton et al., 2004). Although this alteration is conservative, it prevented inactivation of CRTC2 in Ramos B cells subjected to DSBs (Figure 7D,E), perhaps by disrupting AMPK-CRTC2 or CRTC2-14-3-3 interactions. Consistent with this result, TCL1 expression was maintained in 11 of 13 B cell lymphomas that harbored disruptions in the ATM→LKB1→AMPK→CRTC2 signaling pathway (Figure 7F). These results provide the first evidence for aberrant CRTC2 activity in human lymphomas from multiple, distinct defects in a novel DDR pathway.
Figure 7. CRTC2 Inactivation is Disrupted in GC-derived B Cell Lymphomas.
(A) QPCR for ATM in 17 human lymphoma samples and 2 tonsils. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments. Abbreviations: Ton. = tonsil; DLBCL = diffuse large B cell lymphoma; BL = Burkitt lymphoma; FL = follicular lymphoma.
(B) QPCR for LKB1 in 17 human lymphoma samples and 2 tonsils. Values are normalized to 36B4 and are expressed as the mean ± SEM for three independent experiments.
(C) Representative chromatograms showing a CRTC2 496 C→T alteration in human GC B cell lymphomas. Arrow indicates nucleotide 496 in the coding region of CRTC2.
(D) Immunoblot showing expression of CRTC2 and FLAG in Ramos cells 48h post-infection with the indicated lentivirus. β-ACTIN is a loading control.
(E) (Top) Immunoblot showing exogenous CRTC2 localization in Ramos cells, transduced with the indicated lentivirus, after DMSO or Eto (20 µM, 1h) exposure. β-TUBULIN (cytoplasm) and HDAC1 (nucleus) are loading controls. (Bottom) Nuclear/cytoplasmic ratios for CRTC2 are plotted from densitometry.
(F) QPCR for TCL1 expression in 13 human B cell lymphoma samples and in GC B cells at day 3 of culture. Values are normalized to 36B4 and compared to TCL1 expression in differentiated plasma cells at day 7 of culture. Dashes indicate tumors with undetectable TCL1 expression.
DISCUSSION
Here, we describe a mechanism in which exogenous and physiologic DNA damage in GC B cells leads to CRTC2 inactivation, which is required for IG secretion and plasma cell differentiation. Although a prior study showed a BCR signaling requirement for Ab affinity-driven plasma cell development (Phan et al., 2006), the cues that cause GC B cells to differentiate into plasma cells are unknown. Previously we showed that CRTC2 is also inactivated by BCR engagement (Kuraishy et al., 2007), suggesting that CRTC2 inactivation is a response to both genotoxic stress and BCR signaling that terminates the GC reaction. More broadly, these results also implicate LKB1 as a central kinase with the potential to integrate metabolic and now AID-initiated genotoxic stress signaling in a new pathway that can terminate with CRTC2 inactivation to drive terminal cell differentiation.
Our results, along with aspects of two recent studies, provide an unanticipated and important new direction for the DDR by coupling genotoxic stress to non-DNA repair-related physiologic or pathologic cellular maturation. One recent study in pre-B cells demonstrated that RAG-induced DSBs during V(D)J recombination activated transcription by NF-κB, leading to the expression of mature lymphocyte-specific genes (Bredemeyer et al., 2008). However, this study required genetically sustained RAG-induced DSBs to detect mature lymphocyte gene expression, leaving open the question of physiologic relevance. A second recent study showed that genotoxic stress opposed self-renewal in melanocyte stem cells (MSCs) and caused the MSCs to aberrantly differentiate into ectopically pigmented melanocytes, resulting in irreversible hair graying (Inomata et al., 2009). This form of abnormal differentiation resulted from pathologic DNA damage accumulation from the environment, which led to lineage degeneration and aging by an unknown mechanism. In contrast, our study shows a new and surprising mechanism linking physiologic, AID-induced DSBs to ATM and LKB1 signaling in order to inactivate CRTC2, with CRTC2 inactivation required for the differentiation of plasma cells. This regulatory function exceeds the established response to DSBs that maintains genomic integrity, and provides evidence that the DDR influences normal cell development and physiology. A potential ontologic reason for coupling genotoxic stress with differentiation is that the forced elimination of damaged cells from stem or precursor cell pools, such as the GC, may be an intrinsic mechanism to preserve the integrity of pre-terminal cell types and prevent tumorigenesis.
A consistent theme that re-emerges from studies of hematopoietic development is that a block in differentiation seems to promote a malignancy that reflects the stage in development at which the block occurs. Here we show that disruption of the signaling pathway leading to CRTC2 inactivation and plasma cell differentiation occurs in GC-derived lymphomas. As ATM is required for CRTC2 inactivation, defects in this pathway may contribute in part to the IG deficiencies (Nowak-Wegrzyn et al., 2004; Staples et al., 2008) and increased susceptibility to lymphoma (Taylor et al., 1996) observed in patients with A-T. In addition to mutations, aberrant ATM repression in multiple B cell lymphoma subtypes has also been reported (Basso et al., 2005). Like ATM, the LKB1 tumor suppressor is inactivated in a number of human malignancies (Hezel and Bardeesy, 2008; Shaw, 2008). Furthermore, a small population-based case-control study showed that diabetics taking metformin, which activates LKB1 and results in CRTC2 inactivation, had a reduced risk of cancer (Evans et al., 2005). In mice, a hypomorphic mutation in Lkb1 present on a Pten haploinsufficient background markedly accelerated the development of marginal zone B cell lymphoma (Huang et al., 2008). Interestingly, PTEN deficiency is similar to aberrantly sustained TCL1 expression for mature B cells, as both alterations hyperactivate AKT signaling (Teitell, 2005).
In summary, CRTC2 plays a powerful and previously unknown role in normal GC B cell differentiation, and its inactivation by the DDR is critical for downregulation of a genetic program that maintains the GC reaction. These findings place CRTC2 in a regulatory pathway that controls GC exit and plasma cell differentiation during terminal B cell development, and heralds future studies to interrogate the role of CRTC2 as a potential oncogenic factor and therapeutic target in B cell lymphoma.
EXPERIMENTAL PROCEDURES
Cell Culture, Tissues, and Reagents
Wildtype, Atm−/−, Lkb1(T366A), and Lkb1−/− MEFs (N. Bardeesy, Massachusetts General Hospital) were grown in DMEM (Gibco) with 20% FBS plus antibiotics. Nalm-6, Ramos, PBL, and ATM-deficient lymphoblastoid cells were grown in RPMI 1640 (Gibco) with 10% FBS plus antibiotics. Fresh-frozen human tissues and fresh human tonsils were obtained from the UCLA Tissue Procurement Core Laboratory in accordance with institutional guidelines and Institutional Review Board (IRB) approval. Reagents included etoposide, mitomycin C, and metformin (Sigma); and the ATM inhibitor KU55933 (Kudos Pharmaceuticals). α-CD40 (mouse anti-human IgG) was purified from culture medium of G28-5 mouse hybridoma cells (K. Zhang, UCLA).
Primary Human B Cell Culture System
Fresh tonsils were used to isolate naïve B cells as described (Said et al., 2001). Tonsils were minced, and mononuclear cells (MCs) isolated by Ficoll-Paque (GE Healthcare) density centrifugation. MCs were incubated with α-IgD-PE (BD Pharmingen) on ice, washed, incubated with α-PE beads (Miltenyi Biotech), washed, and collected using the MidiMACS system (Miltenyi Biotech). Lentiviral transduction was performed at this stage, as indicated. Cells were seeded 5 × 105 cells/ml and cultured in complete RPMI 1640 plus 20ng/ml IL-4, 20ng/ml IL-10 (BD Pharmingen), and 2µg/ml α-CD40 Ab.
ELISA
ELISA was performed using a human IgG ELISA quantification kit (Bethyl Laboratories).
RNA Analysis
Total RNA was extracted using Trizol (Life Technologies). cDNA was made using the Superscript First-Strand Synthesis System (Invitrogen). QPCR was performed as described previously (Kuraishy et al., 2007). Expression was normalized to a 36B4 mRNA control sequence.
ImmunoBlot, Immunoprecipitation, and Antibodies
Immunoblots were performed as described (Kuraishy et al., 2007). Briefly, 20–50 µg whole cell extract for each sample was separated by SDS-PAGE and transferred to a nitrocellulose membrane. Blocked membranes were incubated with primary Abs in TBS-Tween and 5% milk (or 5% BSA for phospho-specific Abs) overnight (Ab sources provided upon request). Immunoprecipitations were performed with whole cell extracts and primary Ab overnight, followed by precipitation of immune complexes with Protein G beads (Santa Cruz Biotechnologies). Subcellular fractionations were performed using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Scientific) according to the manufacturer’s protocol.
Chromatin Immunoprecipitation
ChIP assays were performed as described (Kuraishy et al., 2007).
Immunofluorescence Microscopy
Ramos or primary B cells were plated on poly-L-lysine cover slips and used for immunofluorescence studies as decribed (Kuraishy et al., 2007).
siRNA Electroporation
LKB1 siRNA (2 µM; Dharmacon) or a scrambled control siRNA (2 µM) were electroporated into Nalm-6 cells using the Amaxa Nucleofector I (program C-05) and Nucleofector kit T (Amaxa, Germany).
Retrovirus and Luciferase Assays
Luciferase assays were performed as described (Kuraishy et al., 2007). A pBABE-FLAG-LKB1 retroviral construct, expressing FLAG-tagged LKB1, was generated by standard methods. Viral supernatant from HEK293T cells was collected 48 and 72h after transfection. PBL cells (1 × 105/well) were incubated with 1 ml of virus supplemented with 2 µl polybrene and centrifuged at 2500 rpm for 1h at 30°C. One day after repeat infection, puromycin (0.5 µg/ml) was added to the media. FLAG-LKB1 expression was determined by Western blot.
Lentivirus
For shRNA, the H1 promoter and RNAi sequences for LKB1, ATM, or scramble were subcloned into FUGW at the PacI site upstream of a ubiquitin promoter-driven EGFP sequence (Lois et al., 2002). For expression vectors, full-length CRTC2 cDNA was cloned into FUGW downstream of the ubiquitin promoter. CRTC2 mutants were generated using the Quikchange site-directed mutagenesis kit (Stratagene). Virus produced by HEK293T cells was concentrated by ultracentrifugation, resuspended in RPMI 1640, and used to spin-infect 2 × 106 cells/well in a 24-well plate for 2h.
In Vitro Proliferation Assays
In vitro proliferative kinetics were assayed using the BrdU flow kit (BD Pharmingen) by the manufacturer’s protocol.
ChIP-on-Chip
ChIP was performed using 2 different Abs against CRTC2 (EMD Biosciences; Cell Signaling). Biological duplicate experiments were performed with each Ab. Array details are in the Supplemental Experimental Procedures.
Gene Expression Arrays
RNA was isolated from Ramos cells without treatment or after 6h of Eto (20 µM) or α-IgM (10 µg/ml) exposure using Trizol, followed by clean-up with the Qiagen RNeasy kit. Array details are in the Supplemental Experimental Procedures.
Gene Ontology Analysis
Gene ontology analysis was performed using Ingenuity Pathways Analysis (http://www.ingenuity.com/products/pathways_analysis.html).
Statistical Analyses
Data are presented as the mean ± SEM. A two-tailed t-test was used for most comparisons, with p < 0.05 considered significant.
HIGHLIGHTS
CREB coactivator CRTC2 is inactivated by DNA breaks via ATM and LKB1 signaling
CRTC2 inactivation is from AID-induced DNA breaks in germinal center B cells
CRTC2 regulates proliferation, plasma cell differentiation, and lymphomagenic genes
Failed CRTC2 inactivation is from defective AID→ATM→LKB1→AMPK→CRTC2 pathway signaling
Supplementary Material
ACKNOLWEDGEMENTS
The authors thank Reuben J. Shaw and Marc Montminy (Salk Institute) for discussions and reagents, and Heather Christofk, Steve Bensinger, Randolph Wall, and Steven Smale (UCLA) for discussions and evaluation of the manuscript. Supported by NIH grants GM07185 (NRSA to MHS and AIK), R01CA90571 (MAT), R01CA156674 (MAT), and by the NIH Roadmap for Medical Research Nanomedicine Initiative (PNEY018228; MAT). MAT is a recent Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All Chip-on-chip and gene expression microarray data have been deposited in the Gene Expression Omnibus (GEO) under the submission number GSE23171.
REFERENCES
- Allman D, Jain A, Dent A, Maile RR, Selvaggi T, Kehry MR, Staudt LM. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- Arpin C, Dechanet J, Van Kooten C, Merville P, Grouard G, Briere F, Banchereau J, Liu YJ. Generation of memory B cells and plasma cells in vitro. Science. 1995;268:720–722. doi: 10.1126/science.7537388. [DOI] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins JB, Weaver BK, et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. Bmj. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes N, Sun Y, Chen S, Paul P, Shaw RJ, Cantley LC, Price BD. DNA damage-induced association of ATM with its target proteins requires a protein interaction domain in the N terminus of ATM. J Biol Chem. 2005;280:15158–15164. doi: 10.1074/jbc.M412065200. [DOI] [PubMed] [Google Scholar]
- Fluckiger AC, Sanz E, Garcia-Lloret M, Su T, Hao QL, Kato R, Quan S, de la Hera A, Crooks GM, Witte ON, et al. In vitro reconstitution of human B-cell ontogeny: from CD34(+) multipotent progenitors to Ig-secreting cells. Blood. 1998;92:4509–4520. [PubMed] [Google Scholar]
- Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fu A, Screaton RA. Using kinomics to delineate signaling pathways: control of CRTC2/TORC2 by the AMPK family. Cell Cycle. 2008;7:3823–3828. doi: 10.4161/cc.7.24.7241. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Greiner A, Muller KB, Hess J, Pfeffer K, Muller-Hermelink HK, Wirth T. Up-regulation of BOB.1/OBF.1 expression in normal germinal center B cells and germinal center-derived lymphomas. Am J Pathol. 2000;156:501–507. doi: 10.1016/S0002-9440(10)64754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27:6908–6919. doi: 10.1038/onc.2008.342. [DOI] [PubMed] [Google Scholar]
- Huang X, Wullschleger S, Shpiro N, McGuire VA, Sakamoto K, Woods YL, McBurnie W, Fleming S, Alessi DR. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem J. 2008;412:211–221. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, Iseki S, Hara E, Masunaga T, Shimizu H, et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- Jansson D, Ng AC, Fu A, Depatie C, Al Azzabi M, Screaton RA. Glucose controls CREB activity in islet cells via regulated phosphorylation of TORC2. Proc Natl Acad Sci U S A. 2008;105:10161–10166. doi: 10.1073/pnas.0800796105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Takemori H, Lin XZ, Tamura M, Muraoka M, Satoh T, Tsuchiya Y, Min L, Doi J, Miyauchi A, et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. Febs J. 2006;273:2730–2748. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H, Freedman A, Inghirami G, Cro L, Baldini L, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracker S, Bergmann Y, Demuth I, Frappart PO, Hildebrand G, Christine R, Wang ZQ, Sperling K, Digweed M, Radbruch A. Nibrin functions in Ig class-switch recombination. Proc Natl Acad Sci U S A. 2005;102:1584–1589. doi: 10.1073/pnas.0409191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- Kuraishy AI, French SW, Sherman M, Herling M, Jones D, Wall R, Teitell MA. TORC2 regulates germinal center repression of the TCL1 oncoprotein to promote B cell development and inhibit transformation. Proc Natl Acad Sci U S A. 2007;104:10175–10180. doi: 10.1073/pnas.0704170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. Embo J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Lumsden JM, McCarty T, Petiniot LK, Shen R, Barlow C, Wynn TA, Morse HC, 3rd, Gearhart PJ, Wynshaw-Boris A, Max EE, et al. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J Exp Med. 2004;200:1111–1121. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Narducci MG, Pescarmona E, Lazzeri C, Signoretti S, Lavinia AM, Remotti D, Scala E, Baroni CD, Stoppacciaro A, Croce CM, et al. Regulation of TCL1 expression in B- and T-cell lymphomas and reactive lymphoid tissues. Cancer Res. 2000;60:2095–2100. [PubMed] [Google Scholar]
- Nowak-Wegrzyn A, Crawford TO, Winkelstein JA, Carson KA, Lederman HM. Immunodeficiency and infections in ataxia-telangiectasia. J Pediatr. 2004;144:505–511. doi: 10.1016/j.jpeds.2003.12.046. [DOI] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- Phan RT, Saito M, Kitagawa Y, Means AR, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat Immunol. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- Phan TG, Paus D, Chan TD, Turner ML, Nutt SL, Basten A, Brink R. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;203:2419–2424. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc Natl Acad Sci U S A. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Said JW, Hoyer KK, French SW, Rosenfelt L, Garcia-Lloret M, Koh PJ, Cheng TC, Sulur GG, Pinkus GS, Kuehl WM, et al. TCL1 oncogene expression in B cell subsets from lymphoid hyperplasia and distinct classes of B cell lymphoma. Lab Invest. 2001;81:555–564. doi: 10.1038/labinvest.3780264. [DOI] [PubMed] [Google Scholar]
- Sapkota GP, Deak M, Kieloch A, Morrice N, Goodarzi AA, Smythe C, Shiloh Y, Lees-Miller SP, Alessi DR. Ionizing radiation induces ataxia telangiectasia mutated kinase (ATM)-mediated phosphorylation of LKB1/STK11 at Thr-366. Biochem J. 2002;368:507–516. doi: 10.1042/BJ20021284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- Shaw RJ. LKB1: cancer, polarity, metabolism, and now fertility. Biochem J. 2008;416:e1, e3. doi: 10.1042/BJ20082023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples ER, McDermott EM, Reiman A, Byrd PJ, Ritchie S, Taylor AM, Davies EG. Immunodeficiency in ataxia telangiectasia is correlated strongly with the presence of two null mutations in the ataxia telangiectasia mutated gene. Clin Exp Immunol. 2008;153:214–220. doi: 10.1111/j.1365-2249.2008.03684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Metcalfe JA, Thick J, Mak YF. Leukemia and lymphoma in ataxia telangiectasia. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- Teitell M, Damore MA, Sulur GG, Turner DE, Stern MH, Said JW, Denny CT, Wall R. TCL1 oncogene expression in AIDS-related lymphomas and lymphoid tissues. Proc Natl Acad Sci U S A. 1999;96:9809–9814. doi: 10.1073/pnas.96.17.9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitell MA. The TCL1 family of oncoproteins: co-activators of transformation. Nat Rev Cancer. 2005;5:640–648. doi: 10.1038/nrc1672. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.