Abstract
ADAR1 catalyzes the deamination of adenosine to inosine in double-stranded RNA. This RNA-editing enzyme is now shown to be involved in hematopoiesis, where it acts to suppress interferon signaling and to block premature apoptosis.
RNA editing is a post-transcriptional processing mechanism that results in an RNA sequence that is different from that encoded by the genomic DNA and thereby diversifies the gene product and function. The type of RNA editing that is most prevalent in higher eukaryotes converts adenosine residues into inosine (A-to-I editing) in double-stranded RNA (dsRNA) through the action of ADAR enzymes (‘adenosine deaminase acting on RNA’)1. One of the three mammalian ADAR family members, ADAR1, is essential, as demonstrated by early embryonic death of ADAR1- null (Adar−/−) mice around gestational days 11.5–12 because of widespread apoptosis and defective hematopoiesis2,3. Further studies of ADAR1 function with conventional Adar−/− mice have thus been hampered and the precise mechanism through which ADAR1 affects hematopoiesis has remained unclear. In this issue of Nature Immunology, Orkin and colleagues report the creation of mutant mouse lines with inducible ADAR1 deficiency and identify an important function for ADAR1 in the maintenance of hematopoietic stem cells (HSCs)4. ADAR1 seems to suppress the deleterious effects of activation of an interferon signaling pathway and to protect HSCs from apoptosis, preventing their destruction in fetal liver as well as adult bone marrow.
Orkin and colleagues first note that the frequency of long-term HSCs (LT-HSCs) and short-term HSCs (ST-HSCs) relative to that of hematopoietic multipotent progenitors (MPPs) is much higher in Adar−/− fetal liver than in wild-type fetal liver. In contrast, both the relative frequency and absolute numbers of more differentiated hematopoietic progenitors are much lower in the mutant mice. These findings indicate not only the dispensability of ADAR1 for the emergence of HCS and their migration to the fetal liver but also the potential disruption of a process affecting the differentiation of HSCs and/or proliferation of progenitors in Adar−/− fetal liver. They next create a mutant mouse line with inducible ADAR1 deficiency (Adarf/−; Mx1-Cre) for transplant studies using fetal liver cells at embryonic day 14.5. When Mx1-Cre expression is induced in transplanted fetal liver–derived cells by dsRNA, the contribution of ADAR1-null cells to peripheral blood leukocytes (all myeloid cells and B and T lymphoid cells) is much lower by 2 weeks and almost disappears by 23 weeks after induction. Bone marrow analyses confirm the loss of the contribution by ADAR1-null cells to multilineage cells as well as the considerable decrease in ADAR1-null-derived HSCs. Using yet another mutant mouse line with tamoxifen-inducible ADAR1 deficiency, the authors further investigate ADAR1 function in adult HSCs. Again, with this mouse model, the frequency of HSCs is much higher and the frequency of MPPs is much lower in ADAR1-null bone marrow.
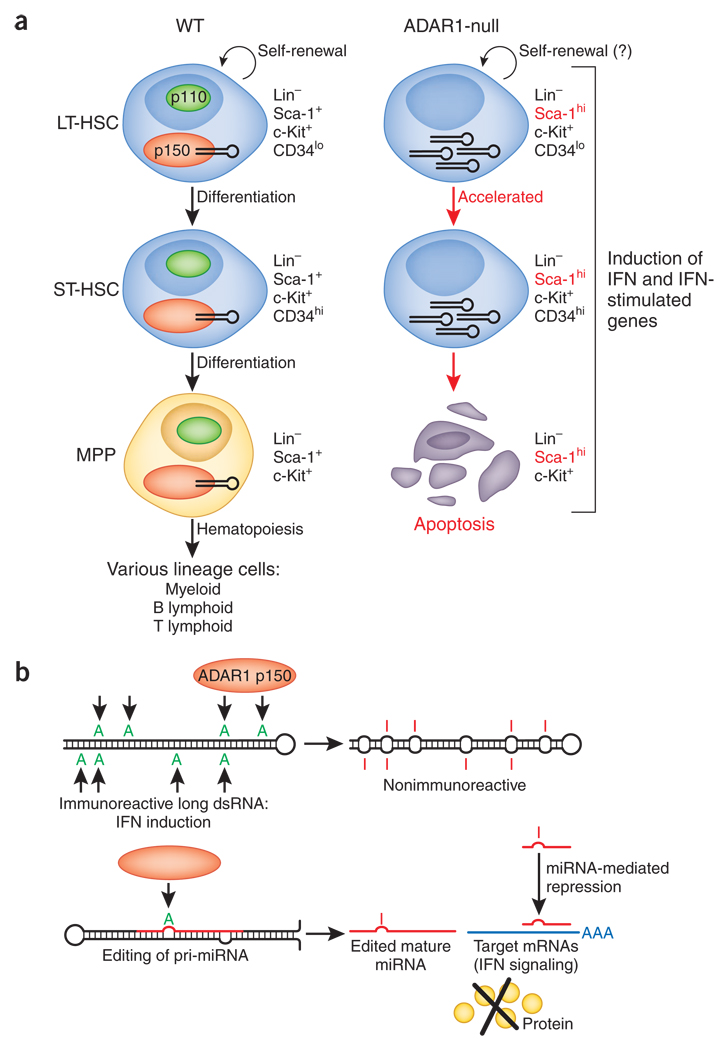
These findings indicate a cell-autonomous requirement for ADAR1 in the maintenance of HSCs in both fetal liver and adult bone marrow. The maintenance of HSCs is regulated by a delicately coordinated balance among self-renewal, differentiation and apoptosis5. Cell cycle analysis of HSCs and progenitor cells shows that resting HSCs undergo slightly more apoptosis but that cycling HSCs undergo much more apoptosis (68%) in ADAR1-null bone marrow than in control bone marrow (3–13%). Notably, cell cycle analysis also shows that ADAR1-null bone marrow has a much larger fraction of S-phase ST-HSCs but not multipotent progenitors. Furthermore, ADAR1-null bone marrow has many fewer LT-HSCs than does control bone marrow. These results suggest exhaustion of LT-HSCs due to continuous activation of the stem cell compartment. Orkin and colleagues propose a model for how ADAR1 functions in HSC biology (Fig. 1a). In this model, ADAR1 is an essential regulator of HSC maintenance but is dispensable for the emergence of LT-HSCs and their self-renewal. However, ADAR1 becomes essential for the survival of HSCs by protecting against apoptosis as these cells progress to the multipotent progenitor stage.
Figure 1.
A-to-I RNA editing of an as-yet-unknown dsRNA by ADAR1 is required for the maintenance of HSCs. (a) In the presence of ADAR1, especially the p150 isoform LT-HSCs are able to self-renew (semicircular arrows) differentiate into ST-HSCs and then into MPPs defined by surface expression of lineage markers (Lin), c-Kit, Sca-1 and CD34. In Adar−/− HSCs and MPPs, more interferon and global upregulation of interferon-stimulated genes, including the gene encoding Sca-1, are detected. Unregulated activation of an interferon signaling pathway in Adar−/− HSCs leads to rapid apoptosis after they differentiate into MPPs. The possibility that ADAR1 is required also for the self-renewal of HSCs is not completely eliminated (right question mark). (b) The ADAR1 target dsRNA critical for suppression of the interferon signaling pathway and consequent protection of HSCs is unknown. Among several possible scenarios, two are presented here: A-to-I editing of noncoding long hairpin dsRNA molecules, such as those made from Alu and long interspersed repetitive elements, may decrease their immunoreactive properties (top); alternatively, A-to-I editing of a specific primary transcript of microRNA (pri-miRNA) and consequent expression of a sequence-altered mature microRNA (edited mature miRNA) may lead to silencing of target genes, perhaps those involved in regulating the interferon signaling pathway (bottom).
Notably, they detect higher expression of interferon-inducible full-length ADAR1 p150 than of the constitutively expressed shorter ADAR1 p110 in wild-type HSCs, which may indicate a possible requirement for ADAR1 p150 in interferon-induced pathways in hematopoiesis. ADAR1 p110 localizes in the nucleus, whereas expression of ADAR1 p150 is detected both in the nucleus and cytoplasm; the presence of ADAR1 p150 substrate RNA specifically in the cytoplasm has been proposed before1. Genome-wide transcriptome analysis of ADAR1-null fetal liver HSCs shows global upregulation of transcripts known to be induced by type I interferon (IFN-α and IFN-β), type II interferon (IFN-γ) or both, including transcripts encoding the transcription factors STAT1 and STAT2, the interferon-regulatory factors IRF1, IRF7 and IRF9, the GTPases Mx1 and Mx2 and the RNA-activated protein kinase PKR. Notably, ADAR1 deficiency also results in upregulation of interferon- inducible transcripts in fetal liver cells of the erythroid lineage but not those of the myeloid lineage. However, the requirement for ADAR1 in specific lineages may depend on the environment (‘niches’) in which HSCs emerge; that is, fetal liver or adult bone marrow. For example, although ADAR1 is shown to be required for differentiation into myeloid cells as well as B and T lymphoid cells in fetal liver, no substantial effects are detected for the differentiation of myeloid and B lineage cells in adult Adar−/− bone marrow, which indicates that more careful analysis is needed to define the absolute cell lineage–specific requirement for ADAR1.
Several important questions remain to be answered. What is the mechanism underlying this requirement for ADAR1 in HSCs? How does ADAR1 suppress the interferon signaling pathway? Given that ADAR1 is a dsRNA-editing enzyme, Orkin and colleagues investigate the possible involvement of an essential dsRNA substrate. As expected, they find that some interferon-inducible genes that are upregulated in ADAR1-null cells are also genes known to be activated by dsRNA, including several encoding dsRNA-binding proteins. Perhaps these results indicate the presence of an immunoreactive dsRNA that elicits the interferon signaling pathway in ADAR1-null HSCs because of lack of A-to-I editing of this as-yet-unknown RNA molecule. Notably, published work has reported interaction of ADAR1 with NF90, a dsRNA-binding protein, as well as involvement of ADAR1 in controlling NF90-regulated genes6. The ADAR1-NF90 interaction is ‘bridged’ by RNA and is independent of the ADAR1 A-to-I RNA editing activity itself6. Orkin and colleagues point out that the ADAR1-null expression ‘signature’ correlates with the transcription profiles of cells that express a variant NF90 isoform, which suggests that the ADAR1-NF90 interaction may be a critical process underlying the ADAR1-null HSC phenotype. This finding indicates that Adar−/− embryos would be ‘rescued‘’ by ‘knock-in’ of an ADAR1 mutant with an intact dsRNA-binding domain but defective catalytic function. It is particularly notable that the interferon-inducible ADAR1 p150 isoform that has high expression in HSCs shows very high affinity for certain short dsRNA molecules such as small interfering RNA1. Furthermore, a published report indicates that ADAR1 p150 acts as a cell-intrinsic sensor of imunoreactive DNA and suppresses interferon synthesis in mouse embryonic fibroblast cells7, which suggests yet another possible mechanism that may also function in HSCs.
Further studies show that ADAR1 deficiency results in the induction of type I interferon but not IFN-γ in fetal liver HSCs and erythroid cells as well as in the remaining embryo body. The authors suggest that this widespread interferon induction is due to the circulation of interferon secreted from fetal liver HSCs and erythroid cells or to other indirect effects of ADAR1 deficiency. This result could explain the reported widespread apoptosis in Adar−/− embryos at gestational day 11.5 (ref. 3). However, those studies have shown that the apoptosis detected in Adar−/− embryos overlaps with the area in which high expression of ADAR1 is detected in wild-type embryos, leading to the proposal that widespread apoptosis results directly from ablation of ADAR1 expression and is not due to indirect effects3.
The most interesting and unresolved issue is the identity of the dsRNA(s) required for the ADAR1-mediated regulation of interferon signaling and maintenance of HSCs. As Orkin and colleagues point out, the critical ADAR1 targets could be transcripts of protein-encoding genes as well as noncoding RNA containing repeats of repetitive elements such as inverted Alu repeats (a class of short interspersed nuclear elements) and long interspersed elements located in introns and untranslated regions8,9. These elements may be immunoreactive and induce interferon if double-strandedness is not diminished through ADAR1-mediated A-to-I RNA editing (Fig. 1b, top). Alternatively, ADAR1 may function as a potent dsRNA-binding factor, as discussed before for its interaction with NF90. Another ‘suspect’ is the primary transcript of microRNA (Fig. 1b, bottom). ADAR1 has been demonstrated to edit highly specific sites of certain primary transcripts of microRNA in various adult tissues and also in developing mouse embryos10–12. A-to-I editing of such transcripts suppresses their processing by Drosha or Dicer10,12 or results in expression of microRNA with altered sequences and silencing of genes different from those targeted by unedited microRNA11. It is plausible that a key gene essential for induction of interferon pathway in HSCs is regulated specifically by an edited microRNA.
References
- 1.Nishikura K. Nat. Rev. Mol. Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartner JC, et al. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, et al. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 4.Hartner JC, Walkley CR, Lu J, Orkin SH. Nat. Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orkin SH, Zon LI. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie Y, Ding L, Kao PN, Braun R, Yang JH. Mol. Cell. Biol. 2005;25:6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, et al. Proc. Natl. Acad. Sci. USA. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Athanasiadis A, Rich A, Maas S. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levanon EY, et al. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 10.Kawahara Y, et al. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawahara Y, et al. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, et al. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]