Summary
Translational control is frequently exerted at the stage of mRNA recruitment to the initiating ribosome. We have reconstituted mRNA recruitment to the 43S preinitiation complex (PIC) using purified S. cerevisiae components. We show that eIF3 and the eIF4 factors not only stabilize binding of mRNA to the PIC, they also dramatically increase the rate of recruitment. Although capped mRNAs require eIF3 and the eIF4 factors for efficient recruitment to the PIC, uncapped mRNAs can be recruited in the presence of eIF3 alone. The cap strongly inhibits this alternative recruitment pathway, imposing a requirement for the eIF4 factors for rapid and stable binding of natural mRNA. Our data suggest that the 5′-cap serves as both a positive and negative element in mRNA recruitment, promoting initiation in the presence of the canonical group of mRNA handling factors while preventing binding to the ribosome via an aberrant, alternative pathway requiring only eIF3.
Introduction
Eukaryotic translation initiation requires over a dozen factors, in contrast to initiation in bacteria, which requires only three. Much of the added machinery in eukaryotes appears to be needed for manipulating the mRNA in order to allow it to bind to the small ribosomal subunit and, subsequently, to enable the ribosomal pre-initiation complex (PIC) to locate the initiation codon. During mRNA maturation and export from the nucleus, eukaryotic mRNAs can fold into stable structures and associate with RNA binding proteins, events that can hinder PIC binding and downstream steps in initiation.
The current model of translation initiation (Jackson et al., 2010; Sonenberg and Hinnebusch, 2009) begins with the assembly of the 43S PIC. A ternary complex (TC) consisting of eukaryotic initiation factor (eIF) 2, GTP and methionyl initiator tRNA (Met-tRNAi) binds to the small (40S) ribosomal subunit in a step facilitated by eIF1, eIF1A and the multisubunit complex eIF3. The PIC is then recruited to the 5′-end of an mRNA via the 7-methylguanosine cap structure. The 5′-cap is bound by eIF4E, a component of the trimeric cap binding complex eIF4F. eIF4F also contains the DEAD-box RNA helicase eIF4A and a multidomain protein, eIF4G, that interacts with a number of other factors and RNA. These interactions are thought to be necessary to bring the PIC to the 5′-end of the mRNA in mammalian systems (Asano et al., 2001; He et al., 2003; Korneeva et al., 2000; Lamphear et al., 1995), however the fact that mRNA can be recruited to the ribosome in the absence of eIF4G in yeast cells suggests that alternative pathways are possible (Jivotovskaya et al., 2006). eIF4A is believed to be responsible for removing structure from the 5′-UTR to allow the ribosome to bind (Svitkin et al., 2001). The ability of eIF4A to unwind short RNA duplexes in vitro is stimulated by eIF4B (Lawson et al., 1989). Once the PIC has been properly recruited to the mRNA it is thought to scan in a 5′ to 3′ direction in search of the start codon, usually the first AUG (Kozak, 1991).
Although general roles for the mRNA recruitment factors have been established, relatively little is known about the actual molecular mechanics of their operation. Previously, we developed a minimal reconstituted translation initiation system using S. cerevisiae components (Algire et al., 2002). This system used an unstructured, model mRNA and therefore did not require the mRNA recruitment factors. We have now recapitulated recruitment of natural mRNAs to the PIC in the reconstituted system. This system has enabled us to analyze the effects of the factors on both the extent and kinetics of mRNA recruitment to the PIC. The eIF4 factors play critical roles in increasing the rate and extent of recruitment of capped, natural mRNAs to the PIC, and eIF3 appears to be the central hub required for mRNA binding to the 43S complex.
We also used the system to examine the role of the 5′-cap structure on the mRNA. Our results suggest that the cap not only stimulates recruitment of natural mRNA via eIF4F, but that it also inhibits an alternative, non-productive recruitment pathway requiring only eIF3. The 5′-5′-triphosphate moiety of the cap is responsible for this inhibition of off-pathway mRNA recruitment. It is possible that this “pathway enforcement” provided by the cap prevents mRNA from entering aberrant PICs lacking the factors required for efficiently performing downstream steps such as scanning.
Results
Initiation Factor Interactions
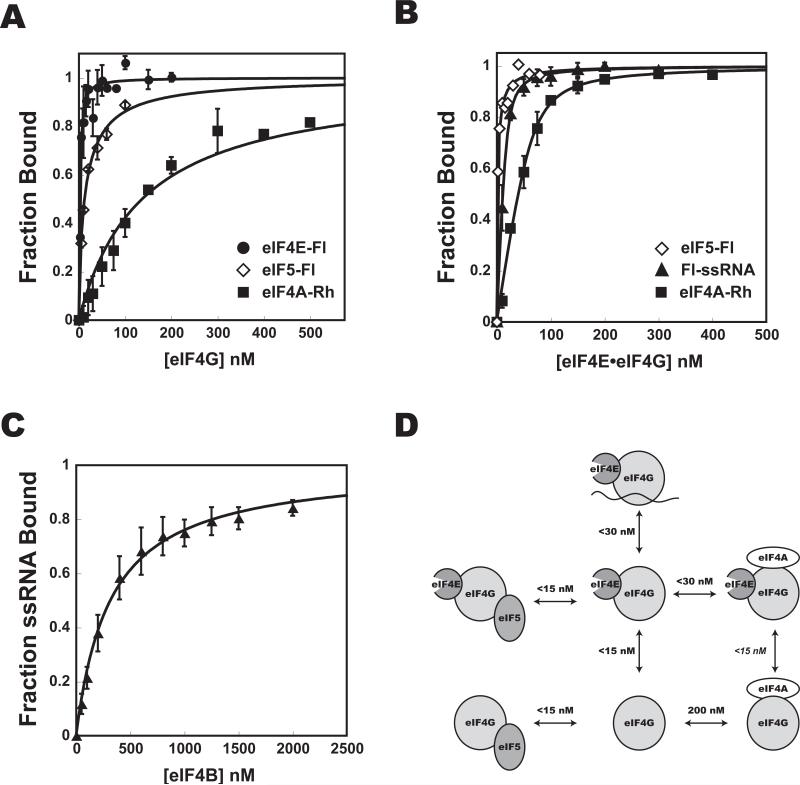
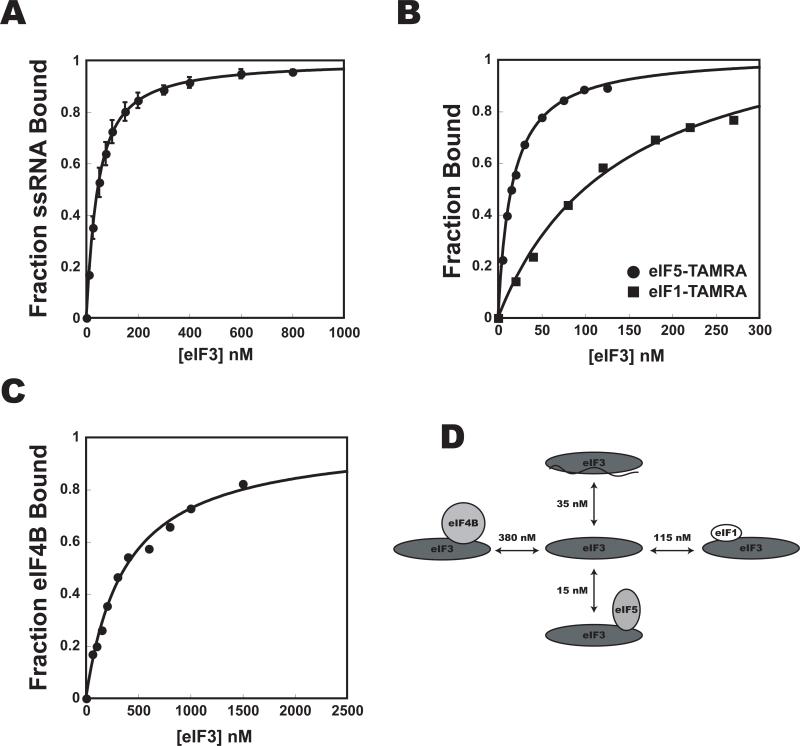
To lay the foundation for a dissection of the molecular mechanics of mRNA recruitment to the PIC, and to determine the concentrations of initiation factors required for complex formation, we began by investigating the interactions among the factors thought to be involved in the process. To do so, we purified the proteins implicated in mRNA recruitment (Fig. S1). Binding affinities were determined by measuring the fluorescence anisotropy of a labeled component while titrating an unlabeled component (Fig. 1, 2). Because of tight binding and the fact that 15-30 nM fluorescently-labeled protein or single-stranded RNA (ssRNA) was required to get sufficient signal, many of the measured Kds are upper limits.
Figure 1. Interaction affinities among eIF4 factors.
Fluorescence anisotropy of eIF4EFl (•), eIF5-Fl (◇), eIF4A-TAMRA (“eIF4A-Rh”;■) or Fl-ssRNA (◇ ) was measured upon addition of increasing concentrations of (A) eIF4G, (B) eIF4E•eIF4G complex, or (C) eIF4B. Kd values are shown in (D). The Kd for the interaction of eIF4E with eIF4A•eIF4G was not measured, but is deduced from the fact that it must be less than the Kd of eIF4E binding eIF4G in the absence of eIF4A. Error bars are average deviations. For all data with error bars n ≥ 2. See Fig. S1 for SDS-PAGE analysis of proteins used.
Figure 2. Interaction affinities between eIF3 and components of the multifactor and pre-initiation complexes.
The affinity of eIF3 for initiation factors and ssRNA was determined by monitoring the fluorescence anisotropy of (A) Fl-ssRNA (Kd = 35 ± 10 nM), (B) eIF1-TAMRA (■, Kd = 115 ± 55 nM), and eIF5-TAMRA (•, Kd = 15 ± 2 nM), and (C) eIF4B-Fl (Kd = 380 nM) as a function of eIF3 concentration. (D): Measured binding affinities. Error bars are average deviations, n ≥ 2.
In general, the data are in good agreement with previous qualitative reports of interactions among yeast initiation factors. Yeast eIF4E binds at least an order of magnitude more tightly to eIF4G1 (hereafter, eIF4G) than does eIF4A (Kds < 15 nM and 200 nM, respectively), consistent with the fact that eIF4E copurifies with eIF4G from yeast but eIF4A does not (Lanker et al., 1992). Interestingly, eIF4A binds more tightly to the eIF4E•eIF4G complex than to eIF4G alone, suggesting that the two factors might bind cooperatively, but this result could be due to the fact that binding of eIF4E induces increased folding of eIF4G (Gross et al., 2003; Hershey et al., 1999). Previous studies have determined that the affinity between mammalian eIF4A and eIF4G HEAT domains is in the μM range (12 μM for HEAT-1 in the absence of adenosine nucleotide, and 1 μM for HEAT-2 in the absence of nucleotide; our measurements were done in the absence of nucleotide; (Marintchev et al., 2009)). The difference in measured affinities could be due to the fact that fragments of eIF4G were used for the measurements with the mammalian factors or to differences between species.
Interactions between eIF3 and eIFs 1 and 5 that stabilize the multifactor complex (MFC) were also of high affinity (Fig. 2), as was the interaction between eIF4G and eIF5 (Asano et al., 2001). This interaction is of particular interest because it has been shown to be important for efficient mRNA and tRNA recruitment and has been suggested to link the yeast ribosome and the mRNA cap (Asano et al., 2001; He et al., 2003). While this role for eIF4G would be contrary to its dispensability in mRNA recruitment in vivo (Jivotovskaya et al.), it may be of importance for downstream events such as scanning or start codon recognition (He et al., 2003). No interaction between eIF3 and eIF4E•eIF4G could be detected, which, although in contrast to the known interaction between mammalian eIF3 and eIF4G (Korneeva et al., 2000), is consistent with previous work in yeast and with the fact that yeast eIF4G lacks the eIF3 binding domain (Asano et al., 2001).
eIF3 plays an essential role in mRNA recruitment
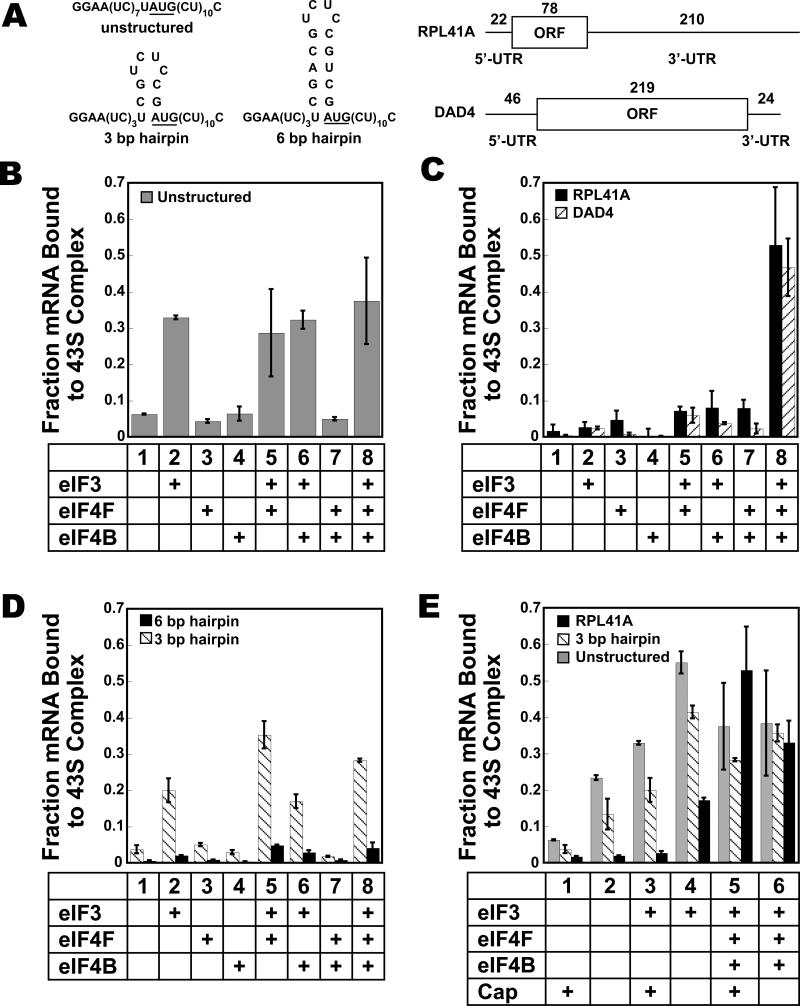
To follow mRNA recruitment, we used a native gel shift assay that can distinguish 43S•mRNA complexes from 43S•eIF3•mRNA complexes and factor-bound mRNAs (see supplementary materials and methods; Fig. S2). Using this assay, we asked which initiation factors are necessary to form a stable 43S•mRNA complex after 30 min of incubation. We first approached this question using an enzymatically capped (see materials and methods), unstructured, model mRNA containing a central AUG start codon (Fig. 3A, “cap-unstructured mRNA”). In all cases 40S ribosomal subunits, TC, eIF1 and eIF1A were included to form 43S complexes. In the absence of additional factors, under these conditions only 6% of cap-unstructured mRNA was stably enough bound to the 43S complex to be observed in the native gels (Fig. 3B, bar 1). Adding eIF3 raised the amount of cap-unstructured mRNA stably bound to the 43S complex by 5.5-fold (Fig. 3B, bar 2). eIF4F and eIF4B did not stimulate unstructured model mRNA recruitment either alone or in the presence of eIF3 (Fig. 3B). (Note that in all experiments using eIF4F there was an excess of eIF4A over eIFs 4E and 4G, mimicking the situation in vivo.) These results indicate that for small, capped, unstructured mRNAs, eIF3 alone is sufficient for maximal binding to the PIC in a manner stable enough to withstand electrophoresis. If the AUG codon was changed to CUC, no stable complex could be observed in the gel assay, regardless of whether eIF3 was present (Fig. S2), indicating that PICs bound to this mRNA locate the start codon and that this interaction is important for stability.
Figure 3. Factor dependence of stable mRNA recruitment to the 43S pre-initiation complex as determined using the native gel shift assay.
The mRNAs are shown in (A). (B) Recruitment of cap-unstructured mRNA; (C) recruitment of cap-RPL41A (black) and cap-DAD4 (striped); (D) recruitment of cap-3 bp (striped) and cap-6 bp (black); and (E) recruitment of capped and uncapped, unstructured mRNA (gray), 3 bp (striped) and RPL41A (black). 30 min points are shown. Error bars are average deviations, n ≥ 2. See Fig. S2 for additional information.
The non-essential, weakly associated eIF3 subunit Hcr1p/eIF3j, which is not visibly present in our eIF3 preparations, inhibited recruitment of mRNA to the PIC (Fig. S2). These data are consistent with the reported negative cooperativity between mammalian eIF3j and mRNA in binding to the 40S subunit (Fraser et al., 2007).
We next investigated the factor requirements for recruitment of real mRNAs to the PIC. Two natural yeast mRNAs were used, DAD4 and RPL41A (Fig. 3A, “cap-RPL41A” and “cap-DAD4”). DAD4 encodes a subunit of the Dam1 complex involved in chromosome segregation and RPL41A encodes a ribosomal protein. RPL41A has been used previously to study mRNA recruitment in vivo in yeast (Jivotovskaya et al., 2006). The 5′-UTR of DAD4 is 46 bases long, only slightly shorter than the average of 52 bases for yeast 5′-UTRs (Cigan and Donahue, 1987), and is predicted to contain two hairpin structures, one of which includes the AUG (Fig. S2). The 5′-UTR of RPL41A is shorter, 22 nucleotides, and is predicted to contain several base-paired regions (Fig. S2).
In the absence of additional factors, neither of these capped mRNAs bound to the 43S complex (Fig. 3C, lane 1). Unlike the case for the model mRNA, addition of eIF3 alone did not increase recruitment of the capped, natural mRNAs (Fig. 3C, lane 2). Similarly, eIF4F and eIF4B did not significantly enhance mRNA recruitment to the PIC either alone, together, or when either factor was combined singly with eIF3 (Fig. 3C, lanes 3-7). However, the combination of eIF3, eIF4F and eIF4B led to a significant stimulation of stable mRNA recruitment to the PIC (Fig. 3C, lane 8). Under these conditions, the natural mRNA was recruited to a level even greater than that of the unstructured model mRNA in the presence of eIF3 (Fig. 3B).
Overall, the factor dependence of recruitment of capped, natural mRNAs is consistent with previous observations in mammalian systems (Benne and Hershey, 1978; Dmitriev et al., 2003; Pestova et al., 1998; Pestova and Kolupaeva, 2002; Trachsel et al., 1977). The important role played by eIF4B is somewhat surprising given the fact that the gene encoding eIF4B is not essential in yeast (Altmann et al., 1993). However, the eIF4B null strain does have a severe growth defect as well as temperature sensitive phenotypes, and mammalian eIF4B has been shown to play a vital role in the translation of mRNAs with structured 5′-UTRs (Dmitriev et al., 2003).
Data described below indicate that these effects of eIF3 and the eIF4 factors on the amount of mRNA bound to the PIC after 30 minutes are due to increases in both the rate and final extent of recruitment.
mRNA structure inhibits recruitment
In order to investigate the origins of the difference in factor requirements between unstructured model mRNA and natural messages, we made two model mRNAs with hairpin structures. If the differences in factor requirements arose solely from the presence of structure in the natural mRNA, we expected that these capped, model mRNAs with three or six base pair hairpins covering the A of the AUG (“cap-3 bp” and “cap-6 bp”; Fig. 3A) would have factor requirements similar to those observed for recruitment of natural mRNA. As expected, in the absence of factors, these structures inhibited stable mRNA recruitment to the 43S complex (Fig. 3D, lane 1). Cap-3 bp was stimulated to bind to the PIC by eIF3, though not to the same extent as cap-unstructured mRNA (compare lanes 2 in Fig. 3B and 3D). Unlike the case for the unstructured mRNA, the addition of eIF4F increased stable recruitment of cap-3 bp approximately two-fold when eIF3 was present (Fig. 3D, lane 5), bringing it to the level of cap-unstructured mRNA recruited in the presence of eIF3 alone (Fig. 3B, lane 2). As with natural mRNA, eIF4F and eIF4B did not promote recruitment of the 3 bp mRNA in the absence of eIF3. In contrast to recruitment of the natural mRNA, eIF4B did not increase the amount of mRNA bound when eIF3 and eIF4F were also present (Fig. 3D, lane 8).
The hairpin structure in cap-6 bp reduced mRNA recruitment to low levels in all cases. What little mRNA recruitment was observed was similar to that for the cap-3 bp in that the greatest amount was bound in the presence of both eIF3 and eIF4F (Fig. 3D, lane 5, black bars). This severe reduction could be due to the placement of the hairpin, which includes the A of the start codon and is only 10 bases from the 5′-end of the mRNA. Although there are differences in the reported effects of the location of mRNA structures in yeast and mammals, both of these positions could increase interference with PIC binding (Kozak, 1989; Lawson et al., 1986; Sagliocco et al., 1993; Vega Laso et al., 1993). In addition, the stability of the 6 bp hairpin is significantly higher than those of the 3 bp hairpin and the structures in RPL41A and DAD4 (ΔG is -0.4 and -7.4 kcal/mol for the 3 and 6 bp hairpins, respectively, and between -2 and -4 kcal/mol per structure for the natural mRNAs; an alternative structure exists for the 3 bp hairpin RNA with a ΔG of -1.4 kcal/mol; Fig. S2). Decreasing the concentration of magnesium in the reactions to 2 or 1 mM did not increase the amount of cap-6 bp that bound the PIC or change the factor requirements (Fig. S2), indicating that 3 mM Mg2+ did not artificially over-stabilize this structure. Overall, the presence of structures in the model and natural mRNAs correlate with the ability of eIF4F to promote their recruitment to the PIC, consistent with a role for this factor in removing structure.
The 5′-7-methylguanosine cap structure blocks natural mRNA recruitment to the PIC in the absence of a complete set of initiation factors
As eIF4F plays an important role in the recruitment of structured model and natural mRNAs to the PIC, we wondered what the contribution of the 5′-cap structure is to this stimulation. We therefore repeated the investigation into factor dependence with uncapped (5′-monophosphate) mRNA. In the absence of any additional factors, the presence of a cap structure had the surprising effect of reducing the amount of structured and unstructured model mRNAs and DAD4 mRNA recruited to the 43S PIC (Fig. 3E, lanes 1-2; Fig. S2). This inhibitory effect of the cap was also seen with both types of model mRNAs as well as with both natural mRNAs in the presence of eIF3 alone (Fig. 3E, lanes 3-4; Fig. S2). Recruitment of uncapped DAD4 mRNA (“uncap-DAD4”) was stimulated by eIF3 to an even greater extent than uncapped RPL41A mRNA (“uncap-RPL41A”; fraction bound of 0.5; Fig. S2). These results suggest that the cap structure destabilizes binding of mRNA to both the 43S complex and the 43S•eIF3 complex. Because the cap should be outside of the footprint of the ribosome when the PIC is located at the start codon, these results imply that the PIC interacts with the 5′-end of the mRNA during recruitment in the absence of the cap-binding complex eIF4F. For the model mRNAs, the addition of eIF4F and eIF4B resulted in approximately the same amount of structured or unstructured mRNA recruited to the 43S complex regardless of the presence of a cap structure (Fig. 3E, lanes 5-6, shaded and striped bars). For RPL41A, the addition of eIF4F and eIF4B not only alleviated the inhibition introduced by the 5′-cap, it further promoted recruitment beyond that in the absence of the cap structure (Fig. 3E, lanes 5-6, black bars). The data also indicate that the cap imposes a requirement for the full set of recruitment factors for stable binding of natural mRNAs to the PIC, a requirement that does not exist in the absence of the cap.
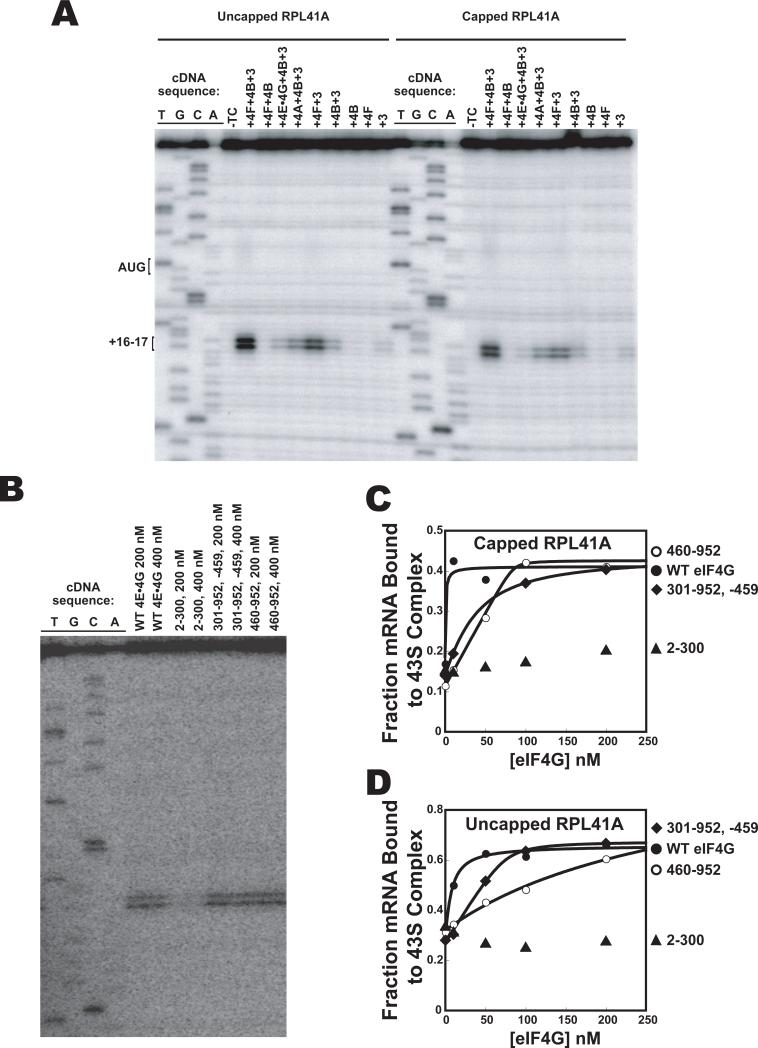
Although the mRNA gel shift assay demonstrates that PICs are bound to the mRNA it does not provide any information about the location of the complexes. To assess the location we used a primer extension assay (“toeprinting”) to locate the position of the ribosome on cap-RPL41A (Hartz et al., 1988). We found that, consistent with work in the mammalian system (Pestova et al., 1998), a TC-dependent stop in primer extension (a toeprint) was observed 16-17 bases downstream of the AUG start codon in the presence of eIFs 1, 1A, 3, 4F and 4B (Fig. 4). In the absence of any of the eIF4 factors a large reduction in toeprint was observed, and leaving out eIF3 eliminated the toeprint entirely. These data confirm that the PICs are capable of locating the start codon and forming a stable complex at this position. Similar results were obtained for capped and uncapped RPL41A (Fig. 4A), consistent with toeprinting results on uncapped mRNAs in mammalian systems (Pestova and Kolupaeva, 2002); phosphatase treating the uncapped mRNA to remove 5′-terminal phosphates did not affect the toeprinting results (Fig. S3; see below). The fact that in the presence of eIF3 alone a significant amount of recruitment of uncapped mRNA to the PIC is observed in the gel shift assay but a toeprint is barely detectable suggests that these complexes are either unable to efficiently locate the start codon or are not stably bound once they find it. This conclusion is consistent with proposed roles for the cap structure and eIF4F factors in scanning and start codon identification (He et al., 2003; Jivotovskaya et al., 2006; Preiss and Hentze, 1998) and with results in mammalian systems showing that eIF4 factors are required for translation of uncapped mRNAs (Ohlmann et al., 1995; Thoma et al., 2004).
Figure 4. Recruitment of uncapped mRNA to the PIC forms complexes unable to stably locate the start codon in the absence of the full complement of factors.
(A) Primer-extension (toeprinting) with uncapped (left) and capped (right) RPL41A mRNA. The first four lanes are dideoxy sequencing lanes. The fifth lane contains 40S ribosomal subunits, eIF1 and eIF1A but no TC. The remaining lanes contain 40S ribosomal subunits, eIF1, eIF1A and TC as well as additional factors as indicated. (B) Toeprinting with uncapped RPL41A mRNA and various fragments of eIF4G. All reactions contain the 43S complex, eIF3, eIF4A and eIF4B. (C) Recruitment of capped RPL41A mRNA to the PIC, monitored using a gel shift assay in the presence of eIF3, eIF4A, eIF4B and the following eIF4G fragments: WT eIF4G and eIF4E (•); 460-952 (○); 301-952 -459 (◇); and 2-300 (◇ ). (D) As in (C), but with uncapped RPL41A mRNA. The presence of eIF4E with the eIF4G fragments did not alter the results for the toeprint or gel shift assays (data not shown).
The C-terminal domain of eIF4G is sufficient to promote mRNA recruitment to the PIC
Yeast eIF4G consists of two major domains. The N-terminal domain binds to eIF4E, PABP and RNA and the C-terminal domain contains RNA and eIF4A binding sites (Lamphear et al., 1995; Tarun and Sachs, 1996). Mammalian eIF4G is larger and contains an eIF3-binding domain and a second eIF4A binding site, both absent from the yeast factor (Korneeva et al., 2000). The C-terminal region promotes translation of uncapped mRNAs in mammalian systems (De Gregorio et al., 1998; Ohlmann et al., 1995; Thoma et al., 2004). We sought to test this activity in the yeast system, as well as to determine which domains of eIF4G are required to promote mRNA recruitment in general. To do so we created three fragments of eIF4G: 460-952 removes the N-terminal regions and does not have PABP or eIF4E binding sites (Hershey et al., 1999; Mader et al., 1995); 301-952, -459 does not have a PABP binding site and has a mutation that blocks interaction with eIF4E (Tarun and Sachs, 1997); and 2-300 removes the C-terminal RNA, eIF4E and eIF4A bindings sites and contains the PABP binding site and an RNA binding site (Berset et al., 2003). We tested the ability of these variants to promote the binding of a PIC at the start codon of capped and uncapped RPL41A mRNA. In the presence of eIFs 3, 4A and 4B, the N-terminal portion of eIF4G (2-300) did not promote a toeprint on either uncapped (Fig. 4B) or capped (not shown) RPL41A. In contrast, both truncations containing the C-terminal domain (460-952; 301-952, -459) produced toeprints similar to that seen with the WT protein on both capped and uncapped mRNA (Fig. 4B and data not shown). The 460-952 fragment did not promote a toeprint in the presence of only eIF3 beyond the weak one observed with eIF3 alone, indicating that eIF4A and eIF4B are still required for functional complex formation with the fragment (Fig. S3).
Similar results were obtained in the mRNA gel-shift assay with both capped and uncapped mRNA (Fig. 4C,D). Interestingly, the concentrations of the 460-952 and 301-952, -459 fragments required to achieve maximal complex formation were higher than needed with WT eIF4G, suggesting that the N-terminal domain increases the affinity of eIF4G for the PIC and/or mRNA (Fig. 4C,D, compare diamonds and open circles with closed circles). Overall, these data indicate that the C-terminal region of eIF4G, after the eIF4E binding site, contains the structures that are functionally important for recruitment of mRNA, independent of interaction with the cap, consistent with previous observations in mammalian systems (Ali et al., 2001; De Gregorio et al., 1998; Ohlmann et al., 1995; Thoma et al., 2004).
Recruitment factors accelerate mRNA binding to the PIC
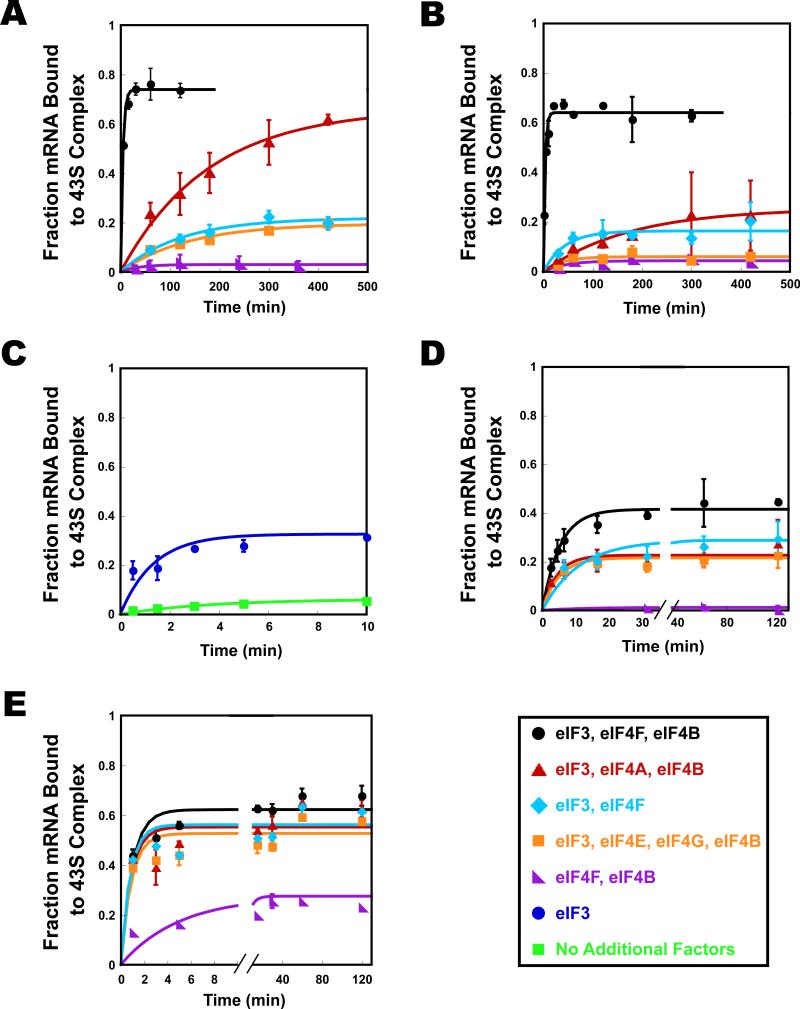
We next measured the kinetics of mRNA recruitment under various conditions. In the presence of eIF3, eIF4F and eIF4B, cap-RPL41A was recruited to the PIC with an observed rate constant (kobs) of 0.25 min-1 and an endpoint of 0.71 (fraction mRNA bound to the PIC; Fig. 5A, black). This rate is significantly slower than in vivo rates of initiation on ovalbumin mRNA measured in chicken oviduct cells (~10 min-1) (Palmiter, 1975). The relatively slow rate in the in vitro system is at least partially due to the low concentrations of 40S subunits and mRNA used, 30 nM and 15 nM respectively, in contrast to the ~10 μM 40S ribosomal subunits in a yeast cell (Von Der Haar and McCarthy, 2002). The kobs for mRNA recruitment in our system changed linearly with 40S subunit concentration (data not shown), and if it continues to scale linearly to 10 μM 40S subunits the extrapolated kobs would be ~80 min-1, well within a kinetically competent range to support initiation in vivo.
Figure 5. Kinetics of stable recruitment of mRNA to the 43S pre-initiation complex.
The fraction of mRNA recruited to the 43S pre-initiation complex was monitored at the times indicated for: cap-RPL41A (A); cap-DAD4 (B); uncap-3 bp (C); uncap-RPL41A (D); and uncap-DAD4 (E). Structures of mRNAs are shown in Fig. 3A. (A) For cap-RPL41A kobs were determined with eIF3, eIF4A, eIF4E•eIF4G and eIF4B (hereafter, “all recruitment factors;” black; kobs = 0.25 ± 0.03 min-1); and in the absence of the following factors: eIF4E•eIF4G (red; kobs = 0.012 ± 0.008 min-1); eIF4B (aqua; kobs = 0.012 ± 0.01 min-1); eIF4A (orange; kobs = 0.009 ± 0.003 min-1); and eIF3 (purple; no recruitment observed). (B) For cap-DAD4 observed rate constants were determined in the presence of all recruitment factors (black; kobs = 0.09 ± 0.02 min-1); and in the absence of the following factors: eIF4E•eIF4G (red; kobs = 0.02 ± 0.01 min-1); eIF4B (aqua; kobs = 0.024 ± 0.03 min-1); eIF4A (orange; kobs = 0.03 ± 0.02 min-1); and eIF3 (purple; no recruitment observed). (C) For uncap-3 bp, observed rate constants were determined in the presence (blue; kobs = 0.83 ± 0.1 min-1) and absence (green; kobs = 0.28 ± 0.05 min-1) of eIF3. Kinetics were followed for a total of 60 minutes. A shorter period is shown here for clarity. (D) For uncap-RPL41A observed rate constants were determined in the presence of all recruitment factors (black; kobs = 0.22 ± 0.02 min-1); and in the absence of the following factors: eIF4E•eIF4G (red; kobs = 0.24 ± 0.04 min-1); eIF4B (aqua; kobs ≥ 0.13 ± 0.06 min-1); eIF4A (orange; kobs ≥ 0.3 ± 0.15 min-1); and eIF3 (purple; no recruitment observed). Kinetics were followed for a total of 500 minutes. A shorter period is shown here for clarity. (E) For uncap-DAD4 observed rate constants were determined in the presence of all recruitment factors (black; kobs = 1.0 ± 0.3 min-1); and in the absence of the following factors: eIF4E•eIF4G (red; kobs ≥ 1.2 ± 0.1 min-1); eIF4B (aqua; kobs ≥ 0.8 ± 0.05min-1); eIF4A (orange; kobs ≥ 0.9 ± 0.5 min-1); and eIF3 (purple; kobs = 0.2 ± 0.05 min-1). Errors are average deviations, n ≥ 2.
In the absence of the eIF4E•eIF4G subcomplex, kobs was reduced 20-fold, to 0.012 min-1, but the endpoint was essentially unchanged (0.66, Fig. 5A, red). In contrast, omitting either eIF4A or eIF4B (Fig. 5A, orange and aqua, respectively) led to a decrease in both the rate of mRNA recruitment (0.009 min-1 and 0.012 min-1, respectively) and the endpoint (0.21 and 0.24). In the absence of eIF3, cap-RPL41A did not bind to the PIC even at very long times (Fig. 5A, purple) indicating that this factor is essential for recruitment of natural, capped mRNA, and actually plays a more integral role than the eIF4F factors, which are most commonly identified with mRNA recruitment. The effects of the factors on the kinetics of mRNA recruitment correlate well with the effects observed in the toeprinting assay (Figs. 4A and S3).
To explore differences among mRNAs in the rate of recruitment to the PIC, we also tested cap-DAD4. In the presence of eIF3, eIF4F and eIF4B, this mRNA bound to the PIC with a rate constant similar to that for cap-RPL41A (Fig. 5B, black; kobs of 0.32 min-1). As with cap-RPL41A, leaving out eIF4E•eIF4G, eIF4A or eIF4B reduced the rate of mRNA recruitment (10-16-fold; Fig. 5B). The effect of eIF4B on the endpoint of recruitment was similar to what was seen with cap-RPL41A (Fig. 5B). Whereas the endpoint of cap-RPL41A recruitment was not affected by eIF4E•eIF4G, cap-DAD4 binding had an endpoint defect in the absence of this subcomplex. The effect of eIF4A was also larger with cap-DAD4 than with cap-RPL41A, suggesting DAD4 may be a “weaker” mRNA, requiring more unwinding of structure than does RPL41A. This is consistent with the fact that DAD4's 5′-UTR is twice as long as that of RPL41A (46 vs. 22 nt) and that the start codon is predicted to be in a more structured region for DAD4 than for RPL41A (Fig. S2). Once again, eIF3 was required for observable mRNA recruitment, even at long times.
These results demonstrate an essential role for eIF3 in the recruitment of capped mRNAs. As mentioned above, some uncapped mRNAs bind the 43S complex in the absence of eIF3 (Fig. 3E). To determine if eIF3 stimulates the kinetics of mRNA recruitment as well as the endpoint, we measured the rate of recruitment of uncapped 3 bp mRNA (“uncap-3 bp”; Fig. 3A) in the presence and absence of eIF3 (Fig. 5C). In the presence of eIF3, this mRNA was recruited to the PIC with a kobs ≥ 0.83 min-1 (Fig. 5C, blue). In the absence of eIF3, kobs was reduced to 0.28 min-1 (Fig. 5C, green). Thus eIF3 increases the rate of recruitment of this mRNA ≥3-fold in addition to a >5-fold enhancement of the endpoint of recruitment (0.34 vs. 0.064). These data indicate that eIF3 plays a role in both enhancing the rate of mRNA binding to the PIC and stabilizing the bound complex.
The 5′-7-Methylguanosine cap reduces the rate of mRNA recruitment in the absence of the eIF4 factors
To further explore the role of the mRNA cap, we determined the kinetics of uncap-RPL41A and uncap-DAD4 recruitment to the PIC (Fig. 5D,E). In the presence of eIF3, eIF4F and eIF4B, kobs for uncap-RPL41A binding was 0.22 min-1, similar to the rate constant for cap-RPL41A (0.25 min-1), indicating that the cap structure primarily increases the endpoint of recruitment (~2-fold; Fig. 5A vs. 5D). Strikingly, in contrast to the situation for cap-RPL41A, where omitting eIF4G•eIF4E, eIF4A or eIF4B had a large (~20-fold) effect on the kinetics of recruitment, leaving out these factors with uncap-RPL41A had little effect on the kinetics (0.24, 0.3 and 0.13 min-1, respectively, compared to 0.22 min-1 with all of the factors). The endpoint of recruitment of uncap-RPL41A was reduced ~2-fold by omission of any of the eIF4 factors. For uncap-DAD4 rates were similarly unchanged in the absence of eIF4G•eIF4E, eIF4A or eIF4B. In addition, for uncap-DAD4 the endpoints were virtually unchanged in the absence of any one of these factors. In the absence of eIF3, the endpoint was reduced approximately 3-fold and the rate of recruitment was reduced 5-fold. Taken together, our data indicate that the 5′-cap structure slows the recruitment of natural mRNAs by over an order of magnitude in the absence of the eIF4 factors. This kinetic inhibition is alleviated by the presence of eIF4F, eIF4A and eIF4B.
eIF3 directly enhances mRNA recruitment to the PIC
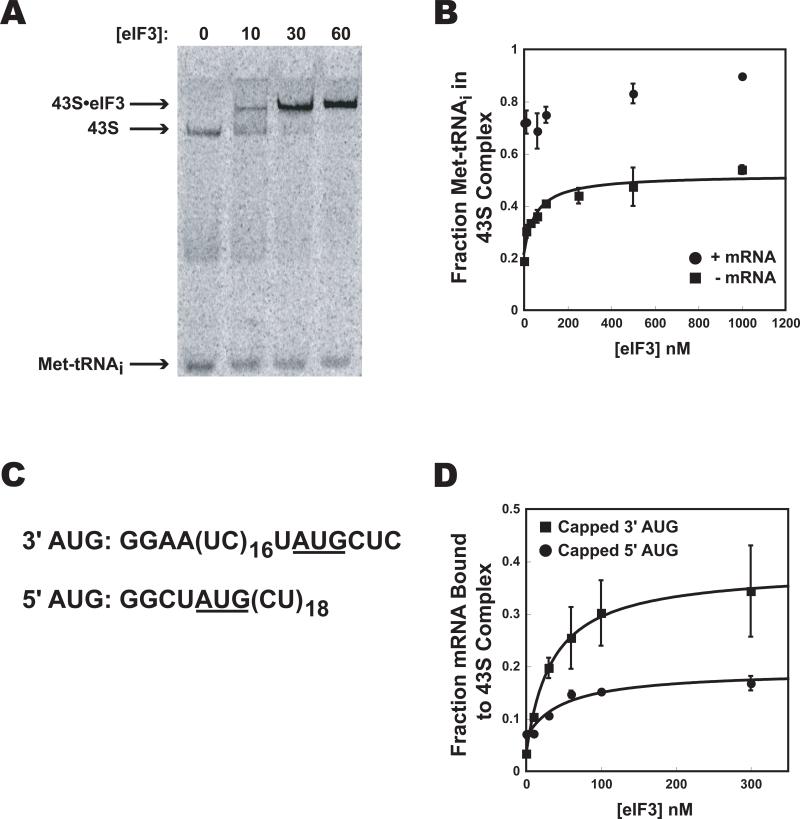
In mammalian systems eIF3 is important for stable, productive binding of the ternary complex to the 40S ribosomal subunit as well as for stable binding of mRNA to the 43S PIC (Benne and Hershey, 1978; Chaudhuri et al., 1999; Trachsel et al., 1977). Because of this, it was difficult to conclude that effects of eIF3 on mRNA recruitment were direct effects rather than indirect ones via promotion of TC binding. In yeast systems, eIF3 is not required for productive binding of TC to the ribosome, although it enhances binding 2-3-fold in vivo (Acker et al., 2007; Algire et al., 2002; Jivotovskaya et al., 2006).
To further investigate the role of eIF3, we quantified its effect on TC recruitment to the yeast 40S subunit. Yeast eIF3 reduced the mobility of pre-formed 43S complex in the native gel shift assay, and bound to this complex tightly, with a Kd of < 30 nM (Fig. 6A). The presence of eIF3 increased the amount of TC bound to 40S subunits in the absence of mRNA (with saturating eIFs 1 and 1A) by 2.5-fold at saturation (Fig. 6B). However, when the AUG-containing unstructured model mRNA was present, eIF3 had only a slight stimulatory effect (~20%; Fig. 6B). These relatively small effects are not consistent with the very large effects of eIF3 on mRNA recruitment to the PIC, and argue strongly against the possibility that eIF3 promotes mRNA recruitment indirectly via enhancement of TC binding. Consistent with this conclusion, in the mRNA recruitment experiments, TC was at a saturating level even in the absence of eIF3, and a five-fold increase in the concentration of TC did not increase the amount of mRNA bound to the PIC in either the absence or presence of eIF3 (data not shown). Addition of eIF5, which binds eIF2 and eIF3 and enhances TC binding to the PIC, also did not increase TC binding or mRNA recruitment, consistent with saturating TC levels in the system.
Figure 6. eIF3 interactions with the PIC.
(A): Reduction in mobility of the 43S complex in native gels caused by eIF3 binding, monitored using 35S-Met-tRNAi. (B): In the absence of mRNA, eIF3 enhances TC binding to 40S subunits (with eIF1 and eIF1A) by ~2.5-fold (■). In the presence of the unstructured, model mRNA (•) 43S complex formation is only slightly enhanced by the presence of eIF3 (<1.3 fold). (C): The sequences of mRNAs used in (D). (D): The effect of eIF3 on binding of capped versions of the two mRNAs shown in (C) to the PIC: 3′-AUG mRNA (■); 5′-AUG mRNA (•). Error bars are average deviations, n ≥ 2.
It has been suggested that eIF3 extends both ends of the mRNA binding channel of the 40S subunit, stabilizing ssRNA association (Kolupaeva et al., 2005; Pisarev et al., 2008; Szamecz et al., 2008). To test these proposals, we used the two unstructured, model mRNAs shown in Figure 6C. These mRNAs are nearly identical, except one has the AUG 3 bases from the 3′-end (“3′-AUG”) and the other has the AUG 4 bases from the 5′-end (“5′-AUG”). As shown in Figure 6D, eIF3 strongly (12-fold) promoted recruitment of capped-3′-AUG mRNA to the 43S PIC, but only weakly (2-fold) promoted recruitment of capped-5′-AUG. The ability of eIF3 to promote recruitment of 3′-AUG mRNA containing the 5′-extension is consistent with eIF3 interacting with mRNA at the exit of the mRNA channel, as previously suggested (Kolupaeva et al., 2005; Pisarev et al., 2008). Because eIF3 also weakly promotes recruitment of 5′-AUG mRNA, which has only a 4 base 5′-extension, not enough to extend out of the exit channel of the 40S subunit when the AUG codon is in the P site (Kozak, 1983; Pisarev et al., 2008), the data are also consistent with an interaction at the entrance of the mRNA channel as previously indicated by genetic data (Szamecz et al., 2008). However, it is also possible that a conformational change in the PIC induced by eIF3 is partly or entirely responsible for these stabilizing effects.
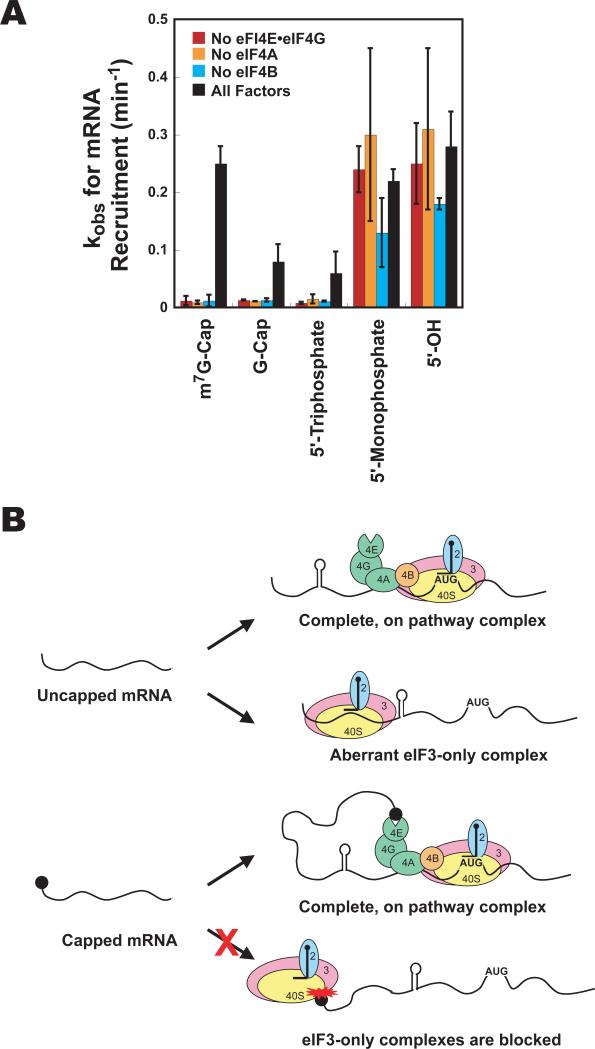
The triphosphate portion of the cap structure is responsible for inhibiting mRNA recruitment
The cap consists of 7-methylguanosine attached via a 5′-5′ triphosphate linkage to the mRNA. To probe how the cap inhibits mRNA recruitment in the absence of the eIF4 factors, we dissected the effect of the components of the structure, investigating recruitment of mRNA with a 5′-G-cap (no methyl group), 5′-triphosphate or 5′-OH. The factor dependence of mRNA recruitment for the G-cap was much like that for the m7G-cap (Fig. 7A). Recruitment was slow in the absence of any single factor, but accelerated in the presence of all factors (eIF4F, eIF4B and eIF3). The stimulation afforded by the complete set of factors is smaller with the G-cap than with the m7G-cap (kobs = 0.08 min-1 vs. 0.25 min-1 for m7G-cap), consistent with the importance of the 7-methyl group for recognition by eIF4E (Marcotrigiano et al., 1997). The 5′-triphosphate also resulted in slow recruitment in the absence of any individual factor (Fig. 7A). Like the G-cap, having the triphosphate alone reduced the ability of the eIF4 factors to increase the rate of mRNA recruitment (kobs = 0.06 min-1). mRNA with no phosphates at the 5′-end (5′-OH) behaved like the monophosphorylated mRNA used in the assays described above. Recruitment was fast regardless of the presence or absence of eIF4 components (Fig. 7A). Despite the efficient binding of mRNA with a 5’-OH to the PIC in the absence of eIF4 factors, only a very weak toeprint was observed under these conditions, indicating that the complexes are unable to scan the mRNA and/or stably interact with the start codon (Fig. S3). As with m7G-capped and triphosphate-capped mRNA, however, a strong toeprint is generated when all the recruitment factors are present. Together, these results indicate that the triphosphate linkage is the portion of the cap structure that inhibits mRNA recruitment in the absence of the full set of factors.
Figure 7. The 5’-5’-triphosphate moiety blocks aberrant mRNA recruitment.
(A) Observed rate constants (kobs) for recruitment to the PIC of mRNAs with various 5′-end structures in the presence of all recruitment factors (black bars) and absence of eIF4E•eIF4G (red bars), eIF4A (orange bars) or eIF4B (aqua bars). Measurements were performed as in Fig. 5. Errors are average deviations, n ≥ 2. (B) Cartoon depicting recruitment of uncapped (top) and capped (bottom) natural mRNAs. Interaction between the cap and the PIC blocks mRNA recruitment and the ability of eIF3 on its own to promote rapid and stable recruitment. Interaction between the cap and the eIF4F complex removes this inhibition and allows productive mRNA binding to the PIC. See Fig. S3 for additional information.
Discussion
eIF3 Functions Directly in mRNA Recruitment
We found that eIF3 is required for binding of mRNA to the PIC, independent of its enhancement of TC binding. Our data indicate that eIF3 increases both the rate and extent of mRNA recruitment. The importance of eIF3 is consistent with recent in vivo studies in yeast (Jivotovskaya et al., 2006) in which a degron system was used to deplete eIF3 or eIF4G. Upon degradation of eIF3, binding of RPL41A mRNA to 43S complexes was dramatically reduced, whereas loss of eIF4G led to RPL41A mRNA accumulation in 43S PICs, which were unable to progress to 80S complexes. These data suggested that eIF3 is critical for mRNA recruitment to the PIC and that eIF4G plays an important role downstream of this event, potentially in scanning. Such an interpretation is consistent with our findings that eIF3 is critical for mRNA recruitment, while eIF4G has a smaller effect on the extent of recruitment but dramatically stimulates the kinetics of this event. Our data further support the importance of eIF3 in mRNA recruitment outside of a role in stabilizing TC binding, as the defect seen in TC binding in the eIF3 degron assay could have been partially responsible for the reduction in mRNA recruitment (Jivotovskaya et al., 2006). The effect of eIF4G on the observed rate of mRNA recruitment in vitro could come, in part, from a role in increasing the efficiency or processivity of scanning. Alternatively, a kinetic effect of eIF4G on mRNA binding itself could have been masked in the in vivo studies, which observed steady-state levels of mRNA in the PIC after degradation of the factor; at steady-state mRNA accumulation in the 43S PIC would be increased by the block in progressing to 80S.
The 5′-7-methylguanosine cap blocks an aberrant mRNA recruitment pathway
An unexpected function of eIF3 observed during the course of this work is its ability to promote recruitment of uncapped, natural mRNA to the ribosome in the absence of the eIF4 factors. This effect is in contrast to the behavior of capped, natural mRNAs, which require eIF4E•eIF4G, eIF4A and eIF4B in addition to eIF3 for stable and rapid recruitment to the PIC. Although it is known that uncapped mRNAs can be translated in vitro (Andreev et al., 2009; Bergmann et al., 1979; De Gregorio et al., 1998; Muthukrishnan et al., 1976; Ohlmann et al., 1995; Preiss and Hentze, 1998; Sonenberg and Shatkin, 1978; Tarun and Sachs, 1995) and there is some evidence that they can also be translated in vivo (Andreev et al., 2009; Gunnery et al., 1997; Lo et al., 1998; Preiss and Hentze, 1998) the cap is clearly stimulatory in both situations.
Here, we have shown that uncapped mRNAs can be recruited to the PIC by eIF3 alone, but the resulting complexes are unable to effectively locate the start codon (Fig. 7B). eIF3 alone did not stimulate recruitment of capped, natural mRNA. The cap structure is well known to stimulate translation in the presence of the full set of factors, but our data indicate that it actually inhibits mRNA recruitment when the eIF4F complex is not available. Our data showed that even in the absence of all the mRNA recruitment factors, the cap structure inhibits mRNA binding to the PIC (Fig. 3E, columns 1 and 2), indicating that the cap interacts negatively with the PIC itself. The observation that eIF3 on its own is able to promote recruitment of uncapped but not capped natural mRNAs suggests that the cap prevents the mRNA from interacting with the PIC in the proper conformation for eIF3 to promote its binding.
The cap decreases both the intrinsic rate and extent of mRNA recruitment to the PIC. The presence of the eIF4 group of factors relieves the kinetic inhibition of mRNA recruitment imposed by the 5′-cap structure, allowing capped mRNA to be recruited at the same rate as uncapped mRNA. These factors appear to play roles both as enhancers of mRNA recruitment and relievers of the inhibition caused by the cap. One model to explain these observations is that the interaction of the cap structure with eIF4F moves the mRNA such that it no longer makes inhibitory contacts with the PIC and can now interact productively with eIF3.
The slow rate of recruitment in the absence of the full set of factors is also observed for mRNAs with a 5′-G-cap or a 5′-triphosphate. Thus inhibition of recruitment correlates with the presence of a 5′-triphosphate moiety. It is possible that the high negative charge density of the 5′-5′-triphosphate linkage of the cap electrostatically inhibits aberrant entry of the mRNA into the 40S subunit's mRNA binding channel. These data are consistent with the proposal by Hentze and colleagues that the 5′-end of uncapped mRNAs must thread through the mRNA entry channel in order to bind the 40S subunit (De Gregorio et al., 1998). Threading might be electrostatically blocked by the 5′-triphosphate group of the cap and the full set of mRNA recruitment factors could act to promote binding of mRNAs via a different mode, for example by inducing a conformational change in the PIC that opens the mRNA binding channel so that the mRNA can be inserted without threading. This role for the 5′-5′-triphosphate moiety of the cap in preventing inappropriate binding of mRNA to the PIC could explain why this unusual structure was incorporated into the cap during evolution.
Although the eIF4F-independent pathway for uncapped, natural mRNA recruitment to the PIC is nearly as efficient as the recruitment of capped mRNA in the presence of eIF4F and eIF4B, it does not appear to lead to productive complexes in their absence. In fact, the absence of a significant toeprint on uncapped mRNA in the absence of the eIF4 factors (Figs. 4 and S3) suggests that this alternative pathway leads to complexes that either inefficiently locate the start codon or are easily displaced by reverse-transcriptase once they do. Inhibition of mRNA recruitment by the 5′-cap structure in the absence of eIF4F may be a mechanism to prevent formation of aberrant ribosomal complexes when the full complement of initiation factors is not available. The ability of the cap structure to inhibit formation of aberrant initiation complexes may help guarantee proper scanning and initiation codon recognition. It has been suggested that eIF4G is important for scanning the mRNA (de Breyne et al., 2009; Jivotovskaya et al., 2006; Pestova and Kolupaeva, 2002; Poyry et al., 2004) and perhaps even plays a role in start site selection (He et al., 2003). The cap has also been implicated in proper start codon recognition (Preiss and Hentze, 1998). Taken together with these observations, our data suggest that the cap acts in “pathway enforcement,” to ensure the presence of eIF4G and other factors required for efficient production of the correct protein. This enforcement of the presence of all of the factors also allows for more effective control by the numerous pathways that target eIF4F to modify the translating pool of mRNAs.
Materials and Methods
Protein and Ribosome Purification
Initiation factors were expressed and purified using standard methods and published procedures (Acker et al., 2007; detailed in Supplementary Materials). 40S ribosomal subunits and other proteins were purified as previously described with minor modifications (Supplementary Materials; Acker et al., 2007; Cong and Shuman, 1995; Neff and Sachs, 1999; Tarun and Sachs, 1997). Purified proteins are shown in Fig. S1 or in (Acker et al., 2007).
RNA Preparation
RNA was transcribed and tRNA was charged as described (Acker et al., 2007). Body-labeled mRNAs (mRNAs with 5′ triphospate or no phosphates) were transcribed with 500 μCi of α-[32P]-CTP per mL of reaction. 5′-OH mRNAs were prepared by treating with Antarctic phosphatase (NEB). See Supplementary Materials for additional cloning information.
RNA was capped enzymatically using the vaccinia virus enzyme. Components (50 mM Tris-HCl, pH 7.8, 1.25 mM MgCl2, 6 mM KCl, 2.5 mM DTT, 1 mM GTP, 100 μM S-adenosylmethionine, 5 μM natural mRNA or 50 μM model mRNA, 2 μCi/μL α-[32P]-GTP (Perkin Elmer), 125 nM capping enzyme, 1 U/μL RiboLock RNase Inhibitor (Fermentas)) were incubated at 37 °C for 60 minutes. For G-capped mRNA SAM was excluded from the reaction. Additional enzyme was added to the reaction after 30 minutes for a final concentration of 250 nM capping enzyme. The reactions were quenched with EDTA. RNA was purified (see Supplementary Materials) and capping efficiency was analyzed by electrophoresis (see Supplementary Materials). Uncapped mRNA was end-labeled with 32P using T4 polynucleotide kinase (NEB).
Fluorescent Labeling
Fluorescent labeling of eIFs 1, 4A, 4B and 5 was performed using expressed protein ligation as described for eIFs 1 and 5 using fluorescein (eIF-Fl) (Acker et al., 2007). eIF4E and eIF4B were also labeled by introducing S200C (Gross et al., 2003) and A130C, respectively, by site directed mutagenesis, and modifying the proteins with fluorescein maleimide (see Supplementary Materials). Single strand RNA was thiophosphorylated with ATPγS using T4 polynucleotide kinase and labeled using fluorescein maleimide (Supplementary Materials).
Biochemical Assays
Fluorescence anisotropy assays were performed as previously described (Maag and Lorsch, 2003). Fluorescently labeled protein or RNA was used at 15 nM (eIF1, eIF5, eIF4E) or 30 nM (eIF4A, ssRNA).
Gel shift assays monitoring [35S]-Met-tRNAi were performed as published (Acker et al., 2007). Final concentrations were 1 mM GDPNP, 200 nM eIF2, 1 nM [35S]-MettRNAi, 1 μM eIF1, 1 μM eIF1A, 30 nM 40S subunits and, when present, 10 μM mRNA. Gel shift assays following [32P]-labeled mRNA were performed as above with several modifications (see Supplementary Materials). Concentrations were the same with the following changes: 200 nM Met-tRNAi, 200 nM eIF3, 1 μM eIF4A, 50 nM eIF4E•4G, 500 nM eIF4B, 15 nM [32P]-labeled mRNA. The concentrations of the initiation factors were saturating (see Supplementary Materials).
All data were fit to a single exponential rate equation for kinetic assays and a hyperbolic or quadratic binding equation for binding assays using KaleidaGraph software (Synergy). All assays were performed at least twice with the exception of eIF5-Fl binding to eIF4E•eIF4G, eIF4B-Fl binding eIF3 and experiments using eIF4G truncations. Errors given are average deviations.
Toeprinting was performed based on the previously described method (Hartz et al., 1989), with minor changes (see Supplementary Materials).
Supplementary Material
Acknowledgements
We thank Patrick Linder, Stewart Shuman and Alan Sachs for providing plasmids; Roy Parker, members of our labs and the reviewers for comments on this manuscript. This work was supported by a grant from the NIH to JRL (GM62128) and the intramural research program of the NIH/NICHD (AGH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- The eIF4 factors and eIF3 accelerate the rate of mRNA recruitment.
- For uncapped mRNA, a recruitment pathway depending only upon eIF3 exists.
- The cap creates a requirement for the eIF4 factors with natural mRNAs.
- The triphosphate moiety of the cap is responsible for blocking aberrant initiation.
References
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- Algire MA, Maag D, Savio P, Acker MG, Tarun SZ, Jr., Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali IK, McKendrick L, Morley SJ, Jackson RJ. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4233–4242. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M, Muller PP, Wittmer B, Ruchti F, Lanker S, Trachsel H. A Saccharomyces cerevisiae Homologue of Mammalian Translation Initiation Factor 4B Contributes to RNA Helicase Activity. EMBO J. 1993;12:3997–4003. doi: 10.1002/j.1460-2075.1993.tb06077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, Dmitriev SE, Terenin IM, Prassolov VS, Merrick WC, Shatsky IN. Differential contribution of the m7G-cap to the 5' end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valassek L, Donahue TF, Hinnebusch AG. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Hershey JWB. The Mechanism of Action of Protein Synthesis Initiation Factors from Rabbit Reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- Bergmann JE, Trachsel H, Sonenberg N, Shatkin AJ, Lodish HF. Characterization of rabbit reticulocyte factor(s) that stimulates the translation of mRNAs lacking 5'-terminal 7-methylguanosine. J Biol Chem. 1979;254:1440–1443. [PubMed] [Google Scholar]
- Berset C, Zurbriggen A, Djafarzadeh S, Altmann M, Trachsel H. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA. 2003;9:871–880. doi: 10.1261/rna.5380903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Chowdhury D, Maitra U. Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40S ribosomal preinitiation complex. J Biol Chem. 1999;273:17975–17980. doi: 10.1074/jbc.274.25.17975. [DOI] [PubMed] [Google Scholar]
- Cigan AM, Donahue TF. Sequence and structural features associated with translational initiator regions in yeast - a review. Gene. 1987;59:1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- Cong P, Shuman S. Mutational analysis of mRNA capping enzyme identifies amino acids involved in GTP binding, enzyme-guanylate formation, and GMP transfer to RNA. Mol Cell Biol. 1995;15:6222–6231. doi: 10.1128/mcb.15.11.6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci U S A. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, Preiss T, Hentze MW. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5' end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC, Shatsky IN. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5' untranslated regions. Mol Cell Biol. 2003;23:8925–8933. doi: 10.1128/MCB.23.24.8925-8933.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JW, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- Gunnery S, Maivali U, Mathews MB. Translation of an uncapped mRNA involves scanning. J Biol Chem. 1997;272:21642–21646. doi: 10.1074/jbc.272.34.21642. [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- He H, von der Haar T, Singh CR, Ii M, Li B, Hinnebusch AG, McCarthy JE, Asano K. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol Cell Biol. 2003;23:5431–5445. doi: 10.1128/MCB.23.15.5431-5445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey PE, McWhirter SM, Gross JD, Wagner G, Alber T, Sachs AB. The Cap-binding protein eIF4E promotes folding of a functional domain of yeast translation initiation factor eIF4G1. J Biol Chem. 1999;274:21297–21304. doi: 10.1074/jbc.274.30.21297. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jivotovskaya AV, Valasek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J Biol Chem. 2000;275:41369–41376. doi: 10.1074/jbc.M007525200. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of Functional Domains in Eukaryotic Protein Synthesis Initiation Factor 4G (eIF4G) with Picornaviral Proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Lanker S, Muller PP, Altmann M, Goyer C, Sonenberg N, Trachsel H. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J Biol Chem. 1992;267:21167–21171. [PubMed] [Google Scholar]
- Lawson TG, Lee KA, Maimone MM, Abramson RD, Dever TE, Merrick WC, Thach RE. Dissociation of double-stranded polynucleotide helical structures by eukaryotic initiation factors, as revealed by a novel assay. Biochemistry. 1989;28:4729–4734. doi: 10.1021/bi00437a033. [DOI] [PubMed] [Google Scholar]
- Lawson TG, Ray BK, Dodds JT, Grifo JA, Abramson RD, Merrick WC, Betsch DF, Weith HL, Thach RE. Influence of 5' proximal secondary structure on the translational efficiency of eukaryotic mRNAs and on their interaction with initiation factors. J Biol Chem. 1986;261:13979–13989. [PubMed] [Google Scholar]
- Lo HJ, Huang HK, Donahue TF. RNA polymerase I-promoted HIS4 expression yields uncapped, polyadenylated mRNA that is unstable and inefficiently translated in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:665–675. doi: 10.1128/mcb.18.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol. 2003;330:917–924. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5' cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan S, Morgan M, Banerjee AK, Shatkin AJ. Influence of 5'-terminal m7G and 2'--O-methylated residues on messenger ribonucleic acid binding to ribosomes. Biochemistry. 1976;15:5761–5768. doi: 10.1021/bi00671a012. [DOI] [PubMed] [Google Scholar]
- Neff CL, Sachs AB. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol Cell Biol. 1999;19:5557–5564. doi: 10.1128/mcb.19.8.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T, Rau M, Morley SJ, Pain VM. Proteolytic cleavage of initiation factor eIF-4 gamma in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD. Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell. 1975;4:189–197. doi: 10.1016/0092-8674(75)90167-1. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- Sagliocco FA, Vega Laso MR, Zhu D, Tuite MF, McCarthy JE, Brown AJ. The influence of 5'-secondary structures upon ribosome binding to mRNA during translation in yeast. J Biol Chem. 1993;268:26522–26530. [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Shatkin AJ. Nonspecific effect of m7GMP on protein-RNA interactions. J Biol Chem. 1978;253:6630–6632. [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5' secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalova L, Munzarova V, Herrmannova A, Nielsen KH, Burela L, Hinnebusch AG, Valasek L. eIF3a cooperates with sequences 5' of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SJ, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Jr., Sachs AB. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZJ, Sachs AB. A common function for mRNA 5' and 3' ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- Thoma C, Bergamini G, Galy B, Hundsdoerfer P, Hentze MW. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol Cell. 2004;15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Trachsel H, Erni B, Schreier MH, Staehelin T. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J Mol Biol. 1977;116:755–767. doi: 10.1016/0022-2836(77)90269-8. [DOI] [PubMed] [Google Scholar]
- Vega Laso MR, Zhu D, Sagliocco F, Brown AJ, Tuite MF, McCarthy JE. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J Biol Chem. 1993;268:6453–6462. [PubMed] [Google Scholar]
- Von Der Haar T, McCarthy JE. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol Microbiol. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.