Abstract
N3-methyladenine (3-mA) is a cytotoxic lesion formed by the reaction of DNA with many methylating agents, including antineoplastic drugs, environmental agents and endogenously generated compounds. The toxicity of 3-mA has been attributed to its ability to block DNA polymerization. Using Me-lex, a compound that selectively and efficiently reacts with DNA to afford 3-mA, we have observed in yeast a mutational hotspot at the 5′-terminus of an A4 tract. In order to explore the potential role of sequence-dependent DNA polymerase bypass of 3-mA, we developed an in vitro system to prepare 3-mA modified substrates using Me-lex. We detail the effects of 3-mA, its stable isostere analogue, 3-methyl-3-deazaadenine, 3-deazaadenine and an THF abasic site on DNA polymerization within an A4 sequence. The methyl group on 3-mA and 3-methyl-3-deazaadenine has a pronounced inhibitory effect on DNA polymerization. There was no sequence selectivity for the bypass of any of the lesions, except for the abasic site, which was most efficiently by-passed when it was on the 5′-terminus of the A4 tract. The results indicate that the weak mutational pattern induced by Me-lex may result form the depurination of 3-mA to an abasic site that is bypassed in a sequence dependent context.
1. Introduction
N3-Methyladenine (3-mA1) is a quantitatively significant DNA adduct formed from the reaction of many methylating agents, including antineoplastic drugs, and environmental and endogenous compounds [1–7]. Because 3-mA is normally formed in combination with a large number of other DNA lesions, including N7-methylguanine [7-mG), O6-methylguanine (6-mG), N7-methyladenine (7-mA) and N1-methyladenine (1-mA) [7–9], it has been difficult to quantify its contribution to the toxicity and mutagenicity of promiscuous methylating agents. Moreover, 3-mA hydrolyzes off DNA at an appreciable rate (t 1/2 = 24 hours) at neutral pH and physiological temperature, yielding abasic sites on the DNA and released 3-mA in solution [10]. The rate of hydrolysis is dependent on whether the adduct is in single- or double-stranded DNA [11], the former being closer to what polymerases encounter at a replication fork. For example, 3-m-dA has a t 1/2 of 35 minutes at pH 7 (37°C) [11]. Therefore, it has not been possible to perform biochemical polymerase studies with single-stranded DNA templates that are site specifically modified with 3-mA.
In a previous study on the potential effect of 3-mA on DNA replication, methyl methanesulfonate (MMS) and dimethyl sulfonate (DMS), which predominantly react with DNA to produce N7-methylguanine, were used to methylate poly(dA-dT) [12]. Under these conditions, the heteropolymer contained a mixture of 3-mA, 1-methyladenine and N7-methyladenine adducts in a ratio of 16 : 3 : 1. Nucleotide incorporation using E. coli DNA polymerase I decreased proportionally to the level of total methyl lesions and/or abasic sites. The authors concluded that 3-mA lesions were likely to be responsible for some of the reduction of DNA polymerization. In another related study, DMS and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) were used to methylate single-strand primed M13mp2 templates and double-stranded templates, and these templates used with E. coli DNA polymerase I or AMV reverse transcriptase [13]. MNNG produces a complex array of lesions, including N7-methylguanine, 3-mA, O6-methylguanine and O4-methylthymine in double- and single-stranded DNA [7–9]. The results of these studies showed that polymerization predominantly stopped one base before adenine residues [13]. Heating of the DNA generated additional termination sites one nucleotide before G, presumably because heating caused depurination of 7-mG to an abasic site that blocked polymerization. In both studies, the DNA is damaged at multiple bases and/or at multiple sites, and there is no control over the depurination.
To overcome some of the ambiguities associated with previous attempts to study the effect of 3-mA on DNA polymerization and to better understand the potential effect of sequence on polymerization past the lesion, we have developed a method to generate 3-mA in situ using {1-methyl-4-[1-methyl-4-(3-methoxysulfonyl-propanamido)pyrrole-2-carboxamido]pyrrole-2-carboxamido}propane (Me-lex), a compound that reacts selectively in the minor groove of double-stranded DNA to efficiently generate 3-mA at A/T rich sequences [14]. We report for the first time the construction of single-strand DNA templates selectively substituted with 3-mA and the effect of this lesion on DNA polymerases within an 5′-CA4C tract previously identified as a hotspot for Me-lex induced p53 mutations in a yeast shuttle vector assay [15, 16].
In a recent study, it was reported that 3-methyl-3-deazaadenine (3-m-c3A), a neutral isostere of 3-mA, represented a significant kinetic block to purified human replicative polymerases Polα and Polδ in a nonrepetitive sequence [17]. However, the human TLS polymerases η, ι, and κ were capable of weak (at least 2 orders of magnitude lower) insertion opposite, and extension from, 3-m-c3-dA. We have extended these studies by determining how 3-m-c3A, 3-deazaadenine (c3A) and a THF abasic site affect polymerization when the lesions are at different positions within an A4-tract.
2. Materials and Methods
2.1. Materials
HPLC purified 5′-Cy5-labeled DNA primers were purchased from MWG Biotech (Table 1 for sequences). The 3-m-c3-dA phosphoramidite was prepared as previously described [18], and the other modified phosphoramidites purchased from Glen Research (Sterling, VA). Unmodified, c3A, 3-m-c3A and THF-modified templates were synthesized and purified at either the University of Nebraska Medical Center or the University of Pittsburgh DNA core facilities. The modified oligomers were analyzed by MALDI-TOF-MS. T7 Sequenase and Klenow fragment, both lacking 3′ → 5′ exonuclease activity, were purchased from USB (Cleveland, OH). Klenow fragment polymerase with exonuclease activity was purchased from Promega (Madison, WI). Yeast Polη was purchased from Enzymax LLC (Louisville, KY).
Table 1.
Sequences of oligomers and primers used in the studies.
| Oligomer name | Sequence | Length | Modification |
|---|---|---|---|
| Primers | |||
|
| |||
| P20 | 5′ CCG TAG GAC TGC CGT AGT GC 3′ | 20 nt | 5′-Cy5 |
| P23 | 5′ CCG TAG GAC TGC CGT AGT GCT GG 3′ | 23 nt | 5′-Cy5 |
| P24 | 5′ CCG TAG GAC TGC CGT AGT GCT GGT 3′ | 24 nt | 5′-Cy5 |
| P25 | 5′ CCG TAG GAC TGC CGT AGT GCT GGT T 3′ | 25 nt | 5′-Cy5 |
| P26 | 5′ CCG TAG GAC TGC CGT AGT GCT GGT TT 3′ | 26 nt | 5′-Cy5 |
| P27 | 5′ CCG TAG GAC TGC CGT AGT GCT GGT TTN 3′ (N = C, G, T or A) | 27 nt | 5′-Cy5 |
|
| |||
| Templates | |||
|
| |||
| Unmodified | 5′ ATC CAA CCT GTA TCA TAC TTA CCA AAA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | — |
| 3-m-c3A/24 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA AAXCCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = 3-m-c3A |
| 3-m-c3A/25 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA AXA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = 3-m-c3A |
| 3-m-c3A/26 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA XAA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = 3-m-c3A |
| 3-m-c3A/27 | 5′ ATC CAA CCT GTA TCA TAC TTA CCX AAA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = 3-m-c3A |
| c3A/24 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA AAX CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = c3A |
| c3A/25 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA AXA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = c3A |
| c3A/26 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA XAA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = c3A |
| c3A /27 | 5′ ATC CAA CCT GTA TCA TAC TTA CCX AAA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = c3A |
| THF/24 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA AAX CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = THF |
| THF/25 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA AXA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = THF |
| THF/26 | 5′ ATC CAA CCT GTA TCA TAC TTA CCA XAA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = THF |
| THF/27 | 5′ ATC CAA CCT GTA TCA TAC TTA CCX AAA CCA GCA CTA CGG CAG TCC TAC GG 3′ | 50 nt | X = THF |
|
| |||
| DFO | 5′-TGGTTTTGGT | 10 nt | |
2.2. Generation of 3-mA Containing Templates
The template DNA (100 nM) was annealed to a 10 nt duplex forming oligomer (DFO) (200 nM) and the primer (100 nM) in 40 mM Tris-HCl (pH 7.5) containing 20 mM MgCl2 and 50 mM KCl (see Table 1 for sequences). After annealing, the reactions were maintained at 4°C and treated with a chilled solution of Me-lex in anhydrous DMSO (final concentration: 0, 0.60, 1.25 and 2.50 mM), with the amount of DMSO not exceeding 10% of the final volume. The reactions were maintained at 4°C for the time specified prior to performing the methylation analyses or polymerase assays. For control purposes in the methylation studies, the DFO was omitted or a full-length complementary strand was included in place of the 10 nt DFO. The sites of Me-lex methylation were determined in a 5′-[32P]-template strand (end labeled with T4 kinase and [γ-32P]-ATP) by generating C, G and ladder marker lanes using THF-OOH [19], DMS [20] and hot formic acid followed by piperidine [20], respectively.
2.3. Nucleotide Polymerase Run-Up Assay
The 5′-Cy5-labeled 20 nt primer (P20, 200 nM) was annealed to unmodified template (Table 1, 100 nM) in the absence or presence of the DFO (200 nM), or to the c3A, 3-m-c3A or THF modified templates (100 nM). The 3-mA lesion was introduced into the unmodified template strand as described above. For T7 Sequenase, 30 μL reactions included 40 mM Tris-HCl (pH 7.5) containing 20 mM MgCl2, 50 mM KCl, 0.1 mM DTT and various concentrations of polymerase and dNTPs. For Klenow fragment (exo−), 30 μL reactions included 50 mM Tris-HCl (pH 7.2) containing 10 mM MgSO4, 0.1 mM DTT and various concentrations of polymerase and dNTP. For Polη, 30 μL reactions included 40 mM Tris-HCl (pH 8), 10 mM MgCl2, 60 mM KCl, 250 μg/mL BSA, 2.5% glycerol, 10 mM DTT and various concentrations of polymerase and dNTPs.
The incubations were initiated by the addition of the dNTP mix at 37°C (unless stated otherwise) and reactions were terminated by the addition of 10 μL of 200 mM EDTA and heating to 90°C for 2 minutes to denature the enzymes. All reactions were concentrated by speed vacuum and then 10 μL of loading buffer (95% formamide, 0.1% bromophenol blue and 20 mM EDTA) added. Reactions were heated at 95°C for 4 minutes and loaded on a 15% denaturing polyacrylamide gel that was run at 60 W for 4 hours. Gels were imaged on a Molecular Dynamics Typhoon fluorescent scanning instrument (Sunnyvale, CA) and analyzed using GE Healthcare ImageQuant Software (Piscataway, NJ).
2.4. Insertion Assays for 3-mA
To measure insertion opposite 3-mA, the lesion was generated as described above. The Cy-5 labeled 25 nt primer (P25, 400 nM) was annealed to the Me-lex methylated template at 30°C for 2 minutes. In these experiments, the 3′-terminus of the primer partially overlaps with the DFO. The primer displaces the DFO upon heating to 37°C when the polymerase and dNTPs are added because of its greater thermal stability relative to the DFO at the higher temperature and because it is present in higher concentration than the DFO. The 30 μL reactions included 40 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 50 mM KCl, 0.1 mM DTT and 500 μM dNTP mix and were run for 5 to 20 minutes at 37°C. Reactions were stopped with 10 μL of 95% formamide, 0.1% brom0phenol blue, and 20 mM EDTA solution. The resulting polymerization product and substrate DNA were separated using an 8 M urea, 15% polyacrylamide denaturing gel. The gels were imaged on a Typhoon fluorescent scanning instrument and analyzed using ImageQuant Software. Polymerase and DNA concentrations were adjusted to achieve less than 30% consumption of the substrate and the DNA concentration was varied from 0 to 1 μM.
2.5. Insertion and Extension Assays with 3-m-c3A, c3A and THF (Abasic Site) Modifications
The insertion studies using T7 Sequenase, Klenow fragment (exo− and exo+) and Polη were done as described above, except the lesions were individually substituted for each of the four A's within the A4 sequence. Primer-20 was used for the runup studies. Primers P23-26 were used to determine the preferential insertion opposite the lesions at position 24–27. For extension from c3A, 3-m-c3A and THF lesions paired with A, G, C or T, 100 nM of the 5′-Cy5 labeled P-27 primer (Table 1) was annealed to 100 nM of unmodified, or c3A, 3-m-c3A or THF substituted templates. In all experiments, polymerase was added and allowed to load for 1 minute, and reactions initiated by the addition of dNTPs. Reactions were stopped by addition of 10 μL of 95% formamide, 0.1% bromphenol blue and 20 mM EDTA. Reactions were heated to 95°C for 4 minutes, immediately chilled on ice and then loaded on a 8 M urea 15% polyacrylamide denaturing gel and run at 60 W for 4 hours. Gels were imaged on a Typhoon (Amersham-Biosciences) fluorescent scanning instrument and analyzed using the ImageQuant Software.
3. Results
3.1. Generation of 3-mA by a Modified Template
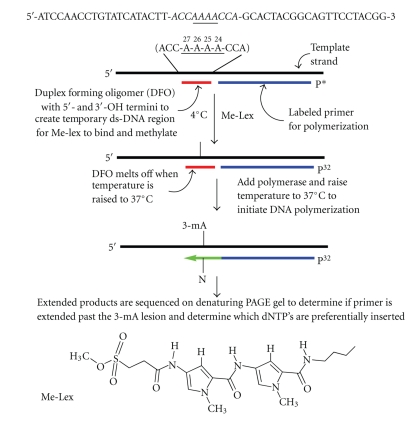
When 3-mA is formed in single-stranded DNA or as the 2′-deoxynucleoside, it is subject to hydrolysis even at neutral pH and room temperature. This instability prevents the preparation of substrates by standard solid phase oligodeoxynucleotide synthesis using phosphoramidite intermediates. However, 3-mA is significantly more stable, almost 40-fold, in double-stranded DNA [11]. Therefore, a scheme was devised to generate a transient double-stranded DNA region on a single-strand DNA template that would serve as a site for methylation by Me-lex and maintain the lesion in a duplex environment until polymerase studies were initiated (Scheme 1). Because Me-lex selectively forms 3-mA lesions, but only reacts to a significant extent with A's that are in A/T rich runs in double-stranded DNA, the template strand was designed with only one Me-lex binding site, that is, an A4 tract. A 10 nt DFO (Table 1) was annealed to the single-stranded DNA template to generate a double-stranded region with a TM of 31°C in the buffer system used to measure DNA polymerase activity. The target sequence included a 5′-CAAAAC track (Scheme 1) that was previously identified as a hotspot for Me-lex induced p53 mutations in a yeast shuttle vector assay [15, 16].
Scheme 1.
Preparation of a single-stranded DNA template modified with 3-methyladenine (3-meA) within an A-tract. The template strand is annealed to a duplex forming oligomer (DFO) that creates a transient 10 base pair duplex centered at the A4 target sequence. This allows efficient methylation at the 3-position of adenine at 4°C by Me-lex, which selectively methylates double-stranded DNA in the minor groove at A/T rich sequences. The two 5′-A's are heavily and equally methylated at the N3-position. The 3-mA lesion is stabilized within the duplex region and is revealed in the single-stranded template needed for the polymerase studies by warming the reaction temperature to 37°C in the presence of the primer, which results in the melting of the DFO and the annealing of the primer. The reaction is initiated by the sequential addition of polymerase and dNTPs.
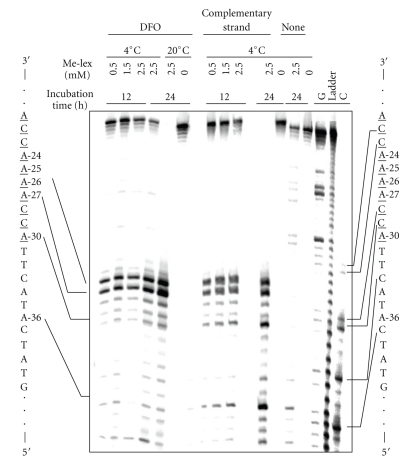
The methylation pattern of Me-lex on the single-stranded DNA template strand in the presence and absence of the DFO, and with a full complementary strand is shown in Figure 1. The location and abundance of the 3-mA adducts was revealed by denaturing PAGE using a 5′-32P-DNA template by initially heating the DNA at pH 7.0 at 90°C for 20 minutes to convert 3-mA lesions into abasic sites and conversion of the abasic sites into strand breaks by heating the DNA with hot piperidine [20]. The G, C and cleavage ladder lanes are included to map the location of the strand breaks.
Figure 1.
The A4 tract sequence is methylated selectively by Me-lex at A-27 and A-26. Maxam-Gilbert chemical sequencing of methylation pattern in 5′-[32P]-labeled unmodified sequence (Table 1) by Me-lex at 4°C and 20°C with 12 or 24 hours incubation times in the presence of: the 10 nt duplex forming oligomer (DFO), which overlaps with underlined bases on the labeled strand, with 0.5, 1.25 or 2.5 mM Me-lex (12 hours) or 0 or 2.5 mM Me-lex (24 hours); the full complementary strand with 0, 1.25 or 2.5 mM Me-lex (12 hours) or 0 or 2.5 mM Me-lex (24 hours); no complementary strand (none) with 0 or 2.5 mM Me-lex (24 hours). Sequencing lanes (G, ladder and C) are on right.
In the absence of the DFO or full complementary strand (Figure 1, identified as none), virtually no methylation of the DNA by up to 2.5 mM Me-lex is observed. With the full complementary strand, the methylation occurs predominantly at the two 5′-A's (A-26 and -27) of the A-tract, but there are bands at other A's (e.g., A-30, A-36) within shorter A/T regions that are in the template sequence. When the 10 nt DFO is used to create a regional double-stranded region on the template, the methylation is more intense at A-26 and A-27, with approximately 33% of the total label being associated with each of the A's using 2.5 mM Me-lex for 12 hours at 4°C. There is approximately 30% of unreacted (i.e., nonmethylated DNA at the top of the gel). The equivalent levels of methylation at A-26 and A-27 indicate that there is only one adduct per template strand. At longer incubation times or higher concentrations of Me-lex, the intensity of the lower band at A-26 starts to predominate because there is slightly more than one methylation event per strand; when there are two lesions on the same strand only the one closest to the 5′-label is observable. It is not known if an A/T sequence that has already been methylated in the minor groove is a good equilibrium-binding site for the Me-lex. We expect not because the methyl group would protrude out into the groove and interfere with the H-bonding and van der Waals interactions between the Me-lex and the DNA minor groove. This is reasonable since the 2-amino group on G has an inhibitory effect on lex dipeptide binding, which accounts for the sequence selectivity of Me-lex for A/T sequences [21, 22]. It should be noted that the Me-lex methylated template may contain small amounts of abasic sites due to spontaneous depurination of 3-mA. Based upon our previous studies, this process would be minimal at 4°C since the lesion has a t 1/2 greater than 24 hours at 20°C [23]. Because of the instability of 3-mA, the methylated templates were prepared immediately before use. Therefore, each methylated template used in the studies may be modified to different extents, although the relative amounts of methylation at different sites within the template will not vary.
In conclusion, conditions required for a single methylation event per strand at either A-26 or A-27 were found. This is a fundamental requirement for the enzymatic studies described below. However, there is nonmethylated DNA in the incubations that results in a background in the polymerase studies on 3-mA modified templates. Because we wanted to probe how 3-mA would effect polymerization within an A-tract, this complication was unavoidable.
3.2. Polymerization of T7 Sequenase, Klenow Fragment (exo−) and Polη Polymerases Past 3-mA
After the 12 hours incubation with solvent (control) or 2.5 mM Me-lex, the template was immediately used in the polymerase studies. This involves the addition of the 5′-Cy5-labeled primer and polymerase, with polymerization initiated by elevation of the reaction temperature to 37°C (unless stated otherwise) and addition of the dNTPs. It should be noted that the primer often runs as a doublet in the presence of polymerase when no dNTPs are added. Without the polymerase, a single band is observed with the same labeled primer.
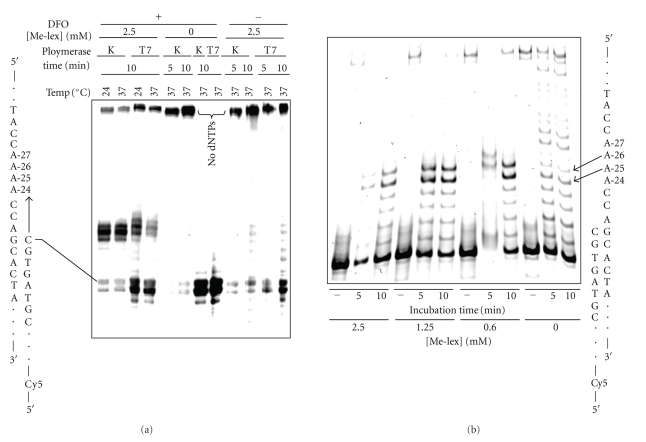
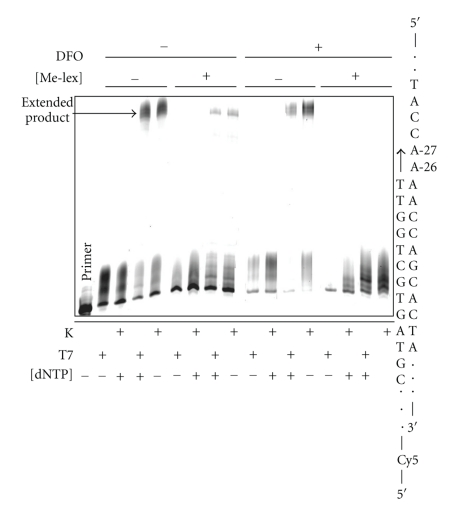
Preliminary incubations were run with unmodified templates (no Me-lex) to demonstrate that the 10 nt DFO, which was designed with a TM of 31°C so it would dissociate off the template at 37°C, did not interfere with polymerization. Accordingly, in these incubations with T7 Sequenase and Klenow fragment (exo−) there is extension product (Figure 2(a)). In contrast, with Me-lex treatment there is a strong block to the polymerase in both the 5 and 10 minutes incubations (Figure 2). The block occurs before each of the two 5′-A's that are targeted for methylation by Me-lex (Figure 2(b)). The bands corresponding to polymerase blocks occur at A-25 and A-26. The band at A-26 arises because either A-26 or A-27 is methylated on each strand (Figure 2(b)). The similar intensities of the bands at A-25 and A-26 are consistent with the equivalent methylation of A-26 and A-27 (Figure 1). As an additional control, the block does not occur if the DFO is not included, or in reactions with the DFO but where Me-lex is omitted (Figure 2(a)). To precisely calculate the extension past the 3-mA lesion it is necessary to accurately know how much of the template is nonmethylated. Since this is not possible in each incubation reaction, a precise kinetic description of the bypass efficiency for T7 Sequenase is not possible. Despite this limitation for precise quantification, 3-mA is clearly a potent block of DNA polymerization.
Figure 2.
The 3-mA lesion blocks DNA polymerization by T7 Sequenase (T7) and Klenow fragment exo− (K) polymerase. (a) DNA template (100 nM) was treated with 0 or 2.50 mM Me-lex for 12 hours at 4°C in the presence or absence of the DFO: 3-mA forms at A-26 and A-27 only when the DFO is present (Figure 1). The runup polymerization assay was initiated by raising incubation temperature to 24 or 37°C and the addition of 5′-Cy5-primer (200 nM) and dNTPs (0 or 100 μM). Polymerization experiments with T7 Sequenase and and Klenow fragment (exo−) were run for 5 or 10 minutes. (b) The Me-lex (0, 0.6, 1.25 or 2.50 mM) treated DNA was used to extend 5′-Cy5-primer in the absence (−) or presence of 100 μM dNTPs for 5 or 10 minutes at 37°C using T7 Sequenase. The polymerase is blocked prior to insertion opposite positions-26 or -27, which are heavily methylated by Me-lex (Figure 1).
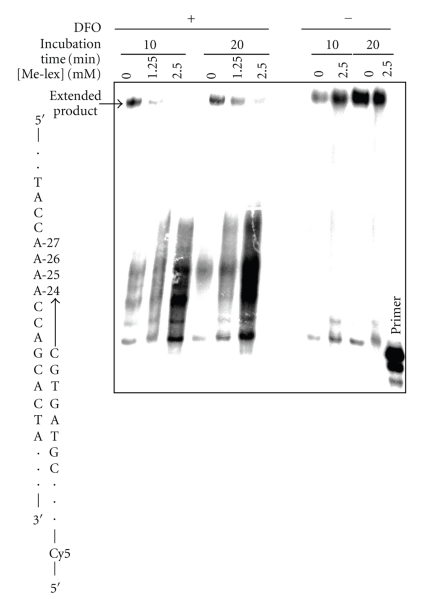
A similar set of experiments was performed using yeast Polη, which is a Y-family polymerase that has a role in the bypass of 3-mA and/or abasic sites based upon the enhanced sensitivity to MMS of yeast lacking both Polη and MAG1, the glycosylase that removes 3-mA [24, 25]. As with T7 Sequenase, full-length product is observed with the Me-lex treated template in the absence of the DFO or with the DFO in the absence of Me-lex (Figure 3). There is a Me-lex dose-dependent block of extended product in the presence of the DFO with 10 or 20 minutes incubation times. Unfortunately the bands near the lesion are poorly resolved so it is not possible to accurately identify where the block occurs. Without Me-lex, there appears to be a weak DFO mediated stalling of Polη extension past the A-tract. This is not observed without the DFO. This raised the possibility that the stalling of the weakly processive Polη could be explained if the formation of 3-mA stabilized the short 10 bp duplex region formed by the 10 nt DFO. To test this issue, we treated the template strand plus DFO with 2.5 mM Me-lex at 5°C and then determined the effect on the thermal stability of the DFO-duplex by measuring the TM (UV using λ 260). The results show that the methylation does not stabilize the duplex formed by DFO (data not shown). In fact, there is evidence for a slight destabilization of the TFO; therefore, it is unlikely that stabilization of the DFO plays a role in the observed stalling of Polη. Regardless, the DFO may partially contribute to the stalling observed with Polη. It is also possible that the TLS polymerase is inefficient in replicating through a repetitive sequence since it primarily functions as an inserter opposite lesions rather than as an efficient extender [26].
Figure 3.
The effect of 3-mA on DNA polymerization by Polη. The DNA template (100 nM) was treated with 0, 1.25 or 2.5 mM Me-lex for 12 hours at 4°C in the presence (+) or absence (−) of the DFO: 3-mA forms at A-26 and A-27 only when DFO is present (Figure 1). The runup polymerization was initiated by raising incubation temperature to 37°C and the addition of 5′-Cy5-primer (200 nM) and dNTPs (0 or 100 μM). Polymerization experiments were run for 10 or 20 minutes. The polymerase is blocked at the methylated adenines, which is also reflected in the level of extended product. Extension is efficient if the DFO is not present during the methylation step (Scheme 1).
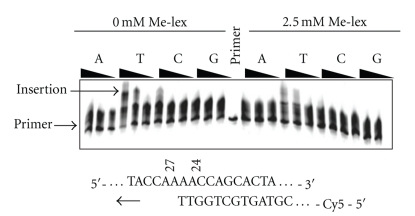
In order to determine whether any dNTP can be inserted opposite 3-mA, a primer (Table 1, P25) was prepared that overlapped with the two A's (A24, A25) that are not methylated by Me-lex (Figure 1). When the methylated template was incubated with a mixture of dNTPs, there is a Me-lex and DFO dependent reduction in extended product for both T7 Sequenase and Klenow fragment (exo−) (Figure 4). It should be noted that the DFO, in the absence of Me-lex treatment, has no effect on the ability of the two polymerases to produce extended product. In addition, Me-lex causes a dose-dependent reduction in insertion of T relative to untreated DNA by T7 Sequenase and there is no evidence for insertion of any other base opposite 3-mA (Figure 5). The apparent weak insertion of dTTP opposite 3-mA is, at least in part, due to the presence of unmethylated DNA (i.e., unmethylated A-26 and A-27) in the incubation. In addition, there is only one 3-mA lesion per strand (i.e., either A-26 or A-27 will be methylated but not both) so some extension up to A-26 was expected to be observed.
Figure 4.
Insertion opposite and extension from 3-mA by T7 Sequenase (T7) and Klenow fragment (K) (exo−) polymerases. The template strand in the absence (−) or presence (+) of the DFO was treated with 0 or 2.5 mM Me-lex as described in Figure 1. A 5′-labeled primer (Table 1, P25) was used that overlaps with the two 3′-A's (A-24, A-25) that are not significantly methylated by Me-lex (Figure 1). T7 Sequenase (T7) or Klenow polymerase exo− (K) was added and the reaction initiated by the addition of the four dNTPs (0 or 500 μM) to determine insertion opposite, and extension from, 3-mA at 37°C. The production of extended product is reduced with Me-lex treatment in the presence of the DFO.
Figure 5.
Insertion opposite 3-mA by T7 Sequenase. The template strand was treated with Me-lex as described in Figure 1. A 5′-Cy5-primer (Table 1, P25) was added that overlaps with the two A's (A24, A25) that are not methylated by Me-lex. T7 Sequenase was added and the reactions initiated by the addition of 5, 50 or 500 μM concentration of the individual dNTPs at 37°C. The insertion of T is inhibited in the 2.5 mM Me-lex treated DNA versus the untreated DNA (0 Me-lex). There is no indication that G, T or C is inserted opposite the lesion.
3.3. Characterization of the Effect of c3A and 3-m-c3A on DNA Polymerization in a Repetitive Sequence
To corroborate our studies with Me-lex on the effect of 3-mA on DNA polymerization, DNA substrates containing an isosteric surrogate of 3-mA were prepared and used for in vitro polymerization assays. Specifically, the phosphoramidites of the synthesized 3-m-c3-dA [18] and the commercially available c3A were incorporated into DNA using solid phase synthesis. The replacement of the minor groove N-CH3 on 3-mA with a C–CH3 in 3-m-c3A eliminates the basic nitrogen and the potential formation of positive charge on the aromatic purine ring; therefore, the glycosidic bond becomes stabilized. The c3A was used as an isostere for A and to determine the impact of the methyl group on 3-m-c3A. The c3A and 3-m-c3A modifications were individually incorporated at each of the A's within the same 5′-CA4C sequence (Table 1) that was used above.
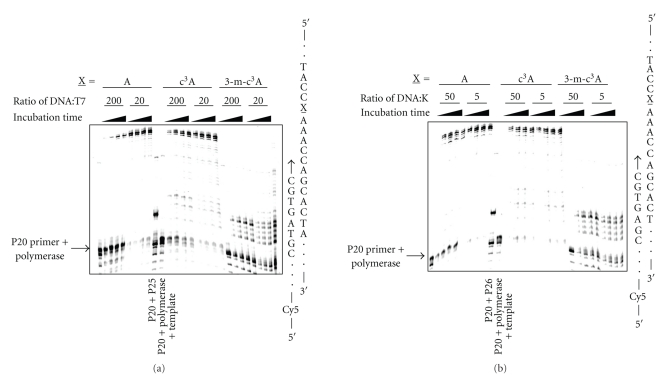
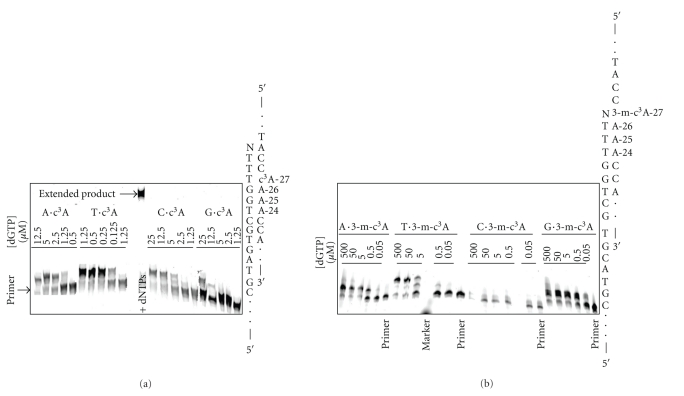
The runup assay using T7 Sequenase shows that the formation of extended product is dependent on the ratio of DNA template to polymerase: the lower the ratio the more full-length product (Figure 6(a)). When the c3A is substituted at A-27 it behaves similar to the unmodified A, although there is some accumulation of stalled bands near the modified residue. In contrast, the 3-m-c3-dA lesion represents a potent block to the polymerase under all conditions tested. The block occurs near the site of the modification and the location of the bands indicates blockage of the polymerase prior to insertion opposite the 3-m-c3A lesion. A similar pattern is observed with Klenow fragment polymerase (exo−) (Figure 6(b)). Clearly, the minor groove methyl group has a significant effect on replication by T7 Sequenase and Klenow fragment polymerase (exo−). The potential insertion of the different dNTPs by T7 Sequenase opposite 3-m-c3A at position 27 with either a 3′- C or G was investigated. There is no significant incorporation in either case, except at the highest concentrations of dNTPs (data not shown). This strongly suggests that it is the insertion and extensions steps that are blocked by the lesion. The effect of the position of the lesions within the A4 tract using T7 Sequenase was determined for the combined dNTPs (Figure 7). The 3-m-c3A does not show any positional effect as it efficiently blocks Sequenase polymerase insertion at all positions in the A-tract. The c3A lesion stalls polymerization but to a much smaller extent than 3-m-c3A.
Figure 6.
The effect of 3-deazaadenine (c3A) and 3-methyl-3-deazaadenine (3-m-c3A) lesions on (a) T7 Sequenase and (b) Klenow fragment polymerization. The template strand with an A, c3A or 3-m-c3A at position-27 (X) was incubated for 2.5, 5, 10 or 20 minutes with polymerase at 37°C using DNA to enzyme ratios of 200:1 or 20:1 for T7 Sequenase, and 50 : 1 or 5 : 1 for Klenow fragment (exo−), respectively. Lanes with primer-20 + polymerase + template, and primers-20 and -25 (no polymerase or template) are included as markers. There is evidence for bands indicating stalling of polymerization with the c3A and a quantitative block with 3-m-c3A, which is also reflected in the extended product.
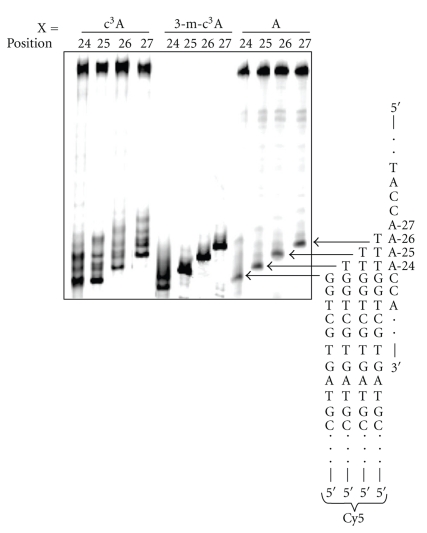
Figure 7.
The effect of the position of the c3A and 3-m-c3A lesions within the A4 tract on the insertion by T7 Sequenase opposite the modification using 10 or 100 μM of the dNTPs. The location of the modification is denoted by the numbering system in Scheme 1. The primers (Table 1, P23, P24, P25 and P26) stop 1 nt before the position of the c3A or 3-m-c3A modification.
Extension from c3A and 3-m-c3A was determined using T7 Sequenase. The efficiency of extension from c3A was highest with c3A·T > c3A·A ~ c3A·C > c3A·G (Figure 8(a)). In contrast, the extension efficiency from 3-m-c3A followed: ~3-m-c3A·T ~ 3-m-c3A·G > 3-m-c3A·C > 3-m-c3A·A (Figure 8(b)). What is unique about the extension of the 3-m-c3A·G terminus, is that only one G is added, while in the other cases there is a two base extension since there are two 3′-C's. This could result from the 3-m-c3A being extrahelical due to the instability of a 3-m-c3A·G pairing, and that the G at the end of the primer pairs with the 3′-C on the template strand, and it is this structure that is extended by one nucleotide.
Figure 8.
Extension from (A) c3A·N and (B) 3-m-c3A·N by T7 Sequenase. The template strand containing a c3A or 3-m-c3A at A-27 was annealed to a 5′-Cy5-primer (Table 1, P27). T7 Sequenase was added and the reactions initiated by the addition of the individual dNTP's. Extension is most efficient from c3A·T termini, while extension is similar from the 3-m-c3A·T and 3-m-c3A·G. Note that for 3-m-c3A·G only one dGTP is added rather than the two observed for 3-m-c3A·T.
3.4. Characterization of the Effect of a THF Abasic Site on DNA Polymerization in a Repetitive Sequence
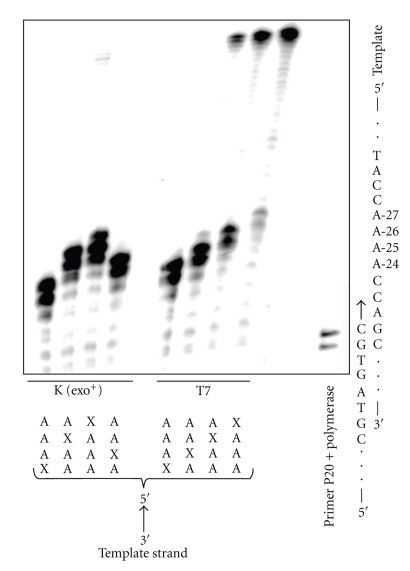
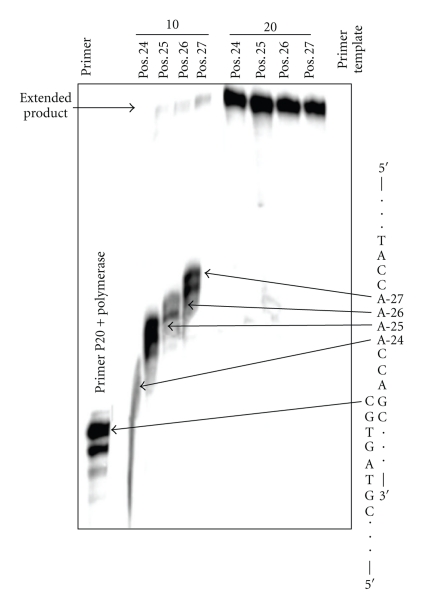
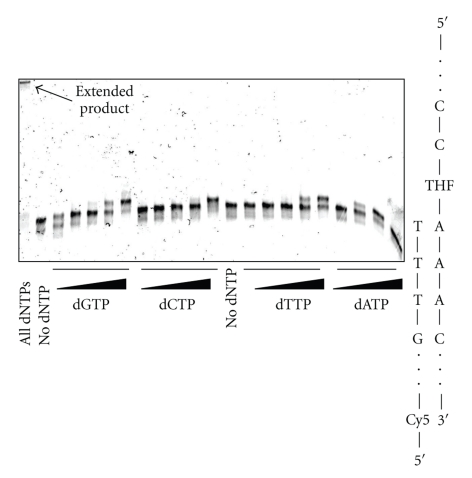
The effect of a THF abasic site was studied because Me-lex affords the same mutational pattern in a variety of DNA repair background yeast strains, which was consistent with the intermediacy of an abasic site being the common mutagenic lesion formed from 3-mA [16]. In contrast to the other lesions, THF abasic site caused a sequence-dependent bypass with T7 Sequenase in the runup assay (Figure 9). In all cases, polymerase extension of the P-20 primer to the lesion is efficient. However, there is virtually no bypass when the lesion is substituted at A-24, which is on the 3′-end of the A4 tract and the first base in the A-tract encountered by the polymerase. As the THF lesion is moved toward the 5′-terminus of the repeat, bypass becomes more efficient. This effect is only observed in the exo− polymerase; with Klenow fragment (exo+) polymerase the THF efficiently stalls the polymerization at each of the four positions (Figure 9). A similar pattern is observed with Polη in that there is enhanced bypass of the THF as it is moved to the 5′-direction of the A-tract at the shorter 10 minutes reaction time (Figure 10). At the longer incubation time, bypass of the THF is similar regardless of position of the lesion within the A-tract. Insertion by T7 Sequenase opposite THF located at the 5′-position of the A-tract is shown in Figure 11. The efficiency of base insertion opposite the THF abasic site at position-27 follows the order: A > G ~ T > C.
Figure 9.
The effect of the position of the THF abasic site lesion within the A4 tract on the insertion and extension by Klenow fragment (exo+) (K) and T7 Sequenase (T7). The location of the THF lesion (X) is shown below the lane. The primer (Table 1, P20) is shown paired to a partial sequence of the template. There is no bypass of the THF-modification using the proofreading Klenow polymerase. When T7 Sequenase is used, the extent of stalling and extended product is dependent on the position of the lesion within the A-tract.
Figure 10.
The effect of the position of the THF abasic site lesion within the A4 tract on the insertion and extension by Polη. Templates containing THF abasic site at different positions within the A-tract were synthesized and extended using yeast Polη for 10 to 20 minutes in the presence of 100 μM dNTPs. In the 10 minutes incubation, the formation of extended product is dependent on the position of the THF lesion. Efficient by-pass of the lesion is observed with the longer incubation time.
Figure 11.
Preferential insertion of the individual dNTPs opposite a THF abasic site located on the 5′-terminus (A-27) of an A4 tract by T7 Sequenase. The dNTP concentrations used were 0.005, 0.5, 5, 50 and 500 μM (for dATP the 500 μM concentration is not shown). Lanes with no added dNTP and with all dNTPs are included. dATP and dGTP are most efficiently inserted opposite the THF lesion.
4. Discussion
Me-lex, which selectively and efficiently reacts with native DNA to form 3-mA, was designed to be a cytotoxic but nonmutagenic DNA alkylating agent, with the hope that it could be the prototype of DNA alkylating drugs that are not carcinogenic [27, 28]. The compound reacts with DNA due to its minor groove binding properties to afford 1,000 fold more 3-mA than an equimolar concentration of MMS or DMS [14, 29]. In addition, the methylation selectivity at N3-A is greater than 90% so that the 3-mA (minor groove) to 7-mG (major groove) ratio exceeds 100 : 1 as compared to the 1 : 10 ratio observed with DMS and MMS [30]. There is a strong correlation between 3-mA formation and cytotoxicity [31, 32]. Moreover, the cytotoxicity is strongly enhanced in cells lacking the 3-methyadenine glycosylase [16, 25, 33] or by treatment of repair proficient cells with antisense oligomers targeted against the glycosylase [34]. These results suggest that 3-mA is directly toxic to cells as are other base excision repair intermediates.
The mutagenicity of Me-lex has been recently evaluated in mammalian Chinese hamster ovary (CHO) cells [35]. The mutation frequency of Me-lex in CHO cells is 2-4 fold above background at a concentration that reduces survival to < 10%. In contrast, MMS at a concentration that causes similar toxicity, results in a 40-fold induction in the mutation frequency [36].
In yeast, the rare point mutations that occur in a shuttle vector system using human p53 cDNA as the target are observed within A/T rich sequences that are methylated by Me-lex [15, 16, 37]. However, mutations are rare even at sites that are heavily methylated by Me-lex so the mutations at the few hotspots are not simply due to the level of methylation. Of particular note is a hotspot at a 5′-CAAAAC sequence in which the two 5-A's that are targeted for methylation by Me-lex are mutated: A→T transversions and A→G transitions [15, 16, 37]. To determine whether 3-mA might be more efficiently bypassed when it is in the 5′-side of an A4 tract, albeit with high infidelity, we designed our polymerase studies using the same 5′-CA4C sequence.
It has been assumed, based upon a number of reports employing DMS, MMS and MNNG, that 3-mA is toxic because it blocks DNA polymerization [12, 13]. Consistent with this assumption, Me-lex causes a cell cycle arrest during S-phase in wild type and AAG−/− ES cells as well as an increase in sister chromatid exchange [33]. The results presented herein confirm the assumption that 3-mA is a potent block of DNA polymerases, including Polη, a Y-family polymerase. The block with 3-mA occurs before the lesion suggesting a problem with polymerase insertion opposite the adduct.
In a recent study, it was reported that 3-m-c3A, a stable isostere of 3-mA, located in a nonrepetitive sequence represented a quantitative kinetic block to purified human replicative polymerases Polα and Polδ [17]. This is similar to what we observe for both T7 Sequenase and Klenow fragment polymerases. However, it was also demonstrated that the human TLS polymerases η, ι, and κ were capable of weak (at least 2 orders of magnitude lower) insertion opposite, and extension from, 3-m-c3-dA. Our studies, using an A-tract sequence, confirm this finding for T7 Sequenase and Klenow fragment (exo−). In terms of 3-mA, if there is any insertion opposite, and extension from this lesion by any of the polymerases, it is certainly inefficient. We appreciate the fact that genetic studies indicate that in vitro experiments may not accurately reflect the ability of polymerases to by pass the lesions in vivo [17, 38].
There has been no study on the effect of c3A on DNA polymerization, although the impact of 3-deazaguanine (c3G) on a number of polymerases has been reported; c3G is a block to Klenow fragment because of the removal of the minor groove nitrogen H-bond acceptor [39–41]. Polη does not require this contact and it can synthesize past c3G, albeit with a much reduced efficiency [42]. In our studies, c3A behaves similar to c3G, but is a significantly less potent block to polymerization than 3-m-c3A, which implies the important steric consequences of the minor groove methyl group.
For reasons mentioned above, we specifically looked at polymerization past 3-m-c3A and c3A lesions when they were placed at different positions within an A-tract sequence. We found no evidence for a positional effect using T7 Sequenase, Klenow fragment (exo−) or Polη in a runup assay. Therefore, our data do not suggest that the mutations produced by Me-lex within the A4 tract are due to sequence dependent bypass of 3-mA. Because our mutation studies with Me-lex in yeast indicated a common pattern with defective base excision repair mutants, including mag1, apn1apn2 and mag1apn1apn2, we hypothesized that abasic sites formed from the hydrolytic or enzymatic depurination of 3-mA were responsible for the mutations [16]. Accordingly, we investigated the sequence-dependent by-pass of a THF abasic site at the different positions within the A4-tract. There is a clear enhancement in bypass by T7 Sequenase and Polη as the lesion is moved from the 3′- to the 5′-side of the A-tract (Figures 9 and 10). This effect was observed with polymerases that lack exonuclease activity; polymerase with exoactivity stalls at the THF site regardless of its position in the A4 tract. As predicted by the A-rule, there is preferential insertion of dATP opposite the THF [26, 43–45]. The more efficient bypass of THF when it is located toward the 5′-side of an A-tract by a Y-family polymerase lacking exoactivity, may be one component of the sequence-specific A→T mutation pattern induced by Me-lex that is the same in yeast lacking Mag1, Apn1/Apn2, and Mag1/Apn1/Apn2 activity [16]. Certainly, the weak, albeit sequence selective mutagenicity observed in vivo, can involve bypass of 3-mA and/or AP sites by other TLS polymerases and polymerase cofactors. Accordingly, the error free and error prone bypass of 3-mA via recombination pathways is under investigation.
In summary, 3-mA, similar to that previously observed for 3-m-c3A [17], is a strong block of DNA polymerization. In comparison, c3A in which the basic nitrogen has been deleted but which lacks the methyl group has less of an effect on DNA polymerization. This indicates that the hydrophobic methyl group may sterically block critical interactions with the polymerases, including the Y-family polymerase Polη. The inability of polymerases to efficiently bypass 3-mA is consistent with the high toxicity and low mutagenicity of Me-lex [15, 16, 32, 37, 38]. The observation that there is a strong sequence-dependency in the bypass of THF within the A4 sequence by polymerases lacking exonuclease activity, for example, Polη, may explain the common sequence specific mutation pattern induced by Me-lex in a number of base excision repair backgrounds. Whether any of the other bypass TLS polymerases may be involved in the weak mutagenicity of Me-lex is also under investigation.
Acknowledgments
This work was supported by NIH Grants RO1 CA29088 (BG) and T32 CA009476 (SS), and AIRC (GF). The authors thank the Molecular Biology Shared Facility at the University of Nebraska Medical Center Eppley Institute (5P30 CA036727) for the preparation of some of the oligomers used in these studies and Silvia Tornaletti for critical comments.
Abbreviations
- c3A:
3-deazaadenine,
- c3G:
3-deazaguanine,
- DFO:
Duplex forming oligomer,
- DMS:
Dimethyl sulphate,
- 3-m-c3A:
3-methyl-3-deazaadenine,
- 3-mA:
N3-methyladenine,
- 7-mG:
N7-methylguanine,
- Me-lex:
{1-methyl-4-[1-methyl-4-(3-methoxysulfonylpropanamido)pyrrole-2-carboxamido]pyrrole-2-carboxamido}propane,
- MMS:
Methyl methanesulfonate,
- MNNG:
N-methyl-N-nitroso-N′-nitroguanidine,
- THF:
Tetrahydrofuran abasic site.
References
- 1.Bennet RA, Pegg AE. Alkylation of DNA in rat tissues following administration of streptozotocin. Cancer Research. 1981;41(7):2786–2790. [PubMed] [Google Scholar]
- 2.Tentori L, Graziani G. Pharmacological strategies to increase the antitumor activity of methylating agents. Current Medicinal Chemistry. 2002;9(13):1285–1301. doi: 10.2174/0929867023369916. [DOI] [PubMed] [Google Scholar]
- 3.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO Journal. 1982;1(2):211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrows LR, Magee PN. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis. 1982;3(3):349–351. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- 5.Bartsch H, Ohshima H, Shuker DEG, Pignatelli G, Calmels S. Exposure of humans to endogenous N-nitroso compounds: implications in cancer etiology. Mutation Research. 1990;238(3):255–267. doi: 10.1016/0165-1110(90)90017-6. [DOI] [PubMed] [Google Scholar]
- 6.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutation Research. 1999;424(1-2):127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 7.Lawley PD. Carcinogenesis by alkylating agents. In: Searle CE, editor. Chemical Carcinogens. 2nd edition. Vol. 1. Washington, DC, USA: American Chemical Society; 1984. pp. 303–484. (ACS Monograph 182). [Google Scholar]
- 8.Beranek DT, Weis CC, Swenson DH. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis. 1980;1(7):595–606. doi: 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- 9.Den Engelse L, Menkveld GJ, De Brij R-J, Tates AD. Formation and stability of alkylated pyrimidines and purines (including imidazole ring-opened 7-alkylguanine) and alkylphosphotriesters in liver DNA of adult rats treated with ethylnitrosourea or dimethylnitrosamine. Carcinogenesis. 1986;7(3):393–403. doi: 10.1093/carcin/7.3.393. [DOI] [PubMed] [Google Scholar]
- 10.Margison GP, O’Connor PJ. Biological implications of the instability of the N glycosidic bond of 3 methyldeoxyadenosine in DNA. Biochimica et Biophysica Acta. 1973;331(1):349–356. doi: 10.1016/0005-2787(73)90021-x. [DOI] [PubMed] [Google Scholar]
- 11.Fujii T, Saito T, Nakasaka T. Purines. XXXIV. 3-methyladenosine and 3-methyl-2’-deoxyadenosine: their synthesis, glycosidic hydrolysis, and ring fission. Chemical and Pharmaceutical Bulletin. 1989;37(10):2601–2609. [Google Scholar]
- 12.Boiteux S, Huisman O, Laval J. 3-methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO Journal. 1984;3(11):2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutation Research. 1985;150(1-2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Chen F-X, Mehta P, Gold B. Groove- and sequence-selective alkylation of DNA by sulfonate esters tethered to lexitropsins. Biochemistry. 1993;32(31):7954–7965. doi: 10.1021/bi00082a017. [DOI] [PubMed] [Google Scholar]
- 15.Kelly JD, Inga A, Chen F-X, et al. Relationship between DNA methylation and mutational patterns induced by a sequence selective minor groove methylating agent. Journal of Biological Chemistry. 1999;274(26):18327–18334. doi: 10.1074/jbc.274.26.18327. [DOI] [PubMed] [Google Scholar]
- 16.Monti P, Campomenosi P, Ciribilli Y, et al. Influences of base excision repair defects on the lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent. Journal of Biological Chemistry. 2002;277(32):28663–28668. doi: 10.1074/jbc.M203384200. [DOI] [PubMed] [Google Scholar]
- 17.Plosky BS, Frank EG, Berry DA, Vennall GP, Mcdonald JP, Woodgate R. Eukaryotic Y-family polymerases bypass a 3-methyl-2′-deoxyadenosine analog in vitro and methyl methanesulfonate-induced DNA damage in vivo. Nucleic Acids Research. 2008;36(7):2152–2162. doi: 10.1093/nar/gkn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irani RJ, SantaLucia J., Jr. The synthesis of anti-fixed 3-methyl-3-deaza-2′-deoxyadenosine and other 3h-imidazo[4,5-c]pyridine analogs. Nucleosides, Nucleotides and Nucleic Acids. 2002;21(11-12):737–751. doi: 10.1081/NCN-120016477. [DOI] [PubMed] [Google Scholar]
- 19.Liang G, Gannett P, Shi X, Zhang Y, Chen F-X, Gold B. DNA sequencing with the hydroperoxide of tetrahydrofuran. Journal of the American Chemical Society. 1994;116(3):1131–1132. [Google Scholar]
- 20.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods in Enzymology. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 21.Kopka ML, Yoon C, Goodsell D, Pjura P, Dickerson RE. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coll M, Aymami J, van der Marel GA, van Boom JH, Rich A, Wang AH-J. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989;28(1):310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- 23.Shah D, Kelly J, Zhang Y, et al. Evidence in Escherichia coli that N3-methyladenine lesions induced by a minor groove binding methyl sulfonate ester can be processed by both base and nucleotide excision repair. Biochemistry. 2001;40(6):1796–1803. doi: 10.1021/bi0024658. [DOI] [PubMed] [Google Scholar]
- 24.Chakravarti D, Ibeanu GC, Tano K, Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. Journal of Biological Chemistry. 1991;266(24):15710–15715. [PubMed] [Google Scholar]
- 25.Engelward BP, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO Journal. 1996;15(4):945–952. [PMC free article] [PubMed] [Google Scholar]
- 26.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase η . Proceedings of the National Academy of Sciences of the United States of America. 2001;98(15):8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker MD, Green SB, Byar DP, et al. Randomized comparisons of radiotherapy and nitrosoureas for the treatment of malignant glioma after surgery. New England Journal of Medicine. 1980;303(23):1323–1329. doi: 10.1056/NEJM198012043032303. [DOI] [PubMed] [Google Scholar]
- 28.Mahaley MS., Jr. Neuro-oncology index and review (adult primary brain tumors). Radiotherapy, chemotherapy, immunotherapy, photodynamic therapy. Journal of Neuro-Oncology. 1991;11(2):85–147. doi: 10.1007/BF02390173. [DOI] [PubMed] [Google Scholar]
- 29.Encell L, Shuker DEG, Foiles PG, Gold B. The in vitro methylation of DNA by a minor groove binding methyl sulfonate ester. Chemical Research in Toxicology. 1996;9(3):563–567. doi: 10.1021/tx9501849. [DOI] [PubMed] [Google Scholar]
- 30.Beranek DT, Weis CC, Swenson DH. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis. 1980;1(7):595–606. doi: 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- 31.Varadarajan S, Shah D, Dande P, et al. DNA damage and cytotoxicity induced by minor groove binding methyl sulfonate esters. Biochemistry. 2003;42(48):14318–14327. doi: 10.1021/bi0353272. [DOI] [PubMed] [Google Scholar]
- 32.Shah D, Gold B. Evidence in Escherichia coli that N3-methyladenine lesions and cytotoxicity induced by a minor groove binding methyl sulfonate ester can be modulated in vivo by netropsin. Biochemistry. 2003;42(43):12610–12616. doi: 10.1021/bi035315g. [DOI] [PubMed] [Google Scholar]
- 33.Engelward BP, Allan JM, Dreslin AJ, Kelly JD, Gold B, Samson LD. 3-Methyladenine DNA lesions induce chromosome aberrations, cell cycle delay, apoptosis and p53. Journal of Biological Chemistry. 1998;273:5412–5418. doi: 10.1074/jbc.273.9.5412. [DOI] [PubMed] [Google Scholar]
- 34.Bobola MS, Varadarajan S, Smith NW, et al. Human glioma cell sensitivity to the sequence-specific alkylating agent methyl-lexitropsin. Clinical Cancer Research. 2007;13:612–620. doi: 10.1158/1078-0432.CCR-06-1127. [DOI] [PubMed] [Google Scholar]
- 35.Russo D, Fronza G, Ottaggio L, et al. High frequency of genomic deletions induced by Me-lex, a sequence selective N3-adenine methylating agent, at the Hprt locus in Chinese hamster ovary cells. Mutation Research. 2009;671(1-2):58–66. doi: 10.1016/j.mrfmmm.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Op het Veld CW, Jansen J, Zdzienicka MZ, Vrieling H, van Zeeland AA. Methyl methanesulfonate-induced hprt mutation spectra in the Chinese hamster cell line CHO9 and its xrcc1-deficient derivative EM-C11. Mutation Research. 1998;398(1-2):83–92. doi: 10.1016/s0027-5107(97)00243-1. [DOI] [PubMed] [Google Scholar]
- 37.Monti P, Iannone R, Campomenosi P, et al. Nucleotide excision repair defect influences lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent in the absence of base excision repair. Biochemistry. 2004;43(19):5592–5599. doi: 10.1021/bi035968x. [DOI] [PubMed] [Google Scholar]
- 38.Monti P, Ciribilli Y, Russo D, et al. Rev1 and Polζ influence toxicity and mutagenicity of Me-lex, a sequence selective N3-adenine methylating agent. DNA Repair. 2008;7(3):431–438. doi: 10.1016/j.dnarep.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spratt TE. Klenow fragment-DNA interaction required for the incorporation of nucleotides opposite guanine and O6-methylguanine. Biochemistry. 1997;36(43):13292–13297. doi: 10.1021/bi971253g. [DOI] [PubMed] [Google Scholar]
- 40.Spratt TE. Identification of hydrogen bonds between Escherichia coli DNA polymerase I (Klenow fragment) and the minor groove of DNA by amino acid substitution of the polymerase and atomic substitution of the DNA. Biochemistry. 2001;40(9):2647–2652. doi: 10.1021/bi002641c. [DOI] [PubMed] [Google Scholar]
- 41.McCain MD, Meyer AS, Schultz SS, Glekas A, Spratt TE. Fidelity of mispair formation and mispair extension is dependent on the interaction between the minor groove of the primer terminus and Arg668 of DNA polymerase I of Escherichia coli. Biochemistry. 2005;44(15):5647–5659. doi: 10.1021/bi047460f. [DOI] [PubMed] [Google Scholar]
- 42.Washington MT, Wolfle WT, Spratt TE, Prakash L, Prakash S. Yeast DNA polymerase η makes functional contacts with the DNA minor groove only at the incoming nucleoside triphosphate. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(9):5113–5118. doi: 10.1073/pnas.0837578100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boiteux S, Laval J. Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry. 1982;21(26):6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- 44.Schaaper RM, Kunkel TA, Loeb LA. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proceedings of the National Academy of Sciences of the United States of America. 1983;80(2):487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauss BS. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair. 2002;1(2):125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]