Abstract
Strategies to control the release rate of bioactive signals from tissue engineering scaffolds are essential for tissue regeneration and tissue engineering applications. Here we report on a strategy to achieve temporal control over nanoparticle release from biomaterials using cell-secreted proteases. This cell-triggered release approach utilizes peptides that are degraded by matrix metalloproteinases (MMPs) at different rates to immobilize nanoparticles directly to the biomaterial surface. Thus, the peptide-immobilized nanoparticles are released with temporal control through the action of cell-released MMPs. We found that release rates of peptide-immobilized nanoparticles were a function of peptide sensitivity to proteases, the number of tethers between the nanoparticle and the surface and the concentration of proteases used to induce release. Cellular internalization of the peptide-immobilized nanoparticles was also a function of the peptide sensitivity to proteases, the number of tethers between the nanoparticle and the surface and MMP expression profile of the cells. Similar trends were observed for peptide-immobilized nanoparticles inside micro-porous hydrogels, indicating protease sensitive tethers are effective in controlling release rate and internalization of nanoparticles. Such a temporal delivery strategy of nanoparticles loaded with therapeutic payloads (e.g. protein, DNA, siRNA) can be an ideal means to guide tissue formation.
Keywords: Controlled release, Nanoparticle, Matrix metalloproteinase, Hydrogel, Cell-mediated release
1. Introduction
Tissue regeneration occurs through a complex sequence of events, which require the presence of various molecules or proteins at different times [1,2]. Regenerative medicine requires that engineered technologies be able to mimic such cellular processes when the body fails to repair itself as a result of disease or serious injury. In cases such as diabetic ulcers, protease over-expression and tissue remodeling overpower the healing process, leaving the body with chronic wounds [3,4]. Bioresponsive scaffolds that provide mechanical, biological, and chemical cues to the diseased tissue have the potential to aid in its regeneration in ways that are not currently possible.
Current state-of-the-art for controlled release of multiple bioactive signals with different release kinetics is achieved through hydrolytically degradable polymer scaffolds. PLGA scaffolds were formed with platelet-derived growth factor (PDGF) embedded into PLGA microspheres to achieve slow release and vascular endothelial growth factor (VEGF) encapsulated into the surrounding polymer to achieve fast release [5]. In addition to being able to provide distinct rates of release for these two growth factors, they were also able to show that the dual and sequential delivery led to significantly enhanced blood vessel formation when compared to delivery of either growth factor on its own. Recent studies have expanded on the idea of hydrolytically degradable scaffolds for controlled release of multiple factors by varying polymer type [6], microparticle encapsulation [7], and microparticle type [8]. Sequential release of surface-coated and encapsulated DNA and DNA polyplexes have also been shown in fibrin hydrogels [9]. However, many of these scaffolds are still limited in their use as the release rates are dictated by the chemistry of the polymer scaffold and are not responsive to the surrounding microenvironment.
Recently researchers have utilized enzymatically degradable polymer scaffolds, which release their payload upon cellular infiltration. Lei and Segura proposed a matrix metalloproteinase (MMP) degradable hydrogel system within which DNA/PEI polyplexes were encapsulated [10]. As cells degraded the matrix through secreted proteases, they were transfected with the polyplexes they encountered during their migration. Although such encapsulation may be favorable in some cases, it still does not allow for distinct release rates of multiple signals within a single system. It may be ideal to immobilize such signals to the biomaterial surface and sequentially release them into the environment upon cellular demand.
Here we report on a strategy to achieve temporal control over nanoparticle release from biomaterials using cell-secreted proteases. This cell-triggered release approach utilizes peptides that are degraded by matrix metalloproteinases (MMP) at different rates to immobilize nanoparticles directly to the biomaterial surface. Release would then be initiated through the action of cell-released proteases and controlled by the sensitivity of the peptide to cleavage by such proteases. Enzymatically degradable tethers have been utilized for the immobilization and release of growth factors [11,12] and small drugs [13], which are only liberated by cleavage caused by cell-secreted proteases, such as MMPs or plasmins, during local tissue remodeling. These proteases are known to be up-regulated during wound healing, microenvironment remodeling, and in diseased states and can, therefore, serve as triggers for bioactive signal delivery [2,14]. Cell responsive systems, however, have yet to be developed for the delivery of multiple factors at distinct rates, which show efficacy in cellular environments.
In this report, we developed a strategy to immobilize nanoparticles through peptide tethers and allow for their release and internalization through a cell-triggered approach. We hypothesized that internalization would depend on both the sequence of the peptide tether and the number of tethers between the nanoparticle and the biomaterial. Further, the internalization rate would be a function of the protease expression profile, with cells that express less proteases resulting in significantly less internalization than cells which over-express proteases.
2. Materials and methods
All supplies and reagents were purchased from Thermo Fisher Scientific unless otherwise noted.
2.1. Peptide modification with NHS-PEG-biotin or NHS-LC-biotin (NHS-PEG-acrylate or NHS-LC-acrylate)
Peptides were purchased from Anaspec (MMPhigh Ac-KRGPQGIWGQDRCGR-NH2, MMPmed Ac-KRGPQGIAGQDRCGR-NH2, MMPlow Ac-KRGDQGIAGFDRCGR-NH2). Peptides were reacted with NHS-PEG-biotin or NHS-LC-biotin (Laysan Bio Inc.) at a 1:2 molar ratio. Sample pH was adjusted to 7.4 and allowed to react at RT for 2 h. The sample was then dialyzed overnight against DI H2O to remove any unreacted substrates, lyophilized and stored at −20 °C until ready for use. Methods and quantities to produce acrylate-modified particles are identical to those just described, with the exception of NHS-PEG-acrylate or NHS-LC-acrylate being used instead of NHS-PEG-biotin or NHS-LC-biotin.
2.2. Particle modification with peptide-PEG-biotin or peptide-LC-biotin or biotinPDA (peptide-PEG-acrylate or peptide-LC-acrylate) tether
Carboxylate-modified, 40 nm polystyrene beads (Invitrogen) were modified to possess peptide tethers. A 2% bead solution (50 µl) was incubated with 50 µl of 5.9 mm EMCH solution in MES buffer for 15 min at RT before adding EDC (10 µL of a 2 mm solution) and adjusting the pH to 6.5 ± .1 with 1 N NaOH/HCl and incubating at RT for 2 h. Samples were kept in the dark to prevent fluorescence quenching. Sample pH (and size) was monitored throughout the reaction period. The sample was dialyzed against DI H2O overnight to remove any unreacted substrates. BiotinPDA (PDA control particles) or peptide samples were reduced using 10 mm TCEP with mixing for 1 h at RT with either 97 mm biotinPDA in DMSO or 9.7 mm of peptide-PEG-biotin/LC-biotin in MES, respectively. Next either 2.5 µl of 9.7 mm biotinPDA in DMSO (10-fold dilution) or peptide-PEG-biotin/LC-biotin in PBS was added to the partially modified bead solution (now ~150 µl after overnight dialysis). Sample pH was adjusted to 7.4 ± .1 and the sample was allowed to react for 2 h at RT while being mixed. The sample was then dialyzed overnight for a second time against H2O. Final particle concentration was determined using a fluorimeter and comparison against a standard curve. Methods and quantities to produce acrylate-modified particles are identical to those just described, with the exception of the secondary conjugation molecules containing acrylate groups instead of biotins.
2.3. Particle modification with NHS-PEG-biotin (NHS-PEG-acrylate or NHS-LC-acrylate) tether
A 2% bead solution (50 µl) was incubated with 50 µl of 5.9 mm ADH solution in MES buffer for 15 min at RT. Next, 10 µl of 2 mm EDC was added. The pH was adjusted to 6.5 ± .1 with 1 N NaOH/HCl and the sample was incubated at RT for 2 h. Samples were kept in the dark to prevent fluorescence quenching. Sample pH (and size) was monitored throughout the reaction period. The sample was dialyzed against DI H2O overnight to remove any unreacted substrates. Next 2.5 µl of 9.7 mm NHS-PEG-biotin (PEG control particles) in PBS was added to the partially modified bead solution (now ~150 µl after overnight dialysis). Sample pH was adjusted to 7.4 ± .1 and the sample was allowed to react for 2 h at RT while being mixed. The sample was then dialyzed overnight for a second time against H2O. Final particle concentration was determined using a fluorimeter and comparison against a standard curve. Methods and quantities to produce acrylate-modified particles are identical to those just described, with the exception of the secondary conjugation molecules containing acrylate groups instead of biotins. Controls containing no PEG were formed using NHS-LC-acrylate.
2.4. Particle characterization
Sample particle size was measured, when required, using DLS with a 10 to 100-fold dilution of the stock bead solution. Particles were also analyzed for degree of modification using elemental analysis for detection of sulfur. Samples (200 µl) were diluted to 1012 particles/ml in DI H2O and analyzed at the UCLA Molecular Instrumentation Center.
2.5. Binding of biotinylated PS nanoparticles to 2D surface
Covalently bound streptavidin to 96-well plates were bought from Nunc Company. Particles were blocked with 1% BSA/1×PBS solution at 4 °C overnight (minimum) in the dark. Wells were washed 3× with 1×PBS-Tween and then either incubated with 100 µl 1×PBS or 100 µl 5 mm free biotin in PBS for samples which tested for specific binding for 1 h. After 1 h the samples in the wells were removed and 100 µl blocked particles were added to each well (3.2×1011 particles/ml). All samples were run in triplicate. The plate was incubated at RT for 3 h after which the wells were again washed 3× with 1×PBS-Tween buffer, the first wash of which sat for ~1 h. Imaging of the plate was conducted using a Typhoon scanner using either a 488 nm blue laser (excitation/emission 505/515) with 520 nm BP 40 nm emission filter at 450 V for yellow-green particles or a 532 nm green laser (excitation/emission 580/605) with 610 nm PB 30 nm emission at 600 V for red particles.
2.6. Release of biotinylated particles from surface
After the plate was bound with particles according to the protocol described above, release of the particles was measured over time. Each well was filled with 100 µl 1×PBS and the plate was incubated at 37 °C for 20–24 h. The PBS in the wells was changed 3–4 times during this period. Collagenase I (Worthington Biochemical Corp.) or mMSC conditioned media was then added to each well and the plate was incubated at 37 °C for usually 120 h, during which several readings were conducted. A single triplicate was left untouched (100% control) and would later be used to normalize the release data. At each time point the solution was removed and the plate was imaged dry using the Typhoon scanner. Release data was all normalized to the 100% control within each individual bead type and then evaluated with respect to the release of particles incubated in PBS (i.e. background release).
2.7. Cell culture and zymography
HEK293T-MMP2 cells were a kind gift from Jeffrey Smith from the Burnham Institute for Medical Research. mMSC, HEK293T, and HEK293T-MMP2 cells were cultured in DMEM medium supplemented with 10% bovine growth serum and 1% penicillin/streptomycin at 37 °C and 5% CO2. The cells were passaged using trypsin following standard cell culture protocols every 2–3 days. To determine the extent to which these cells express matrix metalloproteinases, a gelatin zymogram was used. Cells were plated in 12-well TC treated plates at a density of 100,000 cells/ml and conditioned cell media was collected after exactly 24 h and assayed for total protein concentration using a Bradford assay. Each lane was loaded with 50 µg of total protein and the gel was run in Tris–Glycine SDS running buffer at 110 V. The gel was then placed in renaturing buffer (2.5% Triton-100) for 30 min at room temperature and then incubated in the developing buffer (0.5–1% tris(hydroxymethyl)amino-methane, 1–2% sodium chloride) for 30 min at room temperature with further development in fresh buffer at 37 °C overnight. The gel was stained with coomassie blue for 60 min (0.5% w/v) and de-stained in a methanol, acetic acid, and water solution (50:10:40) until clear bands appeared where active protease was present. Active human matrix metalloproteinase 2 and 9 (5 ng/well, Calbiochem), Collagenase I (20 ng/well), and cDMEM (50 µg/well - background) were run in the gel for comparison.
2.8. Particle internalization assay
Particles were bound and stabilized in PBS as described above. Cells were then plated at a density of 7000 cells/well and allowed to attach and spread at 37 °C. At either 24 or 48 h (separate wells were used for each time point) media was removed and replaced with sterile 1×PBS. Cells were fixed using 4% PFA for 15 min at RT, after which their nuclei were stained for 30 min using HOESCHT dye (Invitrogen) in the dark. Finally, samples were analyzed in PBS using an inverted Zeiss Observer Z1 fluorescence microscope. Flow cytometry analysis was also conducted at these time points to quantify the degree of particle internalization. Cells were washed with PBS and detached using .25% Trypsin for 2 min. They were then re-suspended into a solution of 1% BGS/1×PBS with 10% Trypan blue and remained on ice until they could be analyzed. Trypan blue was used to quench any external fluorescence on the cells so any fluorescence detected by the flow cytometer was due to internalized particles and not by particles nonspecifically bound to the external surface of the cells [15].
2.9. Design template using PMMA microspheres
PMMA microspheres (90–125um, Bangs Laboratories) were bought dry. Approximately 25–30 mg of beads were then added into glass-bottom silicon wells (Electron Microscopy Sciences) and covered with a second glass slide. Unlike previously published techniques (e.g. rotating slide at 250 rpm for 4 h [16]), the beads were then packed by slight tapping for 1–2 min and examined for even packing through phase microscopy. Once satisfactory packing was observed the glass slide was placed into an oven and the beads were sintered for 26 h at 150 °C. The glass slide was then removed from the oven and cooled to room temperature. It was then ready to be used as a template for porous hydrogel formation.
2.10. RGD peptide modification
RGD peptide (Ac-GCGYGRGDSPG-NH2), purchased from GenScript Corp., was reacted at a 2:1 mole ratio with 4-arm PEG-vinyl sulfone (PEG-VS, MW 20,000) for 2 h with mixing at pH 8.5. The final solution was stored at −20 °C until ready for use.
2.11. Porous PEG-acrylate hydrogel formation
A hydrogel precursor solution was made using PEG dimethacrylate (PEG-DA, MW 575), PBS (pH 7.4), and Irgacure 2959 photoinitiator (Ciba) for a final concentration of 10% (w/v) PEG and .05% Irgacure in 35% EtOH. For cell-adhesive gels to be used for in vitro experiments, modified RGD-PEG-VS was also added to the gel mixture for a final concentration of 1000 µm RGD. Approximately 45 µl of gel solution was then added directly on top of a PMMA microsphere template, coved with a second slide, and perfused into the template by centrifugation at 1500 rpm for 3 min. The slide was then exposed to low intensity UV light for 90s to induce polymerization. Once complete, the top slide was removed and the gels were removed from the silicon wells and placed into fresh 100% acetone for 48 h to dissolve the PMMA microsphere template. The gels were then serially re-hydrated into DI H2O and left in fresh water until ready for use.
2.12. Characterization of mechanical properties
Plate-to-plate rheometry was used to determine the mechanical properties of the porous hydrogels. The hydrogels were cut to 6 mm in diameter, which was slightly smaller than the size of the rheometer plate (8 mm) to ensure the gel did not exceed the size limits when under compression, as compression was necessary to avoid slipping. An Anton Parr rheometer under a constant strain of 0.5 and frequency from 0.1 to 10 Hz was used to measure both the storage and loss moduli.
2.13. Gel preparation for SEM imaging
Each porous hydrogel was serially dehydrated in 20, 40, 60, 80, and 100% EtOH for 10 min at a time. Hydrogels were left in 100% EtOH overnight after which they were placed in a dry holder, air dried and, finally, placed under vacuum until time for imaging. The gels were then attached to an SEM holder using carbon tape and coated with a thin layer of gold using a gold sputterer. Images were taken using a JEOL JSM-6700F FE-SEM in the UCLA MCTP core facility. Hydrogels used solely for characterization by SEM did not contain any RGD adhesion peptide.
2.14. Binding acrylate-modified particles to porous hydrogels
Acrylate-modified PS nanoparticles (40 µl of 1E13 particles/ml) with .05% Irgacure 2959 photoinitiator were added directly on top of porous hydrogels in dry glass-bottom silicon well. The hydrogels were incubated for 10 min at RT. Low intensity UV light was shined for 1 min to induce binding of the particles to the hydrogel through surface acrylate groups left unpolymerized by the initial gelling process. The gels were washed using PBS-Tween buffer in the dark and then imaged on glass coverslips with fluorescence microscopy. To test for non-specific binding, unmodified particles were used.
2.15. Cell seeding onto porous hydrogels
Porous hydrogels were sterilized by 3 consecutive 10 min washes in sterile PBS. Gels were then placed into dry low-binding plates. Cells, which may have been previously stained using a fluorescent live cell stain (CFDA cell tracker, Invitrogen) using the manufacturer’s suggested protocol, were then seeded in 2 steps. First 15,000 cells at a density of 15,000 cells/10 µl were added directly on top of each gel, centrifuged down for 3 min at 700 rpm, after which an additional 15,000 cells were added without further centrifugation. The gels were then incubated at 37 °C for 15 min, after which 180 µl of fresh media was added for a final volume of 200 µl/well. The gels were incubated at 37 °C for 24–48 h. Prior to imaging, the media was removed from each well and replaced with sterile PBS. Cell spreading was observed using both phase and fluorescent microscopy.
2.16. Cell spreading and viability assessment
Cell spreading and viability in the porous hydrogel was studied with the LIVE/DEAD viability/cytotoxicity kit (Invitrogen). At various time points each hydrogel was stained with 150 µL of the staining solution for 30 min at room temperature in the dark (as per manufacturer’s protocol) and imaged in PBS using fluorescence microscopy.
2.17. Internalization assay of acrylate-modified particles from porous hydrogels
Once PS nanoparticles were bound to the porous hydrogels, 30,000 unstained cells were seeded onto each gel as described above. The gels were then incubated at 37 °C. At various time points, hydrogels were transferred individually to micro-centrifuge tubes and cells were detached from the gels with 2 min incubation in .25% trypsin followed by an addition of cDMEM and centrifugation for 5 min at 350 × g. The cell solution was then pipetted into a glass coverslip-bottom flexiperm well and incubated at 37 °C for 24 h. Cells were fixed using 4% PFA and cell nuclei were stained using HOESCHT dye as previously described. Samples were imaged in PBS using fluorescence microscopy.
3. Results
Fluorescent polystyrene (PS) nanoparticles were immobilized to a cell binding surface through either protease sensitive peptide tethers or poly(ethylene glycol) (PEG) polymer chains. Terminal biotins on each tether allowed for the immobilization of the modified PS nanoparticles onto a streptavidin surface through robust (Kbinding = 1015) biotin-streptavidin interactions (Fig. 1A). Various peptides, previously reported to have varying degrees of sensitivity towards MMP2 [17] and MMP1 [18], were then used to allow for control over release rates. Particles with varying degrees of tethering were also made to determine whether this could result in an effect on the release kinetics. Cells which express high or low levels of MMPs were then used to study nanoparticle internalization (Fig. 1B).
Fig. 1.
Protease sensitive peptides are used to immobilize model nanoparticles to biomaterials and released as a result of peptide cleavage by cell-released proteases (A); Gelatin zymography was used to determine MMP2 and MMP9 expression of multiple cell types (B). HEK293T cells were shown to express significantly lower levels of MMP2 and 9 compared to mMSCs and HEK293T-MMP2 cells, which specifically and stably produce MMP2; Biotinylated peptide-containing particles were prepared by reacting carboxylate-functionalized polystyrene (PS) nanoparticles with a heterobifunctional cross-linker, namely EMCH, followed by reaction with SH-MMPx-PEG3400-biotin (C); To determine if sufficient biotinylation had been achieved, fluorescent particles were incubated on a streptavidin surface and analyzed for binding using a Typhoon scanner (D); Fluorescence was quantified and showed that in a competitive binding assay of biotinylated particles (1×B) in the prescence of excess free biotin (1×B + B) the degree of binding was significantly reduced to the level of unmodified (−COOH) particles (E); Particles prepared with 10-fold dilutions of SH-MMPmed-PEG3400-biotin (×B) revealed that a specific particle surface density of both PEG and biotin resulted in the highest degree of binding (F). The ** and *** symbols indicate statistical significance at levels of p < .01 and p < .001, respectively, calculated using a multiple comparisons Tukey test. Statistics for Figure F were determined with respect to 1×B particles.
Biotinylated peptide-containing particles were prepared by reacting carboxylate-functionalized PS nanoparticles with a heterobifunctional cross-linker, namely 3,3′-[N-e-Maleimidocaproic acid] hydrazide (EMCH), followed by reaction with SH-MMPx-PEG3400-biotin or SH-MMPx-biotin for tethers with no PEG polymer chain (Fig. 1C). PEG control particles, containing no cleavable peptide, were similarly produced using adipic acid dihydrazide (ADH) for amine functionalization, followed by reaction with NHS-PEG3400-biotin. For controls containing no PEG polymer chain (PDA control particles) the particles were modified with EMCH followed by reaction with reduced biotinPDA. The degree of modification was characterized using elemental analysis for sulfur content (Table 1). Each biotin and each peptide contain a single sulfur atom. Only non-peptide tethered control particles made using NHS-PEG-biotin contained a single sulfur atom per tether. 200 nm commercially available biotinylated particles were also analyzed and showed that the degree of biotinylation of our 40 nm manually modified particles was similar to those commercially manufactured.
Table 1.
The degree of modification per particle was determined using elemental analysis for sulfur.
| Particle Type | Biotin density (biotin/particle) |
Inverse biotin density (nm2/biotin) |
|---|---|---|
| 10×MMPmed-PEG-b | 2607 | 1.93 |
| 1×MMPmed-PEG-b | 1508 | 3.33 |
| .1×MMPmed-PEG-b | 1361 | 3.69 |
| .01×MMPmed-PEG-b | 1022 | 4.92 |
| 1×MMPhigh-b | 1389 | 3.62 |
| 1×MMPmed-b | 1685 | 2.98 |
| 1×MMPlow-b | 1864 | 2.70 |
| 1×bPDA (control) | 2302 | 2.18 |
| 1×PEG-b (control) | 1739 | 2.89 |
| 200 nm biotin (control) | 1,18,364 | 1.06 |
Particle immobilization and release was determined using a typhoon scanner, which can measure the fluorescence intensity of the surface (Fig. 1D). Particles were subjected to a competitive binding assay either in the presence or absence of excess free biotin to ensure particle binding was specific to the interaction of biotinylated particles with surface streptavidin (Fig. 1E). We were also able to determine an ideal PEG-biotin surface density, which resulted in the highest amount of particles binding to the surface (Fig. 1F). Particles averaging less than 1000 tethers (5 biotin/nm2) or greater than ~2500–2600 tethers (~2 biotin/nm2) were unable to adequately bind to the streptavidin surface.
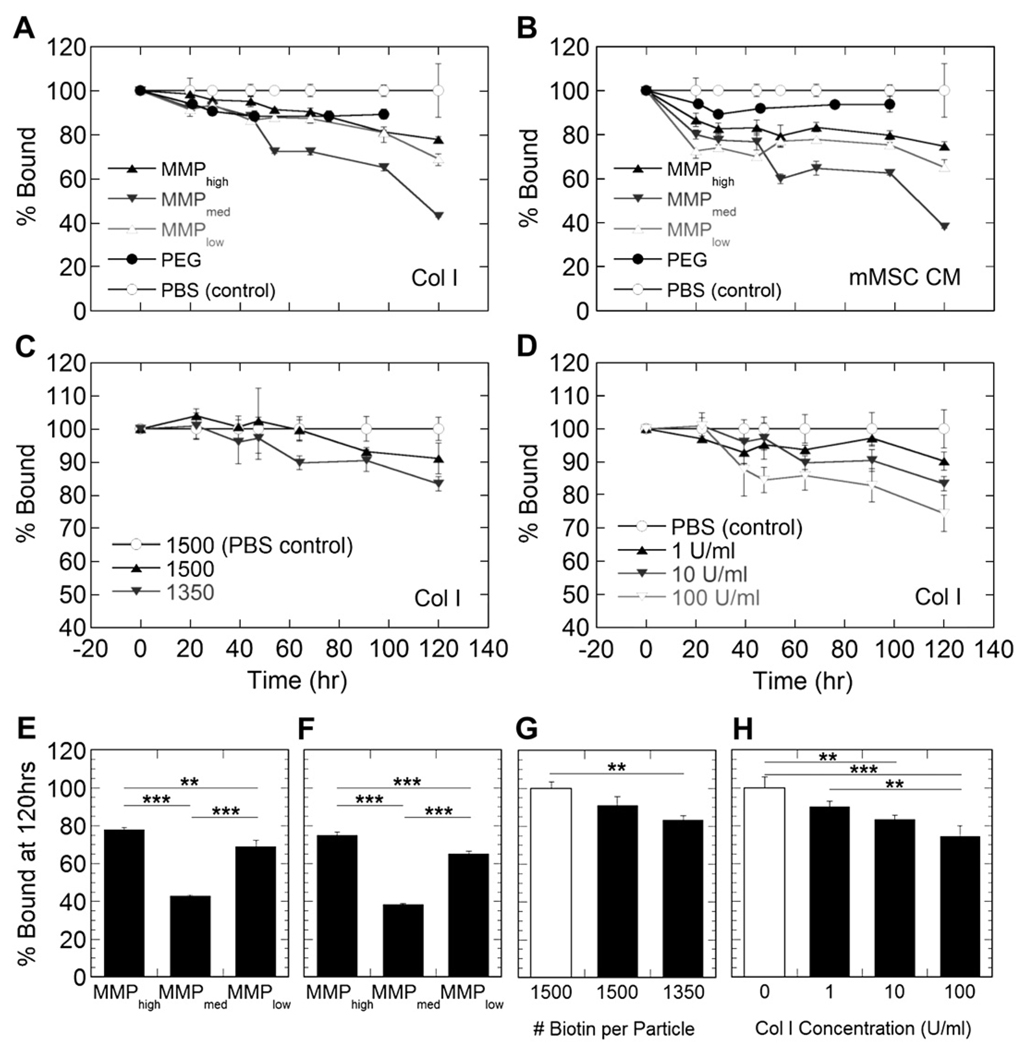
Immobilized particles were then subjected to treatment with Collagenase I (bacterial equivalent to human MMP1) or mouse mesenchymal stem cell (mMSC) conditioned media to assess release kinetics in protease rich environments (Fig. 1B). Importantly, we observed minimal release of control particles (10–12%) with both release mediums indicating that the surface was stable against protease degradation. Peptide-modified particles were then shown to release over a 5-day period with the rate of release being determined by both the peptide sensitivity to Col I (Fig. 2A, E) and proteases in mMSC conditioned media (Fig. 2B, F) in addition to the degree of particle modification and tethering to the surface (Fig. 2C, G). Differences seen with different tethered particles confirm previously published findings for different gene transfer rates for DNA/poly(lysine) particles immobilized through different tether numbers [19]. Particles with fewer tethers and, consequently, fewer interactions with the surface were released more rapidly than those with a greater number of tethers. Additionally, the release rate was dependent on protease concentration (Fig. 2D, H).
Fig. 2.
Release profiles show that nanoparticle release is dictated by peptide sensitivity to Col I (A, E) and those proteases found in complex mMSC conditioned media (B, F). Additionally, release is a function of the degree of particle tethering to the surface (C, G) and protease concentration (D, H). MMPmed particles were used to study the effects of tethering and protease concentration. Figures E–H show statistical differences between release at 120 h. White bars in Figures G, H indicate release in PBS. The ** and *** symbols indicate statistical significance at levels of p < .01 and p < .001, respectively, calculated using a multiple comparisons Tukey test.
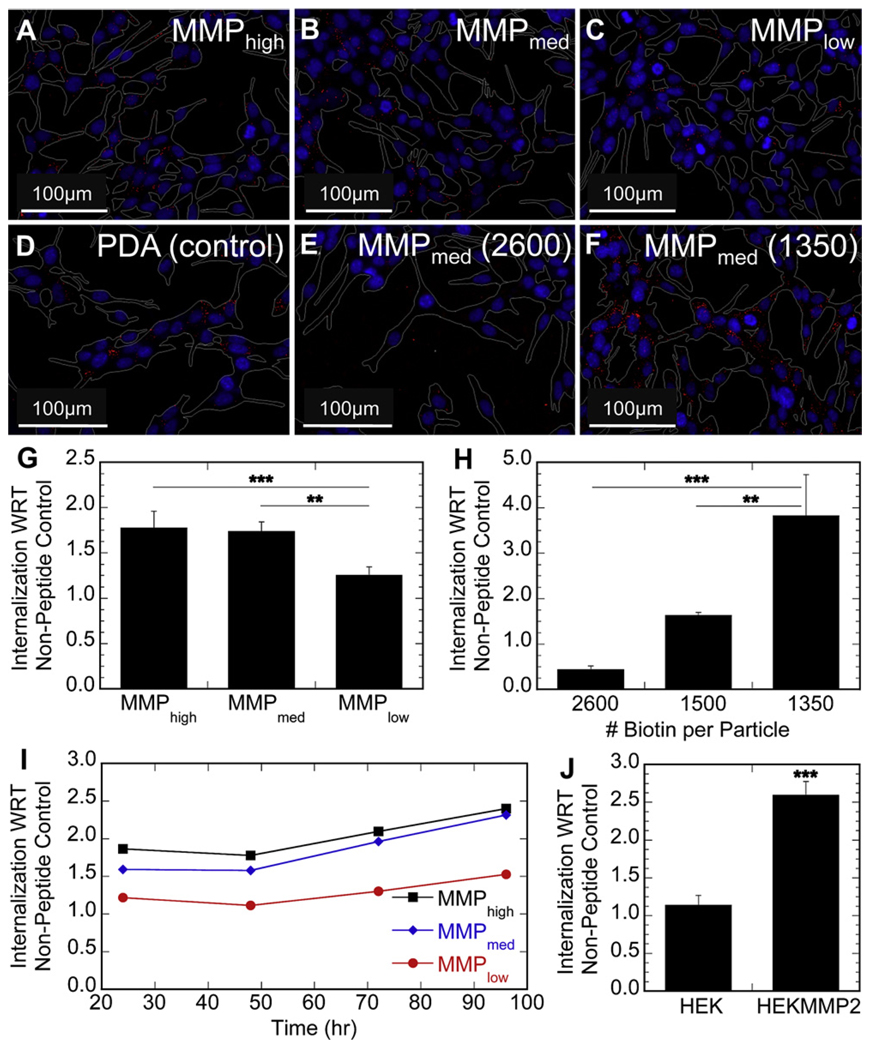
To determine if the cellular internalization rate of the immobilized nanoparticles followed the observed release kinetics, cells that express either high or low levels of proteases were incubated on top of bound particles and particle internalization was assessed using fluorescence microscopy and flow cytometry. Internalization studies were conducted with high protease-expressing mMSCs. A gelatin zymogram was used to characterize the MMP2 and MMP9 expression profile of the cells using conditioned cell media (Fig. 1B). No extensive internalization of nanoparticles immobilized though non-peptide tethers was observed for protease-expressing cells (e.g. mMSCs), while nanoparticles immobilized through peptide tethers were internalized to a significantly greater extent (Fig. 3A–F). Although qualitative differences in internalization between different peptide-modified nanoparticles could be observed using inverted fluorescence microscopy, flow cytometry analysis showed definitively that protease-expressing cells internalized peptide-modified particles in a manner dependant on peptide sensitivity to cell-released proteases, namely MMPs (Fig. 3G). Internalization was also dependant on particle tether density and the observed trend was far more significant when compared to that from the acellular release studies using cell conditioned media (Fig. 3H). Internalization of peptide-tethered particles with respect to non-peptide tether immobilized nanoparticles by mMSCs increased gradually over the entire course of the study (Fig. 3I).
Fig. 3.
2-D in vitro studies were conducted using various modified particles. Fluorescence microscopy was used to qualitatively analyze particle internalization by protease-expressing mMSCs (RED – particle, BLUE – nucleus, WHITE – cell perimeter) (A–F – 20× magnification). Internalization was found to be a function of peptide sensitivity to cell-released MMPs (A–C) and minimal with respect to non-peptide tethered control particles (D). Particle tethering to the surface was a strong determinant for internalization (E–F); Quantitative characterization of internalization by flow cytometry confirmed peptide sensitivity to proteases (G) and the degree of particle tethering, using MMPmed particles (H), dictated release and internalization into protease-expressing mMSCs; Internalization of peptide-modified particle increased with time indicating a sustained release of nanoparticles could be achieved (I); To show that internalization was dictated by protease expression and independent of cell type, HEK293T and HEK293T-MMP2 cells were tested (J). HEK293T-MMP2 cells internalized MMPmed peptide-modified particles 2.6-fold more than control particles, while HEK293T cells internalized only 1.1-fold more. The ** and *** symbols indicate statistical significance at levels of p < .01 and p < .001, respectively, calculated using a multiple comparisons Tukey test. Statistics for Figure J were determined using a single pair comparison Dunnet test.
Genetically modified HEK293T cells were used to highlight the importance of cellular protease expressionin controlling nanoparticle internalization. HEK293T cells were stably transfected to express MMP2 [20] and their internalization of peptide-tethered nanoparticles was compared to that of wild type HEK293T. HEK293T-MMP2 cells internalized 2.6-fold more peptide-tethered nanoparticles with respect to non-peptide tethered particles compared to HEK293T cells, which only internalized 1.1-fold more (Fig. 3J).
To translate our system to three dimensions, we modified a previously described poly(methyl methacrylate) (PMMA) microsphere template method to produce our PEG hydrogels (Fig. 4A) [16]. Gels were characterized using rheometry to test for mechanical properties (Fig. 4B) and phase (Fig. 4C, F) and scanning electron microscopy (SEM) (Fig. 4D, G) to produce micro- and nano-scale structural images, respectively. Next, acrylate-modified PS nanoparticles were covalently bound to the surface of these porous hydrogels through reaction with previously unreacted surface acrylate groups in the presence of Irgacure 2959 photoinitiator with low intensity UV light (Fig. 4E, H).
Fig. 4.
Porous PEG hydrogels were made using a modified PMMA bead template (A). PMMA beads were packed and slightly sintered to allow for μ-sized interconnected pores. Once the PEG-DA monomer was added into the void space and polymerized, the PMMA beads were dissolved in acetone; Plate-to-plate rheometry was used to determine the mechanical properties of the porous hydrogels which were found to be very stiff as the storage modulus was almost constant over the frequency range tested (B); Phase images of porous PEG hydrogels made using a PMMA microparticle template (C, F); SEM images show interconnected pores throughout the gel (D, G); Acrylate-modified particles were covalently crosslinked onto the pore surfaces (E), while non-specific binding of unmodified particles was minimal (H).
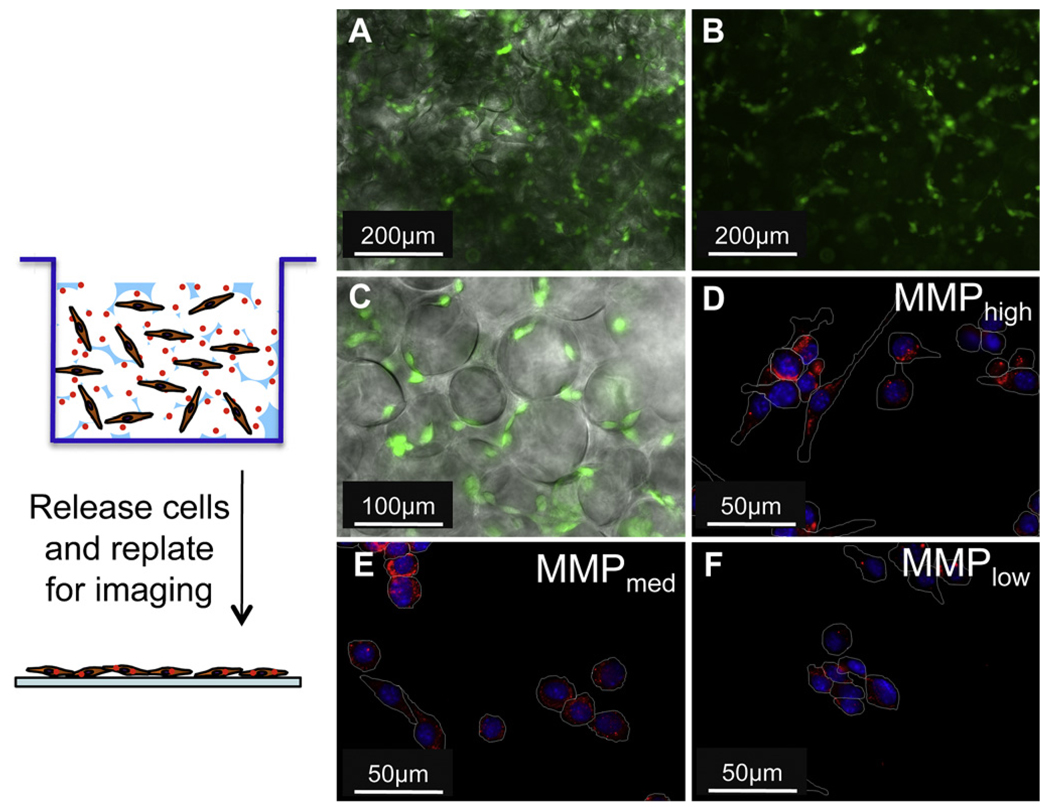
Protease-expressing mMSCs were cultured in hydrogels with peptide-tethered nanoparticles and analyzed for their degree of internalization. Cells were seeded on top of the porous gels and lightly centrifuged to evenly disperse them throughout the gel (Fig. 5A–C). The cells were then incubated at 37 °C for 24 h, after which they were released from the gels by trypsin treatment and replated onto glass coverslips to assess nanoparticle internalization. Without replating cells onto a flat surface, internalization was difficult to visualize using fluorescence microscopy. The observed internalization trend was similar to that seen on the 2-dimensional surface. The extent of internalization of peptide-modified particles by protease-expressing mMSCs was controlled by the sequence of the peptide tether (Fig. 5D–F).
Fig. 5.
Live stained mMSCs were able to spread on porous hydrogels and visualized using fluorescence microscopy (A, B – 10× magnification, C – 20× magnification). Phase image overlaid with fluorescence (B, C); For 3-D in vitro studies, mMSCs were seeded and cultured in the gels for 24 h, after which they were released and replated on glass coverslips to visualize particle internalization via fluorescence microscopy (RED – particle, BLUE – nucleus, WHITE – cell perimeter) (D–F – 40× magnification). Internalization was found to be a function of peptide sensitivity to cell-released MMPs (D–F).
4. Discussion
Strategies to control the release rate of bioactive signals from tissue engineering scaffolds are essential for tissue regeneration and tissue engineering applications. Here we report on a strategy to achieve temporal control over nanoparticle release from biomaterials using cell-secreted proteases. This cell-triggered release approach utilizes peptides that are degraded by MMPs at different rates to immobilize nanoparticles directly to the biomaterial surface. Thus, the peptide-immobilized nanoparticles are released with temporal control through the action of cell-released MMPs. We found that release rates of peptide-immobilized nanoparticles were a function of peptide sensitivity to proteases, the number of tethers between the nanoparticle, and the surface and the concentration of proteases used to induce release (Fig. 2). Cellular internalization of the peptide-immobilized nanoparticles was also a function of the peptide sensitivity to proteases, the number of tethers between the nanoparticle and the surface and MMP expression profile of the cells (Fig. 3). Similar trends were observed for peptide-immobilized nanoparticles inside micro-porous hydrogels (Fig. 4), indicating protease sensitive tethers are effective in controlling release rate and internalization of nanoparticles.
Fluorescent PS nanoparticles were modified with biotinylated peptide or polymer tethers and specifically bound to a streptavidin surface as shown through a competitive binding assay in the presence of excess free biotin (Fig. 1E). Particle and surface binding characteristics identified a narrow tether density range within which the highest degree of surface binding could be achieved (Fig. 1F). We believe this trend resulted from a subtle balance between biotin and PEG for binding and binding inhibition, respectively. Generally highly PEG-ylated particles had difficulty binding due to increased steric hindrance, as has been suggested for biotin-PEG conjugates [21]. High tether density also lead to increased multivalent binding which limited the total number of particles being able to make sufficient interactions for binding on a confined streptavidin surface.
Preliminary acellular release studies were conducted to show potential for controlled release of tethered nanoparticles in protease rich environments (Fig. 2). Non-peptide tethered particles showed minimal release in the presence of Col I and mMSC conditioned media, which indicated that any observed release of peptide-tethered particles is a result of the tether sensitivity to proteases and not the instability of the biotin-streptavidin surface. Peptide-tethered particles showed varying rates of release based on peptide type with both treatments. Although documented sensitivities of the peptides to MMP1 were designated high, medium, and low [18], this same trend was not observed in the presence of Col I (Fig. 2A, E) and mMSC conditioned media (Fig. 2B, F), where MMPmed tethered particles resulted in statistically faster release. This difference may be explained as the documented sensitivities were only shown with respect to pure MMP1, which is inherently different from crude bacterial collagenase, and will differ with respect to a mixture of proteases as is expected to be present in mMSC media. The complex and rich nature of the mMSC media may also explain the accelerated release rates of peptide-tethered particles when compared to their release in Col I. This notion was confirmed when release was tested in varying concentrations of Col I which confirmed that release was dependant on protease concentration (Fig. 2D, H). A higher concentration of proteases would cleave more peptide tethers in a given period of time, increasing the overall rate of release. Tether density was also tested as a means of controlling nanoparticle release rate. We hypothesized that if a single nanoparticle had more or less interactions with the biomaterial, it would take more or less time for a protease or multiple proteases to be able to fully cleave all of its tethers and allow for it to release. Similar studies have been previously conducted which indicated the same principles could dictate the non-specific release of immobilized polyplexes [19]. However, only slight differences in release were observed in the presence of Col I (Fig. 2C, G).
When immobilized nanoparticles were incubated with MMP-expressing mMSCs, the trends were unlike what was observed in our acellular release studies, with the trend high, medium, and low for peptide cleavage rate holding (Fig. 3A–C, G). Protease expressing cells have a much higher local protease concentration directly around their surface than is found in the bulk conditioned media [22], thus protease dependant effects can be amplified when the particles are directly in contact with cells. This phenomenon also explains the significant differences in internalization as a result of differences in tether density (Fig. 3E–F, H), which were clearly observed in our 2-dimensional in vitro study but not our acellular release study. These results show the importance of testing cell-triggered delivery strategies directly with cells rather than relying on acellular studies since the same trends are not always observed.
All peptide-tethered nanoparticles were also shown to release steadily for up to 4 days (Fig. 3I). Such sustained release is essential for a biomaterial, which will eventually be used in vivo where the tissue regeneration process will be on the time scale of days to weeks.
Since the internalization rate of nanoparticles can vary widely among different cell types, HEK293T, which express low levels of proteases [20], were genetically engineered to over-express MMP2 (HEK293T-MMP2). By testing the rate of nanoparticle internalization by HEK293T cells in direct comparison with HEK293T-MMP2 cells, the effect of a cell’s protease expression profile on internalization of immobilized nanoparticles can be studied. The statistically significant difference in overall internalization demonstrated that the protease expression profile of the seeded cells was the main factor in determining the release and internalization kinetics of peptide-tethered nanoparticles (Fig. 3J).
To determine if cell-triggered delivery of peptide-immobilized nanoparticles, which was previously observed on 2-dimensional surfaces, was possible in a 3-dimensional microenvironment we translated our approach for cell-triggered internalization to a 3-dimensional PEG hydrogel. This also allowed for increased surface area and set the stage for potential in vivo applications. Because our strategy for controlling nanoparticle internalization rate requires the immobilization of the nanoparticles to the hydrogel scaffold, we used micro-porous hydrogels rather than nano-porous hydrogels to ensure that no encapsulated nanoparticles were present (Fig. 4). We wanted to limit encapsulated nanoparticles to reduce the background in our internalization experiments. The micro-sized pores allow for increased diffusion and ability to remove nonspecifically bound particles through washing and allow for the attachment of the PS nanoparticles after gel formation. This ensures the nanoparticles are not encapsulated and bound only to the pore surfaces. Interconnected pores in a non-degradable porous PEG hydrogel additionally allow for uninhibited cellular spreading and migration, with particle internalization being a function solely of tether cleavage and not gel degradation.
MMP-expressing mMSCs were once again used to study internalization in the PEG hydrogels. Internalization was determined to be a function of peptide tether sensitivity to MMPs with MMPhigh tethered particles resulting in the most internalization and MMPlow tethered particles resulting in the least internalization after 48 h (Fig. 5D–F). While further optimization to produce 3-dimensional environments to study immobilized nanoparticle internalization will be done in the future, currently our particle-bound porous gels show significant promise for cell-demanded nanoparticle release.
5. Conclusions
Mimicking cellular processes for regenerative medicine through the use of controlled-release systems was limited by the systems’ chemistry and lack of cellular responsiveness. By immobilizing nanoparticles through protease sensitive peptide tethers, release could be tailored specifically for an intended cellular target, which over-expresses such proteases. Release was also shown to vary significantly in the presence or absence of cells, indicating the importance of such cellular release studies, which have generally not been conducted in previous sequential release studies. We observed that the protease (namely MMP) expression profile of the seeded cells was the main factor in determining the release and internalization kinetics of MMP-sensitive peptide-modified particles. Similar trends were observed in 3D micro-porous PEG hydrogels indicating protease sensitive tethers are effective in controlling release rate and internalization, with direct applications to the controlled release of DNA and siRNA polyplexes and other nanoparticles in 3-dimensional hydrogels in vivo.
Acknowledgements
The authors would like to thank Dr. Armando Durazo at the UCLA Molecular Instrumentation Center for running all elemental analysis samples and Dr. Jianjun Zhang and Zheng Chen for their help with SEM imaging. T. Tokatlian would especially like to thank Yuguo Lei, Dr. Quinn Ng, Sean Anderson, and Anandika Dhaliwal for their helpful discussions and advice and Robert Hanusa, Dashel Thompson, and Jonah Schwartz for their time and effort towards this project. The authors acknowledge the NIH (1R21EB007730-01), NSF (CAREER 0747539), and CRCC for funding this work. T. Tokatlian acknowledges the NIH Biotech Training Grant (T32GM067555) for funding. Supporting Information is available online from Wiley InterScience or from the author.
Appendix
Figures with essential colour discrimination. Figs 1, 3, 4 and 5 in this article have parts that are difficult to interpret in black and white. The full colour images can be found in the on-line version, at doi:10.1016/j.biomaterials.2010.07.030.
References
- 1.Schultz GS, Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 2009;17(2):153–162. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 2.Steffensen B, Hakkinen L, Larjava H. Proteolytic events of wound-healing–coordinated interactions among matrix metalloproteinases (MMPs), integrins, and extracellular matrix molecules. Crit Rev Oral Biol Med. 2001;12(5):373–398. doi: 10.1177/10454411010120050201. [DOI] [PubMed] [Google Scholar]
- 3.Pirilä E, Korpi JT, Korkiamäki T, Jahkola T, Gutierrez-Fernandez A, Lopez-Otin C, et al. Collagenase-2 (MMP-8) and matrilysin-2 (MMP-26) expression in human wounds of different etiologies. Wound Repair Regen. 2007;15(1):47–57. doi: 10.1111/j.1524-475X.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 4.Lobmann R, Ambrosch A, Schultz G, Waldmann K, Schiweck S, Lehnert H. Expression of matrix-metalloproteinases and their inhibitors in the wounds of diabetic and non-diabetic patients. Diabetologia. 2002;45(7):1011–1016. doi: 10.1007/s00125-002-0868-8. [DOI] [PubMed] [Google Scholar]
- 5.Richardson TP, Peters MC, Ennett AB, Mooney DJ. Polymeric system for dual growth factor delivery. Nat Biotechnol. 2001;19(11):1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- 6.Holland TA, Tabata Y, Mikos AG. Dual growth factor delivery from degradable oligo(poly(ethylene glycol) fumarate) hydrogel scaffolds for cartilage tissue engineering. J Control Release. 2005;101(1–3):111–125. doi: 10.1016/j.jconrel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Jaklenec A, Hinckfuss A, Bilgen B, Ciombor DM, Aaron R, Mathiowitz E. Sequential release of bioactive IGF-I and TGF-[beta]1 from PLGA microsphere-based scaffolds. Biomaterials. 2008;29(10):1518–1525. doi: 10.1016/j.biomaterials.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Yilgor P, Tuzlakoglu K, Reis RL, Hasirci N, Hasirci V. Incorporation of a sequential BMP-2/BMP-7 delivery system into chitosan-based scaffolds for bone tissue engineering. Biomaterials. 2009;30(21):3551–3559. doi: 10.1016/j.biomaterials.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 9.Saul JM, Linnes MP, Ratner BD, Giachelli CM, Pun SH. Delivery of non-viral gene carriers from sphere-templated fibrin scaffolds for sustained transgene expression. Biomaterials. 2007;28(31):4705–4716. doi: 10.1016/j.biomaterials.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 10.Lei Y, Segura T. DNA delivery from matrix metalloproteinase degradable poly (ethylene glycol) hydrogels to mouse cloned mesenchymal stem cells. Biomaterials. 2009;30(2):254–265. doi: 10.1016/j.biomaterials.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrbar M, Djonov VG, Schnell C, Tschanz SA, Martiny-Baron G, Schenk U, et al. Cell-demanded liberation of VEGF121 from fibrin implants induces local and controlled blood vessel growth. Circ Res. 2004;94(8):1124–1132. doi: 10.1161/01.RES.0000126411.29641.08. [DOI] [PubMed] [Google Scholar]
- 12.Geer DJ, Swartz DD, Andreadis ST. Biomimetic delivery of keratinocyte growth factor upon cellular demand for accelerated wound healing in vitro and in vivo. Am J Pathol. 2005;167(6):1575–1586. doi: 10.1016/S0002-9440(10)61242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tauro JR, Lee B-S, Lateef SS, Gemeinhart RA. Matrix metalloprotease selective peptide substrates cleavage within hydrogel matrices for cancer chemotherapy activation. Peptides. 2008;29(11):1965–1973. doi: 10.1016/j.peptides.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soo C, Shaw WW, Zhang X, Longaker MT, Howard EW, Ting K. Differential expression of matrix metalloproteinases and their tissue-derived inhibitors in cutaneous wound repair. Plast Reconstr Surg. 2000;105(2):638–647. doi: 10.1097/00006534-200002000-00024. [DOI] [PubMed] [Google Scholar]
- 15.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377(1):159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stachowiak AN, Bershteyn A, Tzatzalos E, Irvine DJ. Bioactive hydrogels with an ordered cellular structure combine interconnected macroporosity and robust mechanical properties. Adv Mater. 2005;17(4):399–403. [Google Scholar]
- 17.Seliktar D, Zisch AH, Lutolf MP, Wrana JL, Hubbell JA. MMP-2 sensitive, VEGF-bearing bioactive hydrogels for promotion of vascular healing. J Biomed Mater Res A. 2004;68(4):704–716. doi: 10.1002/jbm.a.20091. [DOI] [PubMed] [Google Scholar]
- 18.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci U S A. 2003;100(9):5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem. 2002;13(3):621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 20.Chen EI, Kridel SJ, Howard EW, Li W, Godzik A, Smith JW. A unique substrate recognition profile for matrix metalloproteinase-2. J Biol Chem. 2002;277(6):4485–4491. doi: 10.1074/jbc.M109469200. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser K, Marek M, Haselgrubler T, Schindler H, Gruber HJ. Basic studies on heterobifunctional biotin-PEG conjugates with a 3-(4-pyridyldithio)propionyl marker on the second terminus. Bioconjug Chem. 1997;8(4):545–551. doi: 10.1021/bc970086u. [DOI] [PubMed] [Google Scholar]
- 22.Lee S-H, Moon JJ, Miller JS, West JL. Poly(ethylene glycol) hydrogels conjugated with a collagenase-sensitive fluorogenic substrate to visualize collagenase activity during three-dimensional cell migration. Biomaterials. 2007;28(20):3163–3170. doi: 10.1016/j.biomaterials.2007.03.004. [DOI] [PubMed] [Google Scholar]