Abstract
Recent whole-genome sequencing efforts led to the identification of IDH1R132 mutations in AML patients. We studied the prevalence and clinical implications of IDH1 genomic alterations in pediatric and adult AML. Diagnostic DNA from 531 AML patients treated on Children’s Oncology Group trial COG-AAML03P1 (N=257), and Southwest Oncology Group trials SWOG-9031, SWOG-9333, and SWOG-9500 (N=274), were tested for IDH1 mutations. Codon R132 mutations were absent in the pediatric cohort, but were found in 12/274 adult patients (4.4%, 95% CI 2.3-7.5%). IDH1R132 mutations occurred most commonly in patients with normal karyotype, and those with FLT3/ITD and NPMc mutations. Patients with IDH1R132 mutations trended towards higher median diagnostic WBC counts (59.2 × 109/L vs. 29.1 × 109/L, P=0.19) than those without mutations, but the two groups did not differ significantly in age, bone marrow blast percentage, overall survival, or relapse-free survival. Eleven patients (2.1%) harbored a novel V71I sequence alteration, which was found to be a germline polymorphism. IDH1 mutations were not detected in pediatric AML, and are uncommon in adult AML.
Keywords: Acute Myeloid Leukemia, IDH1, isocitrate dehydrogenase, pediatric AML, NPM, FLT3
INTRODUCTION
The IDH1 gene encodes isocitrate dehydrogenase 1, a citric acid cycle enzyme that catalyzes the oxidative decarboxylation of isocitrate to alpha-ketoglutarate. Function-altering mutations of IDH1 codon 132 occur frequently (>70%) in low grade gliomas and secondary glioblastoma multiforme.1-5 Screening for IDH1R132 mutations has been performed in a variety of cancers; such mutations are rare in tumors outside the CNS.3,4,6 Although IDH1 mutations were found in 1.7% of patients with B-cell ALL, no mutations were reported in 145 AML patients screened.4,6 Recently, whole-genome sequencing performed on tumor as well as germline DNA from an AML patient with known NPMc+ and NRAS mutations led to the discovery of an IDH1R132 mutation;7 subsequent analyses of 187 patients with AML treated on multiple clinical protocols detected IDH1R132 mutations in 8.5% (16/188) of adult patients, corresponding to 16.3% (13/80) of those with normal cytogenetics. The study further suggested a possible adverse outcome for those with mutated IDH1 and wild-type NPM. To define the prevalence and potential clinical implications of IDH1 mutations in unselected AML patients, we genotyped diagnostic marrow DNA obtained from 1 pediatric and 3 adult AML trials for identification of IDH1 exon 4 mutations.
PATIENTS, MATERIALS, AND METHODS
Newly diagnosed patients with de novo AML enrolled on either the pediatric AML trial COG-AAML03P1, or 1 of 3 SWOG adult AML trials (SWOG-9031, SWOG-9333, and SWOG-9500), were eligible for this study. SWOG-9333 patients randomized to induction chemotherapy with mitoxantrone and etoposide were excluded, as the results of that trial demonstrated somewhat poorer outcomes for the mitoxantrone / etoposide treatment arm.8 COG-AAML03P1 enrolled patients aged 0 to 21 years; SWOG-9031 and SWOG-9333 enrolled “older” adult AML patients (> 55 years), while SWOG-9500 enrolled “younger” adult AML patients (18-55 years). Patients with acute promyelocytic leukemia (M3 AML) were excluded. Details of these clinical trials have been published.8-11 Genomic DNA extracted from diagnostic marrow specimens was available from 257 COG and 274 SWOG patients. In accordance with the Declaration of Helsinki, consent was obtained from all study participants for treatment on their respective clinical trials and evaluation of tissue samples. Institutional review board approval was obtained from the Fred Hutchinson Cancer Research Center prior to mutation analysis, and this study was approved by the Myeloid Disease Biology Committees of the COG and SWOG.
IDH1 Molecular Analysis
We amplified the entire coding region of IDH1 exon 4, containing codon R132, by performing polymerase chain reaction (PCR) using the following primer pair: IDH1F (5’-CTCAGAGCCTTCGCTTTCTG- 3’) and IDH1R (5’-GCAAAATCACATTATTGCCAAC-3’). Thermocycler conditions were as follows: 94°C for 5 minutes; 35 cycles at 94°C for 30 seconds, 58°C for 45 seconds, and 72°C for 1 minute; and a final extension step at 72°C for 7 minutes. Purified PCR products were sequenced using the BigDye® Terminator sequencing reaction and run on an ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA).
Statistical Methods
This study included all eligible patients on studies COG-AAML03P1, SWOG-9031, SWOG-9333, and SWOG-9500 for whom DNA was available for analysis. Data regarding clinical characteristics and treatment outcomes were collected and evaluated according to the standard practices of the COG and SWOG for their respective studies. Overall survival (OS) was calculated as time from study entry until death, with observation censored at the date of last contact for patients last know to be alive. Relapse-free survival (RFS) was calculated as time from complete remission (CR) until relapse or death, whichever occurred first, with observation censored at the date of last contact for patients last known to be alive without report of relapse.
RESULTS
Patient Population
Cryopreserved diagnostic specimens were available from 257 (76%) of the 340 eligible pediatric patients enrolled on COG-AAML03P1, and 274 (56%) of the 487 eligible adult patients enrolled on SWOG-9031, SWOG-9333 and SWOG-9500. Demographics, laboratory and clinical characteristics, and outcome for those with and without available specimens were compared. SWOG patients included in this study had significantly higher (P<0.001) median diagnostic white blood cell (WBC) count (29.8 × 109/L vs. 4.6 × 109/L), peripheral blood blast percentage (42% vs. 12%), and marrow blast percentage (71% vs. 59%) than the 213 patients without available samples. The two groups did not differ significantly in induction CR rate, although SWOG patients included in this study had somewhat improved OS (17% vs. 9%, P=0.094) and RFS (22% vs. 8%, P=0.012) compared to adult patients whose samples were not studied.
IDH1 Mutation Analysis
We aimed to determine the prevalence and clinical implications of IDH1 mutations in both pediatric and adult AML. Of the 531 (257 COG, 274 SWOG) diagnostic AML specimens screened, 70 contained nucleotide substitutions involving 3 distinct codons (Figure 1). R132 missense mutations were identified in 12 patients, in which arginine was replaced with 1 of 4 different amino acids (8 patients with R132H, 2 with R132L, 1 with R132G and 1 with R132C). In addition, 58 patients had silent G105G polymorphisms; 11 patients with G105G harbored an additional, novel V71I sequence alteration (Figure 1). Overall, IDH1R132 mutations were found in 12/531 (2.3%, 95% CI 1.2-3.9%) patients in our study cohort, while IDH1V71I,G105G was present in 11/531 patients (2.1%, 95% CI 1.0-3.7%).
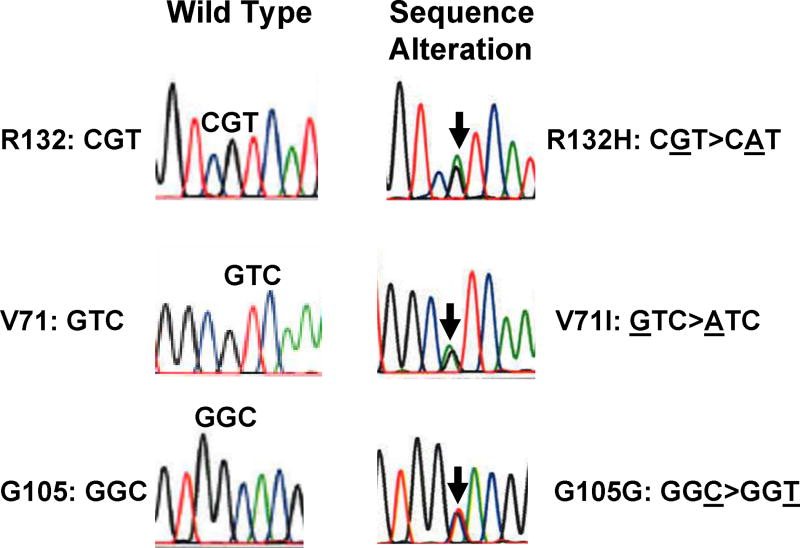
Figure 1. Representative sequence alterations of the IDH1 gene, as shown on electropherogram.
Wild-type and heterozygous mutant sequences are shown for the affected amino acid residues: R132, V71, and G105.
Pediatric AML
In the COG cohort (median age 9.8 years), 37 nucleotide substitutions were identified in 30/257 (11.7%; 95% CI, 8.0%-16.2%) patients tested. G105G silent mutations accounted for 30 of these, and 7 patients (2.7%, 95% CI 1.1%-5.5%) harbored the additional V71I alteration. Previously described IDH1R132 mutations were not detected (0.0%, 95% CI 0.0%-1.4%). Cytogenetic data were available for 235 (91%) patients, of whom 47 (20%) lacked abnormalities, 62 (26%) had core-binding factor (CBF) translocations, and 49 (21%) had 11q23 abnormalities. Since function-altering mutations in neoplasia have only been previously described at codon R132 of IDH1, we sought to determine whether V71I might represent a novel germline polymorphism. Remission specimens from 3 patients with IDH1V71I,G105G were available and were examined by direct sequencing. In 3/3 cases, IDH1V71I,G105G was detected in paired germline DNA obtained from remission specimens, confirming that IDH1V71I,G105G is a naturally occurring polymorphism. Thus, disease-associated mutations were not identified in childhood AML.
Adult AML
In the 274 adult AML patients (median age 63 years) treated on the 3 SWOG clinical trials, we detected 44 nucleic acid substitutions in 40 patients; 12 patients (4.4%, 95% CI 2.3-7.5%) harbored R132 mutations, 4 patients (1.5%, 95% CI 0.4-3.7%) had both V71I and Gl05G, and 24 patients (8.8%, 95% CI 5.7-12.8%) had G105G alone. Characteristics of the adult AML patients are shown by R132 mutation status in Table 1. Evaluation of IDH1 mutations by age category in adult AML demonstrated R132 mutations were detected in 3/82 (3.7%, 95% CI 0.8-10.3%) of younger adults enrolled on SWOG-9500, compared to 9/192 (4.7%, 95% CI 2.2-8.7%) of older adults enrolled on SWOG-9031 and SWOG-9333.
Table 1.
Characteristics of 274 adult AML patients, by R132 mutation status.
| R132 Mutation (N =12) | R132 Wildtype (N = 262) | |||||
|---|---|---|---|---|---|---|
| Patients | % | Patients | % | P a | ||
| Gender | Female | 4 | 33% | 122 | 47% | 0.56 |
| Male | 8 | 67% | 140 | 53% | ||
| Race | Asian | 1 | 8% | 6 | 2% | 0.34 |
| Black | 0 | 0% | 26 | 10% | ||
| Native American/ Alaskan | 0 | 0% | 1 | 0.4% | ||
| White | 11 | 92% | 226 | 86% | ||
| Unknown | 0 | 0% | 3 | 1% | ||
| Karyotype (10 R132+, 193 WT) b | Normal | 6 | 60% | 79 | 41% | 0.33 |
| +8 | 1 | 10% | 27 | 14% | 1.00 | |
| -7/del(7q) | 0 | 0% | 16 | 8% | 1.00 | |
| CBF abnormality | 0 | 0% | 22 | 11% | 0.61 | |
| Abn(11q23) | 0 | 0% | 9 | 5% | 1.00 | |
| Other clonal abnormalities | 2 | 20% | 44 | 23% | 1.00 | |
| Nonclonal abnormality | 1 | 10% | 6 | 3% | 0.30 | |
| FLT3/ITD (12 R132+, 243 WT) | Present | 6 | 50% | 83 | 34% | 0.35 |
| Absent | 6 | 50% | 160 | 66% | ||
| Median | Min-Max | Median | Min-Max | P a | ||
| Age (yrs) | 61 | 34-81 | 63 | 18-88 | 0.59 | |
| BM Blasts (%; 11 R132+, 246 WT) | 80% | 38-99% | 71% | 0-99% | 0.59 | |
| WBC (×109/L) | 59.2 | 1.2-98.2 | 29.1 | 0.7-298.0 | 0.19 | |
| Circ. Blasts (%; 10 R132+, 255 WT) | 48% | 4-99% | 42% | 0-99% | 0.52 | |
| Platelets (×109/L; 11 R132+, 262 WT) | 53 | 10-189 | 57 | 2-1052 | 0.98 | |
| Hemoglobin (g/dL; 12 R132+, 256 WT) | 9.5 | 6.0-12.2 | 9.2 | 4.3-14.3 | 0.67 | |
| Estimate | 95% CI | Estimate | 95% CI | P a | ||
| Response to induction chemotherapy (%) | Complete response | 75% | 43-95% | 51% | 45-57% | 0.14 |
| Resistant disease | 17% | 2-48% | 29% | 24-35% | 0.52 | |
| Other, not evaluable | 8% | --- | 20% | --- | ||
| Overall survival at 5 years (%) | 25% | 5-57% | 17% | 12-21% | 0.55 | |
| Relapse-free survival at 5 years (%) | 33% | 7-70% | 22% | 14-29% | 0.51 | |
P-values based on Fisher’s exact test (sex, karyotypes, FLT3/ITD, response); Pearson chi-squared test (race; exact calculation); Wilcoxon rank-sum test (continuous variables); or logrank test (overall and relapse-free survival). Results based on all 274 patients except where indicated.
Total of karyotype categories in R132 Wildtype column exceeds 193 because some patients have multiple clonal abnormalities.
Cytogenetic data were available for 203 (74%) SWOG patients, 85 (41.9%) of whom had normal karyotype. Six of ten IDH1R132 patients with known cytogenetic data had normal karyotype. The prevalence of IDH1R132 mutations in patients with a normal karyotype was 7.1% (6/85, 95% CI 2.6-14.7%). Clonal cytogenetic abnormalities in patients with IDH1R132 included +8, +13, and inv(3) in one patient each (Table 1). We evaluated 255 adult patients for FLT3/ITD,13,14 which was detected in 6/12 (50.0%) patients with IDH1R132 mutations, and in 83/243 (34.2%) patients without such mutations. We then screened patients with mutated IDH1R132 for other AML-associated mutations. NPM exon 12 mutations were common, occurring in 9/12 (75.0%) of patients with IDH1R132 mutations; NRAS exon 1 mutations were detected in 2/12 (16.7%) patients, while CEBPA bZip mutations were detected in 1/12 patients (8.3%), and WT1 mutations were not detected. Nine (75.0%) of the 12 adult AML patients with IDH1R132 achieved CR; 7 of these patients have relapsed or died and 2 remain in long-term remission, (9+ years), for an estimated median RFS of 19 months (95% CI 8-36 months). Two patients (16.7%) with IDH1 R132 had resistant disease (RD) and died within 30 days, while one died at day 19 of induction toxicity and had indeterminate response to induction.
DISCUSSION
In summary, IDH1R132 mutations were detected in 12/531 (2.3%) of all AML specimens screened, while another 11 patients (2.1%) harbored the IDH1V71I,G105G polymorphism. The R132 mutation was not detected in the pediatric cohort; its prevalence was 4.4% in adult AML patients. IDH1R132 mutations occurred most frequently in patients with normal karyotype, and showed significant overlap with FLT3/ITD and NPMc. Interestingly, IDH1R132 mutations were not detected in any of the 257 pediatric patients tested, and were in fact observed only in patients 34 years of age or older. A similar phenomenon has been observed in brain tumors, where, despite the high mutation rate in adult gliomas and supratentorial PNETs, IDH1R132 mutations are almost never observed in pediatric tumors of similar histology.2,4,12 These somatically acquired mutations may uncover a biologically distinct, temporally-acquired pathway to neoplasia that is exclusive to adult patients.
Given the low prevalence of IDH1 mutations, our study did not have adequate power to determine either significant differences in outcome between patients with and without such mutations, or the interaction between IDH1 mutations and other disease-associated mutations. It is notable that 7 of 12 patients with IDH1R132 mutations died of refractory disease or after relapse from CR; one additional patient died of induction toxicity, and two patients died in CR (one of consolidation toxicity, the other of stroke after 8 years). Nonetheless, the small number of patients with IDH1 mutations in our study appeared to have higher rates of CR, OS, and RFS than their wild-type counterparts. As IDH1 mutations are be highly associated with NPM exon 12 mutations, any improvement in outcome in the IDH1-mutated cohort may be related to the favorable prognostic effect of harboring NPMc. Larger prospective studies will need to be performed to define the prognostic relevance of IDH1 mutations in AML, as well as their interaction with other known molecular markers of prognosis.
Finally, the role of IDH1 mutations in AML pathogenesis is not clear. Recent data showed that IDH1R132 mutations result in dominant-negative inhibition of isocitrate dehydrogenase catalytic activity, resulting in the induction of the HIF-1α pathway.15 HIF signalling has been linked to the regulation of myeloid cell survival,16 and HIF-1α expression is increased by activation of the PI3K/Akt pathway,17 a signaling cascade known to be dysregulated in AML. It remains to be determined whether IDH1 mutations contribute to myeloid pathogenesis via HIF-1α activation, or whether these mutations are simply secondary events unrelated to the leukemic process. If the former is true, then antineoplastic agents currently in development that target HIF-1α18 may someday provide a novel therapeutic approach for AML patients with IDH1R132 mutations.
The discovery by Mardis et al7 of an AML-associated mutation in IDH1, a metabolic enzyme not previously implicated in leukemia, highlights the tremendous potential of whole-genome sequencing to further elucidate the molecular biology of AML. As next-generation sequencing technologies become more readily available, their application to the various cytogenetic and molecular subtypes of this heterogeneous disease will allow the discovery of novel genomic alterations. These discoveries should improve disease classification and risk stratification, enhance our understanding of leukemogenesis, and provide novel targets of therapy. Cooperative groups such as COG and SWOG will be provided the opportunity to determine the prevalence and prognostic significance of mutations in newly-recognized genes through retrospective studies, with the ultimate goal of identifying clinically-relevant mutations for upfront risk identification and therapy allocation in future trials.
TABLE 2.
Clinical profile of 12 adult AML patients with IDH1R132 mutations.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | R132H | R132H | R132H | R132H | R132L | R132H | R132H | R132H | R132G | R132L | R123H | R132C |
| Sex | F | M | M | M | F | F | F | M | M | M | M | M |
| Age at diagnosis | 65 | 60 | 64 | 40 | 57 | 62 | 34 | 71 | 64 | 58 | 81 | 52 |
| WBC (×109/L) | 31.9 | 98.2 | 81.7 | 56.3 | 62.1 | 21.9 | 68 | 63.4 | 55 | 94 | 1.2 | 6.3 |
| BM blasts (%) | 80 | 82 | 94 | 82 | NA | 38 | 99 | 44 | 71 | 64 | 80 | 40 |
| Cytogenetics 1 | Nonclonal abn 1 | Normal | NA | Normal | NA | inv(3) | Normal | Normal | Normal | Normal | +13 | +8 |
| NPM exon 12 mutation | + | + | + | + | + | + | + | + | + | |||
| FLT3/ITD | + | + | + | + | + | + | ||||||
| NRAS exon 1 mutation | + | + | ||||||||||
| CEBPA bZip mutation | + | |||||||||||
| Response to induction chemotherapy | CR | CR | Ind 2 | CR | CR | CR | CR | CR | CR | RD | RD | CR |
| Relapse-free survival 3 | Relapse d14 | Relapse d399 | NA 2 | Died in CR d91 | Alive CCR d3696 | Relapse d255 | Alive CCR d3451 | Died in CR d2900 | Relapse d568 | NA 2 | NA 2 | Relapse d1106 |
| Overall survival 4 | Died d113 | Died d688 | Died d19 | Died d139 | Alive d3727 | Died d496 | Alive d3497 | Died d2923 | Died d1203 | Died d20 | Died d22 | Died d1518 |
Normal cytogenetics is defined by the absence of clonal abnormalities. Patient #2 had a single nonclonal abnormality: +18 in one of 50 metaphase cells examined.
Ind = indeterminate (patient died before response assessment); NA = not applicable.
For relapse-free survival, time to event (relapse or death in CR) or last contact is measured from date of CR.
For overall survival, time to death or last contact is measured from date of entry into study.
Acknowledgments
We are grateful to the patients and families who consented to the use of biologic specimens in these trials. We thank the AML Reference Laboratories of the COG and SWOG for providing diagnostic specimens, and Dr. Cherise Guess for scientific editing. This work was supported by the National Institutes of Health (grants No. K23 CA92405, CA18029, CA32102, CA114563, R01 CA114563-01, R21 CA102624, R21 CA10262-01, U10 CA032102-30, CA38926, CA20319, CA27057, CA12213 and Children’s Oncology Group Chair’s grant NIH U10 CA98543).
Footnotes
AUTHORSHIP PH designed research, performed research, analyzed data, and wrote the manuscript.
TAA and KJK served as senior statisticians, performed statistical analyses, and edited the manuscript.
KLM and JK performed research and edited the manuscript.
RBG performed statistical analyses and edited the manuscript.
SCR, BAH, VO, CAH, JLF, ASG, SHP, JEA, GHR, LB, CLW, IDB, JPR, FRA, and DLS analyzed data and edited the manuscript.
SM designed research, analyzed data, and wrote the manuscript.
Conflict of interest disclosure: The authors declare no competing financial interests.
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 3.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 4.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 6.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 7.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring Mutations Found by Sequencing an Acute Myeloid Leukemia Genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson JE, Kopecky KJ, Willman CL, Head D, O’Donnell MR, Luthardt FW, et al. Outcome after induction chemotherapy for older patients with acute myeloid leukemia is not improved with mitoxantrone and etoposide compared to cytarabine and daunorubicin: a Southwest Oncology Group study. Blood. 2002;100:3869–3876. doi: 10.1182/blood-2001-12-0354. [DOI] [PubMed] [Google Scholar]
- 9.Ho PA, Alonzo TA, Gerbing RB, Pollard J, Stirewalt DL, Hurwitz C, et al. Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children’s Oncology Group. Blood. 2009;113:6558–6566. doi: 10.1182/blood-2008-10-184747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leith CP, Kopecky KJ, Godwin J, McConnell T, Slovak ML, Chen IM, et al. Acute myeloid leukemia in the elderly: assessment of multidrug resistance (MDR1) and cytogenetics distinguishes biologic subgroups with remarkably distinct responses to standard chemotherapy. A Southwest Oncology Group study. Blood. 1997;89:3323–3329. [PubMed] [Google Scholar]
- 11.Petersdorf SH, Rankin C, Head DR, Terebelo HR, Willman CL, Balcerzak SP, et al. Phase II evaluation of an intensified induction therapy with standard daunomycin and cytarabine followed by high dose cytarabine for adults with previously untreated acute myeloid leukemia: a Southwest Oncology Group study (SWOG-9500) Am J Hemato. 2007;82:1056–1062. doi: 10.1002/ajh.20994. [DOI] [PubMed] [Google Scholar]
- 12.Hayden JT, Fruhwald MC, Hasselblatt M, Ellison DW, Bailey S, Clifford SC. Frequent IDH1 mutations in supratentorial primitive neuroectodermal tumors (sPNET) of adults but not children. Cell Cycle. 2009;8:1806–1807. doi: 10.4161/cc.8.11.8594. [DOI] [PubMed] [Google Scholar]
- 13.Stirewalt DL, Kopecky KJ, Meshinchi S, Appelbaum FR, Slovak ML, Willman CL, et al. FLT3, RAS, and TP53 mutations in elderly patients with acute myeloid leukemia. Blood. 2001;97:3589–3595. doi: 10.1182/blood.v97.11.3589. [DOI] [PubMed] [Google Scholar]
- 14.Stirewalt DL, Kopecky KJ, Meshinchi S, Engel JH, Pogosova-Agadjanyan EL, Linsley J, et al. Size of FLT3 internal tandem duplication has prognostic significance in patients with acute myeloid leukemia. Blood. 2006;107:3724–3726. doi: 10.1182/blood-2005-08-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walmsley SR, Chilvers ER, Whyte MK. Hypoxia, hypoxia inducible factor and myeloid cell function. Arthritis Res Ther. 2009;11:219–226. doi: 10.1186/ar2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang BH, Liu LZ. PI3K/PTEN signaling in angiogenesis and tumorigenesis. Adv Cancer Res. 2009;102:19–65. doi: 10.1016/S0065-230X(09)02002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onnis B, Rapisarda A, Melillo G. Development of HIF-1 inhibitors for cancer therapy. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00876.x. e-pub ahead of print on 8 August 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]