Abstract
Mammalian cytochrome P4501 (CYP1) genes are well characterized, but in other vertebrates only the functions of CYP1A genes have been well studied. We determined the catalytic activity of zebrafish CYP1A, CYP1B1, CYP1C1, CYP1C2 and CYP1D1 proteins using 11 fluorometric substrates and benzo[a]pyrene (BaP). The resorufin-based substrates, 7-ethoxyresorufin, 7-methoxyresorufin, and 7-benzyloxyresorufin, were well metabolized by all CYP1s except CYP1D1. CYP1A metabolized nearly all substrates tested, although rates for non-resorufin substrates were typically lower than resorufin-based substrates. Zebrafish CYP1s did not metabolize 7-benzyloxyquinoline, 3-[2-(N,N-diethyl-N-methylamino)ethyl]-7-methoxy-4-methylcoumarin or 7-methoxy-4-(aminomethyl)-coumarin. CYP1B1 and CYP1C2 had the highest rates of BaP metabolism. 3-hydroxy-BaP was a prominent metabolite for all but CYP1D1. CYP1A showed broad specificity and had the highest metabolic rates for nearly all substrates. CYP1C1 and CYP1C2 had similar substrate specificity. CYP1D1 had very low activities for all substrates except BaP, and a different regioselectivity for BaP, suggesting that CYP1D1 function may be different from other CYP1s.
Keywords: cytochrome P450, zebrafish, substrate specificity, expression system, CYP1 family
1. Introduction
Cytochrome P450s (CYPs) are a large superfamily of enzymes that primarily catalyze mixed-function oxidation reactions [1]. CYPs are capable of metabolizing a wide range of substances including steroids [2], pharmaceuticals [3], and xenobiotic compounds [4]. The CYP1 family is important for the metabolism of cyclic structures including polycyclic aromatic hydrocarbons (PAHs) and aromatic amines [5]. The induction of CYP1 catalytic activity, often measured as ethoxyresorufin-O-deethylase (EROD) activity, and expression (protein and transcript) have been widely used as a biomarker of exposure and effects to planar halogenated aromatic hydrocarbons in vertebrates.
Vertebrate CYP1 subfamilies include the CYP1A, CYP1B, CYP1C and CYP1D subfamilies, but not all vertebrate lineages contain all subfamilies or an orthologous CYP1 gene complement. Mammals have 3 CYP1 genes: CYP1A1, CYP1A2, and CYP1B1. Non-mammalian vertebrates have genes from all the CYP1 subfamilies [6]. Similar to mammals, birds have two CYP1A genes while amphibians and fish have a single CYP1A gene, except for some polyploid species including the salmonids and some frogs [6]. Like their mammalian counterparts, non-mammalian vertebrates have a single CYP1B gene, CYP1B1. The CYP1C subfamily contains the paralogous CYP1C1 and CYP1C2 genes in fish [6]; a single CYP1C gene is thought to be present in birds, amphibians and reptiles [6]. The CYP1D subfamily has recently been identified in non-mammalian vertebrates, and contains a single CYP1D1 gene [7].
Mammalian CYP1 enzymes are highly inducible by the aryl hydrocarbon receptor [AHR, 8]. CYP1A1 is constitutively expressed at very low levels, but has the highest induction profile by AhR ligands in liver [9]. CYP1A2 and CYP1B1 have substantial hepatic constitutive expression and are induced by AhR ligands, but less then CYP1A1 [5, 10]. In zebrafish, CYP1A is the highest constitutively expressed CYP1 in liver [11]. Zebrafish have less constitutively expressed CYP1B1, CYP1C1, CYP1C2, and CYP1D1 in the liver, gill and kidney compared to CYP1A [7, 11]. All zebrafish CYP1s, with the exception of CYP1D1, are upregulated through the AHR [7, 11]. Studies of basal expression and induction of CYP1D1 are lacking in other species.
The function of mammalian CYP1s has been well documented, but less is known of other vertebrate CYP1s, outside CYP1A. Mammalian CYP1A1 and CYP1B1 are both able to metabolize PAHs [5] and estrogens [12]; CYP1B1, but not CYP1A1, can metabolize some aromatic amines [5]. CYP1A2 can metabolize estrogens [12] and many aromatic amines [5]. Less overall metabolism is seen from CYP1B1, but the metabolites formed tend to be more highly reactive [5, 12, 13].
Previously, we determined estradiol metabolism by zebrafish CYP1s and CYP3A65 [14] in a study that was, to our knowledge, the first to examine functional differences between non-mammalian vertebrate CYP1s. Zebrafish CYP1A and CYP1B1 metabolized estradiol with similar regioselectively to their mammalian orthologs [14]. Zebrafish CYP1A, CYP1C1, and CYP1C2 metabolized estradiol with a similar regioselectivity, but CYP1C2 metabolized estradiol at a lower rate than CYP1A and CYP1C1 [14]. Heterologous expression of the CYP1s was optimized for our research using the prototypical CYP1 substrate 7-ethoxyresorufin (ER). Zebrafish CYP1A, CYP1B1, CYP1C1, and CYP1C2 were all able to metabolize ER [14]. CYP1D1 did not metabolize estradiol or ER at a high catalytic rate [14].
Catalytic activity and substrate specificity of individual CYP isoforms can be determined [13, 15] using synthetic substrates that produce fluorescent metabolites [16] or analytical detection of metabolites produced from model compounds. For CYP1s, model compounds such as BaP [17] and fluorescent assays based on the alkoxyresorufin compounds, including ER, are appropriate as they are known mammalian CYP1 substrates [18]. As the function of the non-mammalian CYP1 genes CYP1C1, CYP1C2 and CYP1D1, are not clear, inclusion of atypical mammalian CYP1 substrates may be appropriate to discern novel function not seen in mammalian species where these genes are lacking. Here, we present the first detailed catalytic assessment of the substrate specificity of CYP1s from a non-mammalian vertebrate. Zebrafish CYP1A, CYP1B1, CYP1C1, CYP1C2 and CYP1D1 have previously been cloned and coexpressed with human CYP reductase [14]. We have tested the metabolic capabilities of CYP1s using BaP and 11 synthetic fluorescent-based CYP substrates. The fluorescent substrates are comprised of 4 resorufin based compounds, 5 coumarin compounds, a fluorescein substrate, and a quinoline. This data will help to determine the functional roles of non-mammalian CYP1s and suggest whether the novel CYP1Cs and CYP1D proteins have overlapping or novel functions.
2. Methods
2.1 Cloning, expression and purification of zebrafish CYPs
Zebrafish CYP1A, CYP1B1, CYP1C1, CYP1C2, and CYP1D1 were cloned and coexpressed with human cytochrome P450 reductase in E. coli JM109 cells, and purified bacterial membrane fractions were isolated as previously reported [14]. Bacterial expression utilized a strategy for expression without 5' modification [19]; each CYP gene was cloned with the ompA+2 leader sequence to allow targeting of the protein to the bacterial outer membrane. This sequence is excised after insertion of the protein into the membrane allowing expression of full-length vertebrate membrane proteins in bacteria [19]. Expression was optimized for each cloned CYP with the addition of 0–1 mM δ-aminolevulinic acid (Ala; MP Biomedicals, Solon, OH) and functional proteins confirmed with CO-difference spectra and catalytic assays. Optimal Ala concentrations for each insert were determined to be 0.1 mM for all CYP constructs except CYP1A and CYP1B1 where Ala was added at 0.5 mM and 1.0 mM, respectively [14].
2.2 Fluorescent based catalytic assays
All substrates and metabolites analyzed, as well as fluorescent wavelengths and concentrations used are shown in Table 1. Reactions involving the resorufin based substrates 7-ethoxyresorufin (ER), 7-methoxyresorufin (MR), 7-pentoxyresorufin (PR), and 7-benzyloxyresorufin (BR) were in 50 mM Tris, 0.1 M NaCl, pH 7.8. Reactions involving 7-benzyloxy-4-(trifluoromethyl)coumarin (BFC), 7-methoxy-trifluoromethylcoumarin (MFC), 7-benzyloxyquinoline (BQ), dibenzylfluorescein (DBF), 3-cyano-7-ethoxycoumarin (CEC), 3-[2-(N,N-diethyl-N-methylamino)ethyl]-7-methoxy-4-methylcoumarin (AMMC), and 7-methoxy-4-(aminomethyl)-coumarin (MAMC) were in 0.5 M potassium phosphate buffer, pH 7.5. Reactions were initiated with 1.33 nM NADPH (Sigma, St. Louis, MO) and all were assayed at 30°C. All resorufin based substrates (ER, MR, PR, BR) were from Sigma (St. Louis, MO) and BFC, MFC, BQ, DFB, CEC, AMMC and MAMC were from BD Gentest (Woburn, MA). Catalytic activity was determined using a kinetic assay and normalized for total P450 content.
Table 1.
Fluorescent cytochrome P450 assay conditions. The concentrations used and metabolites detected are given for each substrate. Bolded lettering in brackets represents the substrate abbreviations. The excitation (Ex) and emission (Em) conditions are shown as the wavelength/bandwidth of filters used and the wavelength was always within 10 nm of the optimal wavelength for each metabolite.
| Substrate | Concentration (μM) | Metabolite | Ex (nm) | Em (nm) | Human CYP Specificity |
|---|---|---|---|---|---|
| 7-ethoxyresorufin (ER) | 2 | resorufin | 540/35 | 590/20 | 1A1>1A2=1B1 |
| 7-methoxyresorufin (MR) | 5 | resorufin | 540/35 | 590/20 | 1A2 |
| 7-benzyloxyresorufin (BR) | 5 | resorufin | 540/35 | 590/20 | 1A, 2B, 3A |
| 7-pentoxyresorufin (PR) | 5 | resorufin | 540/35 | 590/20 | 2B |
| 7-benzyloxy-4-(trifluoromethyl)coumarin (BFC) | 1000 | 7-hydroxy-4-(trifluoromethyl)coumarin (HFC) | 400/30 | 528/20 | 3A4 |
| 7-methoxy-trifluoromethylcoumarin (MFC) | 1000 | 7-hydroxy-4-(trifluoromethyl)coumarin (HFC) | 400/30 | 528/20 | 2C9, 2C19, 1A2 |
| 7-benzyloxyquinoline (BQ) | 1000 | 7-hydroxyquinoline (HQ) | 400/30 | 528/20 | 3A4 |
| dibenzylfluorescein (DBF) | 10 | fluorescein | 485/20 | 528/20 | 2C8, 2C9, 3A4 |
| 3-cyano-7-ethoxycoumarin (CEC) | 10 | 3-cyano-7-hydroxycoumarin (CHC) | 400/30 | 460/40 | 2C9, 2C19, 1A2 |
| 3-[2-(N,N-diethyl-N-methylamino)ethyl]-7-methoxy-4-methylcoumarin (AMMC) | 50 | 3-[2-(N,N-diethylamino)ethyl]-7-hydroxy-4-methylcoumarin hydrochloride (AHMC) | 400/30 | 460/40 | 2D6 |
| 7-Methoxy-4-(aminomethyl)-coumarin (MAMC) | 50 | 7-hydroxy-4-(aminomethyl)-coumarin (HAMC) | 400/30 | 460/40 | 2D6 |
2.3 Benzo[a]pyrene metabolism
Incubations contained 0.1 M sodium phosphate, pH 7.4; 0.25 mg ml−1 human microsomal epoxide hydrolase (BD Biosciences, San Jose, CA); 1 mM NADPH, membrane fractions containing 100 mg total protein of recombinant zebrafish CYPs coexpressed with human CYP reductase; and benzo[a]pyrene (BaP; Accustandard, New Haven, CT) at a final concentration of 0, 20, 40, 80, or 120 μM. BaP was dissolved in DMSO such that the final concentration of DMSO was 0.5%. Reactions without epoxide hydrolase were assayed at 40 μM BaP only.
Reactions were allowed to proceed at 28°C for 30 minutes until terminated with 250 μl of cold acetone and incubation on ice for 10 minutes. The internal standard 6-hydroxychrysene (Accustandard, New Haven, CT) was added, metabolites extracted and the sample was dried down according to Kim et al [10]. Evaporated samples were resuspended by sonication in 100 μl acetonitrile (Caledon, Georgetown, Canada) and stored under argon until analysis. Samples were analyzed by UPLC/MS for the presence of BaP-7,8,9,10-tetrol; BaP-9,10-dihydrodriol; BaP-7,8-dihydrodiol; BaP-1,6-dione; BaP-3,6-dione; BaP-6,12-dione; 9-OH-BaP; 3-OH-BaP; the parent compound BaP; and the internal standard, 6-hydroxychrysene, following methods from Zhu et al [20]. The limits of quantification were 0.01 ng ul−1 for all metabolites except BaP-7,8,9,10-tetrol, which was 0.08 ng ul−1; the limit of quantification for BaP was 0.327 ng ul−1. Catalytic activity was normalized for total P450 content.
3. Results
3.1 CYP heterologous expression
The optimization and description of these expressed proteins has been discussed in detail elsewhere [14]. Since expression of CYP proteins, which are membrane bound, is problematic in bacterial expression systems, we used the approach of Pritchard et al. [19] to clone a leader sequence (ompA2+) that would effectively target the zebrafish proteins to the bacterial outer membrane. This approach prevented the need for 5' modification of the gene; the leader sequence is cleaved after targeting allowing for the proper folding and expression of full-length, unmodified vertebrate protein in bacteria. Each CYP gene was co-expressed with the human reductase gene; reductase is a co-enzyme required for appropriate electron transfer from NADPH to the CYP enzyme during catalysis. Reductase levels were high and similar amongst the expressed proteins [14]. The total P450 content was determined in each preparation using CO difference spectra and all catalytic data were normalized for P450 content.
3.2 Catalyic activity of zebrafish CYP1 proteins in fluorescent assays
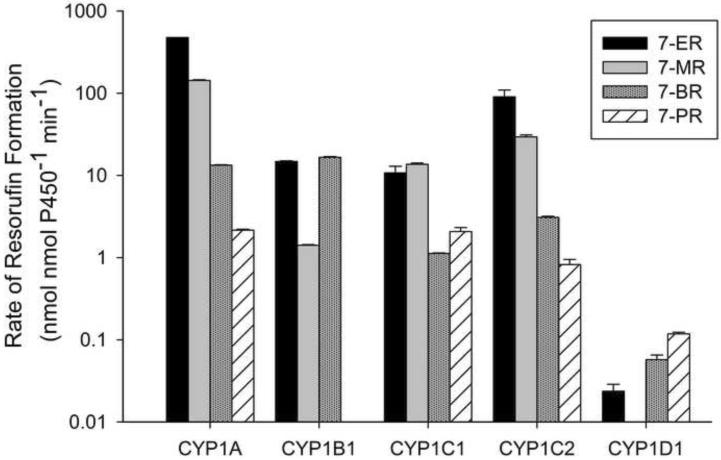
The catalytic activity of each expressed CYP were assessed for 11 synthetic substrates (Table 1) and the model PAH, benzo[a]pyrene (BaP). None of the CYPs tested were able to metabolize the quinoline BQ or the methylcoumarins AMMC and MAMC. All of the CYP1s metabolized ER and BR (Figure 1). CYP1A had the highest rate of ER metabolism; catalytic activity was 1–4 orders of magnitude larger for this protein than the other CYP1s. CYP1C1, CYP1C2 and CYP1B1 all had rates of resorufin production from ER greater than 10 nmol nmol P450−1 min−1, although CYP1C2 had catalytic activity for ER approximately 1 order of magnitude larger than CYP1C1 and CYP1B1. CYP1D1 had the lowest rate of metabolism for ER and all other substrates tested.
Figure 1.
Metabolism of alkoxyresorufins by expressed zebrafish CYP1s. Dealkylation was detected by quantification of the fluorescent metabolite resorufin (see materials and methods, and Table 1 for full details). Results are Mean ± SEM, n=2. PR metabolism by CYP1B1 and MR metabolism by CYP1D1 were below the limits of detection.
The catalytic profiles for ER and MR were similar (Figure 1) and except for CYP1C1, ER metabolism was always higher. CYP1A had the highest rate of resorufin formation from MR, and CYP1B1 and CYP1C1 had similar and more moderate rates of catalytic activity. Metabolism of MR by CYP1C2 was approximately 1 order of magnitude larger than CYP1C1. Metabolism of BR was similar for CYP1A1 and CYP1B1; less activity was seen for this substrate by the CYP1C proteins. As for all resorufin substrates, CYP1D1 had much lower rates of BR metabolism compared to the other CYP1s. PR was metabolized by all CYP1s except CYP1B1 although overall CYP1 activity for PR was lower compared to the other alkoxyresorufins (Figure 1).
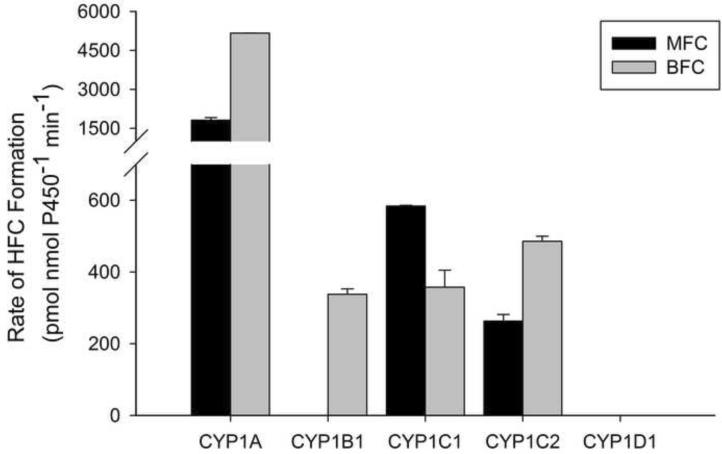
The trifluoromethylcoumarin substrates displayed similar metabolic profiles (Fig. 2). MFC and BFC were each metabolized the most by CYP1A, and moderately by each of the CYP1Cs. CYP1B1 was unable to metabolize MFC, and CYP1D1 did not metabolize either MFC or BFC (Fig. 2).
Figure 2.
Metabolism of trifluoromethylcoumarins by expressed zebrafish CYP1s. Dealkylation was detected by quantification of the fluorescent metabolite HFC from either BFC or MFC (see materials and methods, and Table 1 for full details). Results are Mean ± SEM, n=2. MFC metabolism by CYP1B1, and MFC and BFC metabolism by CYP1D1 were below the limits of detection.
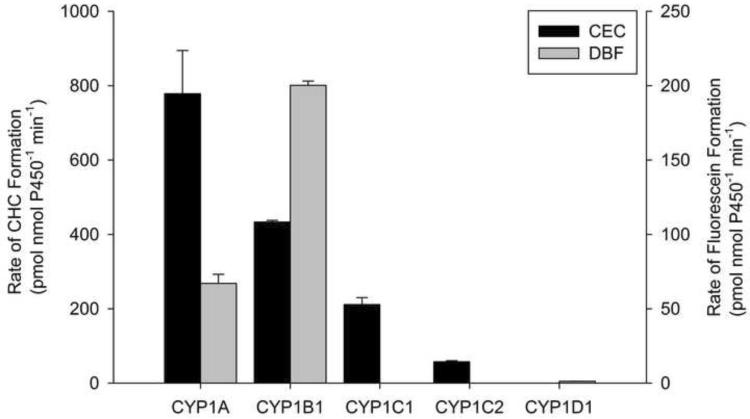
DBF was only metabolized by CYP1A, CYP1B1 and CYP1D1 (Fig. 3), but at rates that were much less than that seen for the alkoxyresorufins. CEC was metabolized the most by CYP1A and not at all by CYP1D1 (Fig. 3). CYP1C1 metabolized CEC at a 4-fold higher rate than CYP1C2. The catalytic rates of CYP1s for DBF and CEC were significantly less than those seen for the alkoxyresorufins (nmol compared to pmol).
Figure 3.
Metabolism of CEC and DBF by zebrafish. Metabolism was quantified by detection of the fluorescent metabolite CHC or fluorescein for CEC and DBF, respectively (see materials and methods, and Table 1 for full details). Results are Mean ± SEM, n=2. Metabolism of DBF by CYP1C1 and CYP1C2 and CEC metabolism by CYP1D1 were below the limits of detection.
3.3 BaP metabolism by zebrafish CYP1 proteins
At the two lowest BaP concentrations tested, 20 and 40 μM, most metabolites were detectable, yet below the limit of quantification. With only quantifiable data for the two largest BaP concentrations, 80 and 120 μM, it was not possible to calculate kinetic parameters (Km or Vmax) for the total metabolism of BaP by zebrafish CYP1s. Total metabolism of BaP was calculated as the sum of all quantifiable BaP metabolites. Total metabolism of BaP and the regioselectivity of BaP metabolism by CYP1s with 80 μM BaP are shown in Table 2. All metabolites were detected in at least one of the samples and quantifiable amounts of BaP-1,6-dione and BaP-3,6-dione were formed by all CYP1s (Table 2). The 3-hydroxy BaP was the most prominent metabolite of CYP1B1 and CYP1C2, accounting for 45.0 and 53.5 % of the total BaP metabolism, respectively. CYP1A and CYP1C1 predominantly produced BaP-7,8-dihydroxydiol and 3-hydroxy BaP, in nearly equal quantities of 20–25% of total BaP metabolism. The BaP -6,12- and -1,6- dione metabolites were the major products of CYP1D1 metabolism. The least formed BaP metabolite was BaP-7,8,9,10-tetrol (Table 2).
Table 2.
Benzo[a]pyrene metabolites produced by expressed zebrafish CYP1s. Rate of formation of each metabolite is given as pmol nmol P450−1 min−1 in reactions with 80 μM BaP and human epoxide hydrolase at 28°C.
| CYP Isoform | 7,8,9,10 tetrol | 9,10 diol | 7,8 diol | 1,6 dione | 3,6 dione | 6,12 dione | 9-OH | 3-OH | Total Metabolism |
|---|---|---|---|---|---|---|---|---|---|
| CYP1A | * | 6.03 | 15.29 | 7.66 | 10.00 | 4.21 | 3.51 | 13.71 | 60.41 |
| CYP1B1 | * | * | 40.63 | 29.64 | 24.48 | 9.69 | 3.03 | 87.87 | 195.34 |
| CYP1C1 | * | 2.53 | 24.17 | 18.13 | 13.66 | 13.07 | 23.70 | 21.18 | 116.45 |
| CYP1C2 | * | 2.95 | 60.38 | 8.02 | 11.12 | * | + | 94.70 | 177.17 |
| CYP1D1 | * | ND | ND | 9.35 | 7.37 | 11.06 | 6.11 | * | 33.89 |
Metabolite detected but below limit of quantification
Metabolite detected at other substrate concentrations tested
ND - Metabolite was not detected
4. Discussion
Though the metabolic capacity of the CYP1 family has been extensively investigated in mammals [12, 15, 21], less is known about the metabolic capacity of CYP1s in fish, and especially the more recently discovered CYP1Cs and CYP1D1. We began by investigating the catalytic activity of each CYP using a variety of specific fluorescent substrates, some of which have been used in non-mammalian vertebrates [22, 23] and represent both typical and atypical mammalian CYP1 substrates. Here we have begun to show differences in substrate specificity between the piscine CYP1 isoforms, using heterologously expressed zebrafish proteins.
4.1 Resorufin-based substrate activity (ARODs)
The alkoxyresorufins ER and MR are widely used as biomarkers in fish and mammals for CYP1 activity. ER and MR are widely believed to be predominantly metabolized by CYP1A in fish [24, 25] as they are well correlated in hepatic microsomes from AhR ligand exposed fish [26]. Though mammalian CYP1A1 is most responsible for ER metabolism, CYP1A2 and CYP1B1 are capable of significant ER metabolism [27], suggesting that this substrate is a common CYP1 substrate in at least mammals. Zebrafish CYP1A was the dominant CYP responsible for ER metabolism (Figure 1) with at least an order of magnitude higher activity for this substrate than the other CYP1s, except CYP1C2. CYP1B1 had activity that was about 30-fold less then CYP1A, similar to the difference seen between mammalian CYP1A and CYP1B1 [18]. The CYP1Cs were able to metabolize ER to significant levels; CYP1C1 had activity similar to CYP1B1, and CYP1C2 had only a five-fold lower catalytic rate than CYP1A, suggesting that ER is a broad CYP1 substrate in fish. The rates of ER metabolism across the CYP1s suggest that ER activity in fish and possibly other non-mammalian vertebrate livers are the result of overall CYP1 activity, particularly from CYP1A and CYP1C2, and not solely CYP1A activity. Differences in the expression of CYP1 isoforms will likely determine their relative roles in vivo. CYP1D1 appears to be the sole CYP1 in fish with little capacity for ER metabolism.
Mammalian CYP1A2 activity is commonly measured by MR activity, though it is also strongly metabolized by CYP1A1 [18]. Zebrafish CYP1A had the highest activity for MR; CYP1B1 metabolized MR at a 100-fold lower rate (Figure 1). Mammalian CYP1B1 had a low rate of MR metabolism compared to either mammalian CYP1A1 or CYP1A2 [18]. A linear relationship between metabolism of ER and MR has been reported in scup liver microsomes [25], tilapia liver S9 preparations [28], trout exposed to β-naphthoflavone [a proto-typical AHR ligand; 26], and AHR induced fish hepatocytes from multiple fish species [29] suggesting that both ER and MR activity were catalyzed by CYP1A [29]. Our data shows that CYP1A is likely the dominant CYP1 responsible in vivo for both ER and MR metabolism in liver where CYP1A has the highest expression compared to other CYP1s [7, 11], but that significant contributions by other CYP1s, particularly CYP1C2, are likely to be biologically relevant. The biological relevance of CYP1-mediated ER and MR metabolism may be higher in extrahepatic organs. ER was metabolized three times faster than MR by both zebrafish CYP1A and CYP1C2, a difference similar to the ER:MR metabolism ratios of 2.5 to 4 seen in fish hepatocytes [29], supporting the application of our in vitro data to in vivo functional differences.
PR activity, a marker of CYP2B in mammals, has been detected in fish [30], though the specific CYPs responsible for PR metabolism have not been identified. Phenobarbital, a major mammalian CYP2B inducer, inhibited PROD activity in fish [31]. PR metabolism was neither correlated with ER or MR metabolism in fish liver S9 preparations [28] nor in liver microsomes from trout treated with β-napthoflavone [26], suggesting that it is not a substrate for CYP1A. Our data does show that CYP1A, CYP1C1 and CYP1C2 are all able to metabolize PR at similar rates, but much lower than any other alkoxyresorufins, suggesting some but not all PR metabolism may be a result of CYP1 related activity. However, PR was metabolized at a rate that was 100 times less than ER by CYP1C2, and 250 times less than ER by CYP1A (Figure 1). Tilapia S9 preparations had from 3 to 60 fold less PR activity [28], turbot S9 preparations had 20 fold less PR activity [30], killifish had between 30 and 60 fold less PR activity [26] and rainbow trout had 50 fold less PR activity [26] than ER metabolism. Since our rates of PR metabolism compared to ER for purified CYP1s are much lower than those reported for total hepatic preparations, it is likely that the major CYPs responsible for PROD activity in fish hepatic microsomes are not CYP1 proteins.
BR is believed to have a broader enzyme specificity than the other alkoxyresorufins; BR was metabolized by CYP1A, CYP2B and CYP3A in mammals [30]. BR and ER were the only fluorometric substrates to be metabolized by each zebrafish CYP1. Like our zebrafish CYP1A and CYP1B1 proteins, mammalian CYP1A1 and CYP1B1 metabolize BR at similar rates [18]. Fish exposed to CYP1 inducers have increased BR metabolism, but not to the extent that is seen with ER [26, 30, 31] or MR [26, 30]. Our data suggests that the CYP1s, mostly CYP1A and CYP1B1, may be responsible for some BR metabolism in fish, but supports the notion that other CYPs are responsible for most of the in vivo BR activity in fish [30, 31].
4.2 Non-AROD based fluorescent assays
MFC (Figure 2) and CEC (Figure 3), both substrates for human CYP2C9, CYP2C19 and CYP1A2, are metabolized by zebrafish CYP1A, and by a lesser extent from CYP1C1, and CYP1C2. Since both MFC and CEC are metabolized by mammalian CYP1A2, it is not surprising that they are metabolized by zebrafish CYP1A. However, mammalian CYP1B1 can metabolize both CEC and MFC [32], yet zebrafish CYP1B1 was only able to metabolize CEC and not MFC. Overall the CYP1s had a slightly higher preference for CEC over MFC, but neither is likely a specific CYP1 substrate and these compounds are likely broader CYP substrates in fish similar to mammals.
BFC, a vertebrate CYP3A substrate not usually associated with CYP1 activity, was metabolized by CYP1A at an order of magnitude higher than the other CYP1s (Figure 2). CYP1A metabolized BFC at a faster rate than zebrafish CYP3A65 [14] suggesting that the use of BFC metabolism as a marker of CYP3A activity in fish [33] may be problematic. DBF, another mammalian CYP3A4 substrate, was only minimally metabolized by zebrafish CYP1s (Figure 3). Yet, DBF is also a CYP2C8 and CYP2C9 substrate in mammals. Overall, CYP1A has the broadest substrate profile of any zebrafish CYP1 tested, and broader then would be predicted based on mammalian CYP1A enzymes.
4.3 BaP metabolism
Hepatic CYP1 enzymes catalyze the first step in the metabolism of BaP, a model polycyclic aromatic hydrocarbon (PAH), that can result in activation to mutagenic and carcinogenic intermediates [34]. The dominant BaP metabolite found in mammals is usually 3-hydroxy-BaP (3-OH-BaP) [34], which was the most common zebrafish metabolite found from CYP1B1 and CYP1C2 and one of the dominant metabolites of CYP1A and CYP1C2. Our zebrafish CYP1A had a four-fold higher proportion of 3-OH-BaP than 9-hydroxy-BaP (9-OH-BaP) similar to that seen in heterologous zebrafish CYP1A expressed in yeast [35]. However, 3-OH-BaP was produced at much higher levels relative to other metabolites in yeast expressed zebrafish CYP1A [35] than our bacterial expressed CYP1A. Different expression systems can have an effect on rates of BaP metabolism [10], so whether fish CYP1A produces 3-OH-BaP at much higher rates than other metabolites in vivo is unclear at this time.
Contrary to the phenol metabolites, the diol epoxide metabolites are particularly genotoxic, of which 7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene (BaPDE) can most easily form DNA adducts [36]. BaPDE is formed from BaP through a series of reactions starting with the CYP catalyzed formation of BaP-7,8-epoxide, followed by epoxide hydrolase (EH) catalyzed formation of BaP-7,8-dihydrodiol (BaP-7,8-diol), and then CYP formation of BaPDE [36]. Mammalian CYP1A1 forms the most BaP-7,8-diol, the first step in production of the genotoxic diol epoxide [10]. Mammalian CYP1A2 has a very low rate of BaP metabolism, and forms almost no BaP-7,8-diol. Zebrafish CYP1C2 formed the most BaP-7,8-diol but no CYP1 produced quantifiable rates of the BaP-7,8,9,10-tetrol. CYP1D1 formed the fewest total metabolites detected and no BaP-7,8-diol, similar to mammalian CYP1A2 [10]. The toxic intermediate primarily responsible for DNA adduct formation is the BaP-r-7,t-8-dihydrodiol-t-9,10-epoxide(anti). Our methods did not distinguish the stereochemistry associated with the diol and tetrol metabolites, therefore, without additional studies with isoform selective inhibitors and DNA binding studies, it is difficult to predict the in vivo significance of BaP-7,8-diol formation.
Contrary to all fluorescent substrates tested, CYP1D1 had rates of BaP metabolism that were more similar to rates seen with the other CYP1 proteins. Considering that we had both measurable peaks in the CO-difference spectra and similar BaP metabolism, this suggests that CYP1D1 was not inactive in our expression system but that there are major functional differences between CYP1D1 and the other CYP1 proteins. CYP1D1 was the only CYP to not produce detectable amounts of all BaP metabolites measured and had a regioselectivity for BaP that was distinct from the other zebrafish CYP1s (Table 2). CYP1D1 produced no detectable BaP-7,8-diol, and very little 3-OH-BaP, which were major metabolites of all other zebrafish CYP1s. This suggests that CYP1D1 may produce a distinct spectrum of BaP metabolites in vivo.
4.4 Conclusions
This is the first report of the catalytic profile of purified protein from the entire CYP1 family of a fish species. We have demonstrated that ER and MR, known mammalian CYP1A1 and CYP1A2 substrates respectively and commonly used in fish as a marker of CYP1A activity, are highly metabolized by CYP1A, but CYP1C1 and CYP1C2 likely contribute to this activity in vivo. In fact, knockdown of CYP1C1 significantly reduces EROD activity in Fundulus heteroclitus embryos (K. Willett, unpublished data), which strongly supports our interpretations of the in vitro data. Two other alkoxyresorufins, PR and BR, are likely not specific indicators of CYP1 activity in vivo. BFC, a mammalian CYP3A substrate, is strongly metabolized by CYP1A, and more than CYP3A65, such that it may contribute to BFC activity in vivo. Yet, BFC metabolism is neither significantly induced in hepatic microsomes from rainbow trout exposed to β-napthoflavone nor are BFC metabolism and EROD or MROD well correlated [26]. The potential contribution of CYP1s toward BFC metabolism in vivo remains unclear. Fish CYP1A was the broadest and most active CYP for the substrates tested. Both zebrafish CYP1A and CYP1B1 metabolized ER, BR, and MR similar to mammalian CYP1A1 and CYP1B1, respectively. CYP1C1 and CYP1C2 had similar substrate specificity, though with differences in activity for most substrates. Overall, CYP1D1 seems to have very minimal, if any, activity for all of the substrates tested except BaP, suggesting the function of CYP1D1 is different from other CYP1s.
Acknowledgements
We would like to thank Dr. Thomas Friedberg for supplying the human cytochrome P450 reductase construct and Dr. Frederick W. Dahlquist for supplying the pCW expression vector. We thank Emily Smith and Eric Hirsch for the optimization of fluorescent assays. This study was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC; Discovery Grant #328204 to JYW) and the Canadian Foundation for Innovation and Ontario Research Fund. The Department of Biology, McMaster University, provided partial support for M.L.S. We would like to acknowledge the technical help of Dr. Bonnie Avery and Sai Boddu and support from NIH (NIEHS grant number R01ES012710 to KLW) for the BaP metabolite detection research.
Abbreviations
- AHR
aryl hydrocarbon receptor
- BaP
benzo(a)pyrene
- CYP
cytochrome P450
- PAH
polycyclic aromatic hydrocarbon
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Guengerich FP. Chem. Res. Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- [2].Zhang Y, Gaikwad NW, Olson K, Zahid M, Cavalieri EL, Rogan EG. Metabolism. 2007;56:887–894. doi: 10.1016/j.metabol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- [3].McGinnity DF, Griffin SJ, Moody GC, Voice M, Hanlon S, Friedberg T, Riley RJ. Drug Metab. Dispos. 1999;27:1017–1023. [PubMed] [Google Scholar]
- [4].Blom MJ, Wassink MG, Kloosterboer HJ, Ederveen AG, Lambert JG, Goos HJ. Drug Metab. Dispos. 2001;29:76–81. [PubMed] [Google Scholar]
- [5].Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. J. Biol. Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- [6].Goldstone JV, Goldstone HM, Morrison AM, Tarrant A, Kern SE, Woodin BR, Stegeman JJ. Mol. Biol. Evol. 2007;24:2619–2631. doi: 10.1093/molbev/msm200. [DOI] [PubMed] [Google Scholar]
- [7].Goldstone JV, Jonsson ME, Behrendt L, Woodin BR, Jenny MJ, Nelson DR, Stegeman JJ. Arch. Biochem. Biophys. 2009;482:7–16. doi: 10.1016/j.abb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nakajima M, Iwanari M, Yokoi T. Toxicol. Lett. 2003;144:247–256. doi: 10.1016/s0378-4274(03)00216-9. [DOI] [PubMed] [Google Scholar]
- [9].Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Biochem. Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- [10].Kim JH, Stansbury KH, Walker NJ, Trush MA, Strickland PT, Sutter TR. Carcinogenesis. 1998;19:1847–1853. doi: 10.1093/carcin/19.10.1847. [DOI] [PubMed] [Google Scholar]
- [11].Jonsson ME, Orrego R, Woodin BR, Goldstone JV, Stegeman JJ. Toxicol. Appl. Pharmacol. 2007;221:29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee AJ, Cai MX, Thomas PE, Conney AH, Zhu BT. Endocrinology. 2003;144:3382–3398. doi: 10.1210/en.2003-0192. [DOI] [PubMed] [Google Scholar]
- [13].Shimada T, Gillam EM, Oda Y, Tsumura F, Sutter TR, Guengerich FP, Inoue K. Chem. Res. Toxicol. 1999;12:623–629. doi: 10.1021/tx990028s. [DOI] [PubMed] [Google Scholar]
- [14].Scornaienchi ML, Thornton C, Willett KL, Wilson JY. In Submission.
- [15].Gautier JC, Lecoeur S, Cosme J, Perret A, Urban P, Beaune P, Pompon D. Pharmacogenetics. 1996;6:489–499. doi: 10.1097/00008571-199612000-00002. [DOI] [PubMed] [Google Scholar]
- [16].Crespi CL, Stresser DM. J. Pharmacol. Toxicol. Methods. 2000;44:325–331. doi: 10.1016/s1056-8719(00)00112-x. [DOI] [PubMed] [Google Scholar]
- [17].Yang SK, Selkirk JK, Plotkin EV, Gelboin HV. Cancer Res. 1975;35:3642–3650. [PubMed] [Google Scholar]
- [18].Murray GI, Melvin WT, Greenlee WF, Burke MD. Annu. Rev. Pharmacol. Toxicol. 2001;41:297–316. doi: 10.1146/annurev.pharmtox.41.1.297. [DOI] [PubMed] [Google Scholar]
- [19].Pritchard MP, McLaughlin L, Friedberg T. Methods Mol. Biol. 2006;320:19–29. doi: 10.1385/1-59259-998-2:19. [DOI] [PubMed] [Google Scholar]
- [20].Zhu S, Li L, Thornton C, Carvalho P, Avery BA, Willett KL. J. Chromatogr. B. 2008;863:141–149. doi: 10.1016/j.jchromb.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guo Z, Gillam EM, Ohmori S, Tukey RH, Guengerich FP. Arch. Biochem. Biophys. 1994;312:436–446. doi: 10.1006/abbi.1994.1330. [DOI] [PubMed] [Google Scholar]
- [22].Kashiwada S, Hinton DE, Kullman SW. Comp. Biochem. Phys. C. 2005;141:338–348. doi: 10.1016/j.cca.2005.07.006. [DOI] [PubMed] [Google Scholar]
- [23].Stegeman JJ, Woodin BR, Singh H, Oleksiak MF, Celander M. Comp. Biochem. Phys. C. 1997;116:61–75. doi: 10.1016/s0742-8413(96)00128-4. [DOI] [PubMed] [Google Scholar]
- [24].Klotz AV, Stegeman JJ, Woodin BR, Snowberger EA, Thomas PE, Walsh C. Arch. Biochem. Biophys. 1986;249:326–338. doi: 10.1016/0003-9861(86)90009-3. [DOI] [PubMed] [Google Scholar]
- [25].Schlezinger JJ, Stegeman JJ. Aquat. Toxicol. 2001;52:101–115. doi: 10.1016/s0166-445x(00)00141-7. [DOI] [PubMed] [Google Scholar]
- [26].Smith EM, Wilson JY. Aquat. Toxicol. 2010 doi: 10.1016/j.aquatox.2010.01.005. DOI:10.1016/j.aquatox.2010.1001.1005. [DOI] [PubMed] [Google Scholar]
- [27].Shimada T, Yamazaki H, Foroozesh M, Hopkins NE, Alworth WL, Guengerich FP. Chem. Res. Toxicol. 1998;11:1048–1056. doi: 10.1021/tx980090+. [DOI] [PubMed] [Google Scholar]
- [28].Parente TE, De-Oliveira AC, Silva IB, Araujo FG, Paumgartten FJ. Chemosphere. 2004;54:1613–1618. doi: 10.1016/j.chemosphere.2003.09.027. [DOI] [PubMed] [Google Scholar]
- [29].Smeets JM, Wamsteker J, Roth B, Everaarts J, van den Berg M. Chemosphere. 2002;46:163–172. doi: 10.1016/s0045-6535(01)00054-6. [DOI] [PubMed] [Google Scholar]
- [30].Hartl MG, Kilemade M, Sheehan D, Mothersill C, O'Halloran J, O'Brien NM, van Pelt FN. Mar. Environ. Res. 2007;64:191–208. doi: 10.1016/j.marenvres.2007.01.002. [DOI] [PubMed] [Google Scholar]
- [31].Addison RF, Sadler MC, Lubet RA. Biochem. Pharmacol. 1987;36:1183–1184. doi: 10.1016/0006-2952(87)90435-7. [DOI] [PubMed] [Google Scholar]
- [32].Stresser DM, Turner SD, Blanchard AP, Miller VP, Crespi CL. Drug Metab. Dispos. 2002;30:845–852. doi: 10.1124/dmd.30.7.845. [DOI] [PubMed] [Google Scholar]
- [33].Hegelund T, Ottosson K, Radinger M, Tomberg P, Celander MC. Environ. Toxicol. Chem. 2004;23:1326–1334. doi: 10.1897/03-155. [DOI] [PubMed] [Google Scholar]
- [34].Gelboin HV. Physiol. Rev. 1980;60:1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- [35].Miranda CL, Chung WG, Wang-Buhler JL, Musafia-Jeknic T, Baird WM, Buhler DR. Aquat. Toxicol. 2006;80:101–108. doi: 10.1016/j.aquatox.2006.07.018. [DOI] [PubMed] [Google Scholar]
- [36].Peltonen K, Dipple A. J. Occup. Environ. Med. 1995;37:52–58. doi: 10.1097/00043764-199501000-00008. [DOI] [PubMed] [Google Scholar]