Abstract
Cardiolipin is a unique phospholipid of the mitochondrial inner membrane. Its peroxidation correlates with release of cytochrome c and induction of apoptosis. The phthalocyanine photosensitizer Pc 4 binds preferentially to the mitochondria and endoplasmic reticulum. Earlier Förster resonance energy transfer studies showed colocalization of Pc 4 and cardiolipin, which suggests cardiolipin as a target of photodynamic therapy (PDT) with Pc 4. Using liposomes as membrane models, we find that Pc 4 binds to cardiolipin-containing liposomes similarly to those that do not contain cardiolipin. Pc 4 binding is also studied in MCF-7c3 cells and those whose cardiolipin content was reduced by treatment with palmitate. Decreased levels of cardiolipin are quantified by thin-layer chromatography. The similar level of binding of Pc 4 to cells, irrespective of palmitate treatment, supports the lack of specificity of Pc 4 binding. Thus, factors other than cardiolipin are likely responsible for the preferential localization of Pc 4 in mitochondria. Nonetheless, cardiolipin within liposomes is readily oxidized by Pc 4 and light, yielding apparently mono- and dihydroperoxidized cardiolipin. If similar products result from exposure of cells to Pc 4-PDT, they could be part of the early events leading to apoptosis following Pc 4-PDT.
Keywords: liposomes, cardiolipin, affinity constant, phthalocyanine, Pc 4, photodynamic therapy
Introduction
Phthalocyanines belong to a class of porphyrin-like macrocycles currently employed as photosensitizers in experimental photodynamic therapy (PDT), a minimally invasive and highly localized cancer treatment.1, 2 In PDT, a photosensitizing drug is excited to the singlet state with an appropriate wavelength of visible light and undergoes intersystem crossing to the triplet state. Energy is transferred from the triplet state to ground-state molecular oxygen to produce primarily singlet oxygen (1O2), which can react with a variety of cellular molecules, such as lipids, proteins, and nucleic acids, producing an oxidative stress in the host cells.2, 3 It is assumed that type II photochemistry, which produces singlet oxygen, is the dominant mechanism for PDT in cells and tissues.1, 4 The oxidative stress in the cells can induce a variety of cell death pathways. Cells that have been treated with PDT can undergo apoptosis, necrosis, and∕or autophagy, with the dominant mechanism dependent on the cellular site of the photosensitizer, the overall dose, and the genetic composition of the cells.2, 5 Whereas a specific singlet oxygen-generated product of cholesterol has been demonstrated6, 7 to be formed when the plasma membrane and other cholesterol-rich membranes are damaged by PDT, the immediate critical molecular target(s) of singlet oxygen in mitochondria have not been identified.
Pc 4, a phthalocyanine photosensitizer first synthesized at Case Western Reserve University8 and now in clinical trials at University Hospitals Case Medical Center, shows promise for treatment of cancer.9 Pc 4 has been shown to bind preferentially and with high affinity to both mitochondrial and endoplasmic reticulum (ER)∕Golgi membranes.10, 11, 12, 13, 14 Numerous reports have implicated mitochondria as important targets for PDT. Photosensitizers that localize in mitochondria can be highly efficient in killing cells.15, 16 In addition to preferential distribution in specific membranes, Pc 4 exerts photodamage to certain membrane-associated molecules, such as Bcl-2, which points to the possibility of some specificity in the binding of the photosensitizer.17 Pc 4-PDT generates reactive oxygen species (ROS) in the mitochondria, which leads to opening of the permeability transition pore (PTP), loss of mitochondrial membrane potential, and release of cytochrome c (Cyt-c) and Smac∕DIABLO from mitochondria.11, 18, 19 Although these changes in mitochondria are implicated in most apoptotic pathways, the initial trigger remains in dispute.
Earlier studies from our laboratory implicated cardiolipin (CL) as a possible binding site and target for Pc 4. In those studies, Förster resonance energy transfer (FRET) was observed from the CL ligand 10-N-nonyl-acridine orange (NAO) to Pc 4, suggesting possible colocalization of Pc 4 and CL in mitochondrial membranes.12 CL is confined almost exclusively (>80%) to the inner mitochondrial membrane. Within that membrane, CL is distributed20 between the inner and outer leaflets at a ratio of 60:40, and is associated with many proteins needed for mitochondrial functions, such as respiration.21, 22 CL comprises 14% of the mitochondrial lipids with slight variations in different species,23 and its four constituent fatty acids are comprised of 90 to 95% of linoleic acid24 (C-18:2), which makes it highly sensitive to oxidation by 1O2. During apoptosis the asymmetric distribution of CL collapses, and the level of CL in the outer mitochondrial membrane increases to 40% of the total content, whereas 60% of it remains in the inner membrane.20 Thus, CL is an attractive candidate for one initial target of PDT with Pc 4 or any photosensitizer with similar binding properties. The four unsaturated fatty acids of CL may act as quenchers∕targets of 1O2, generating oxidized CL that may be a trigger of apoptotic events. Because of its high reactivity, 1O2 reacts within a very short range (of the order of 10 to 20 nm) of its site of formation, which must be at the site of localization of the photosensitizer.25
In recent studies in our laboratories, it was found that Pc 4-PDT-generated 1O2 efficiently oxidizes Cyt-c in simple solution, generating a variety of oxidized peptides, including at least two products of oxidized histidine that are26 unique to 1O2.26 These products were also found27 when Cyt-c was incorporated into liposomes before PDT or when isolated rat heart mitochondria were exposed to Pc 4-PDT. CL of liposomes or mitochondria was also oxidized under the same conditions; this could have occurred initially by direct interaction with 1O2 and∕or secondarily via the peroxidase activity of Cyt-c, which is known20 to be activated by CL. It is also possible that some of the oxidation of CL resulted from radical chain reactions initiated at sites removed from the final product.28, 29
Ostrander et al. demonstrated a highly significant correlation between Cyt-c release from mitochondria and a decrease in mitochondrial CL, the latter resulting from acylation of CL precursors with palmitate.30 Wilson et al. demonstrated the ability of NAO to competitively inhibit the uptake of the photosensitizing preparation Photofrin® into mitochondria, indicating that some photosensitizers might bind to CL of the inner mitochondrial membrane.31 Kriska et al.32 demonstrated oxidation of CL following exposure of human breast cancer cells in culture to photosensitization by protoporphyrin IX. Kagan et al.20 showed that Cyt-c peroxidase activity propagates the oxidation of CL and that this reaction is pivotal in the triggering of apoptosis (via mitochondrial membrane permeabilization and release of proapoptotic factors from mitochondria). Moreover, accumulation of peroxidized CL may act as a molecular switch that initiates the development of proapoptotic events when autophagy fails to effectively eliminate damaged mitochondria. Thus, CL-mediated signaling may be a key point in regulation of both autophagy and apoptosis.20 Among the many contributions of Hasan and her coworkers to understanding PDT mechanisms, one highlight is her work on targeting photosensitizers to molecules specifically overexpressed in cancer cells; the goal of studies such as Verma et al.33 is to increase the extent and specificity of the photodynamic response. In general, PDT targeting of the cellular compartment causes direct tumor cytotoxicity, while PDT with photosensitizers confined within the tumor vasculature result in extensive vascular damage and shutdown.33 The purpose of this study was to test whether Pc 4 might preferentially bind to CL. For the first part of the study, liposomes of different lipid compositions were used as organelle membrane models to examine the interaction of Pc 4 and CL by spectroscopic techniques. For the second part, human breast cancer MCF-7c3 cells were treated with palmitate with the expectation that it would decrease the cellular CL content.30 The relative level of CL was estimated by thin-layer chromatography34 (TLC). The ability of the palmitate-treated cells to bind Pc 4 was then measured by flow cytometry and compared with nontreated cells.
Materials and Methods
Chemicals
Pc 4 was synthesized by methods previously reported.35 Cholesterol (CHOL), L-α-dimyristoylphosphatidylcholine (DMPC), and L-α-dimyristoylphosphatidylethanolamine (DMPE) were obtained from Avanti Polar Lipids (Alabaster, Alabama). We obtained 10-N-nonyl-acridine orange (NAO) from Invitrogen (Carlsbad, California). CL, 9,10-anthraquinone-2-sulfonate sodium salt (AQS−), sodium palmitate, phosphate-buffered saline (PBS), fatty-acid-free bovine serum albumin (BSA), and carbonyl cyanide m-chlorophenylhydrazone (CCCP) were purchased from Sigma-Aldrich (St. Louis, Missouri). Tetramethylrhodamine methylester (TMRM) was from Molecular Probes, Inc. (Eugene, Oregon). RPMI 1640, fetal bovine serum, and penicillin∕streptomycin were from Hyclone (South Logan, Utah). All other compounds were the highest grade commercially available. Water was treated in a Millipore Milli-Q system (Billerica, Massachusetts).
Estimation of the pKa of Pc 4
The pKa of Pc 4 was calculated using V8.0 Batch pKa Prediction Software (Advanced Development, Toronto, Ontario, Canada), with the assumption that the OSi(CH3)2(CH2)3N(CH3)2 group of Pc 4 is its most basic group and that CH3OSi(CH3)2(CH2)3N(CH3)2 is a good model for this group.
Liposome Preparation and Pc 4 Incorporation
Large unilamellar vesicles were prepared with a Mini-Extrudor from Avanti Polar Lipids, Inc. Typically, an aliquot of lipid solution in absolute ethanol was evaporated to form a thin film of lipid. All excess ethanol was removed during evaporation. PBS (pH=7.4), composed of 0.1 M NaH2PO4, 0.1 M K2HPO4, and 0.15 M NaCl, was added to hydrate the film. Aliquots of the mixture were extruded through a membrane of pore diameter ≅100 nm (Whatman®, Clifton, New Jersey) at 15°C above the lipid-transition temperature36 (Tc) of DMPE (Tc=49°C). To avoid the loss of Pc 4 through its absorption to the extrusion membrane, Pc 4 was incorporated into liposomes after extrusion from a stock solution of Pc 4 in tetrahydrofuran (THF)∕ethanol (EtOH) (1:1), and the liposome-Pc 4 complex was incubated at 15°C above the Tc of DMPE for 30 min. The resultant liposomes were determined to be 125 to 200 nm in diameter by dynamic light scattering on a Brookhaven Instruments Corporation 90 Plus Particle Sizing Analyzer (Holtsville, New York).
Stock Solutions of Pc 4
Stock solutions were prepared in 1:1 THF∕EtOH for the liposome studies and in dimethylformamide (DMF) for the cellular studies. The solutions were stored at 4°C and protected from ambient light.
Spectroscopic Studies
Absorption spectra were recorded on a Varian Cary 50 Bio UV-Vis Spectrophotometer (Varian, Palo Alto, California). Fluorescence spectra were recorded on a Varian Cary Eclipse Fluorescence Spectrophotometer. Pc 4 was excited at 610 nm (Q band), and the emission spectra were recorded between 620 and 800 nm. Measurements on liposomes were performed above the lipid transition temperature using a Quartz SUPRASIL (QS) controlled-temperature cell with a 10-mm path length (Hellma USA, Plainview, New Jersey).
Binding Experiments
Measurements were made at a fixed concentration of Pc 4 (6 μM) and increasing liposome concentrations from 0 mM to the concentration at which maximum association was attained. In water, Pc 4 is insoluble, highly aggregated, and nearly nonfluorescent. On the addition of liposomes, only Pc 4 monomers bind and become fluorescent. Association constants for Pc 4 were calculated as
| (1) |
where Ka is the equilibrium association constant between the aggregated dye in the aqueous phase and the monomers in the membrane-bound phase, (F−F0)∕(F∞−F0) is the fraction of phthalocyanine associated with lipid L as monomer, F is the fluorescence measured after each addition of liposomes, F0 is the fluorescence measured in the absence of lipid, F∞ is the maximum fluorescence attained, and [L] is the lipid concentration.37 All fluorescence values are those at the peak of the emission spectrum.
Fluorescence-Quenching Experiments
Quenching of Pc 4 fluorescence was studied in liposomes with or without CL, as described elsewhere.37 Briefly, increasing concentrations of AQS− (0 to 13 mM) were used to quench Pc 4 fluorescence in liposomes without or with 20% CL. Experiments were carried out at [Pc 4]=6 μM, [lipid]=2.1 mM. The lipid concentration used was that corresponding to the maximum incorporation of Pc 4 into liposomes as monomer (in the plateau region of association plots). Pc 4 was excited at 610 nm and the fluorescence emission was recorded in the range of 620 to 800 nm. These determinations were performed at 15°C. The bimolecular quenching rate constant Kq was calculated from
| (2) |
where kq is the bimolecular quenching constant, τ is the fluorescence lifetime in the absence of the quencher for a silicon phthalocyanine38 (≅5.8 ns), [Q] is the concentration of AQS−, and F and F0 are the intensity of fluorescence at the maximum of the emission spectrum in the presence or absence of AQS−, respectively.
Cell Culture
MCF-7c3 cells were grown in RPMI (Roswell Park Memorial Intitute) 1640 medium containing 10% fetal bovine serum and 1% penicillin∕streptomycin in an atmosphere of 5% CO2∕95% air at 37°C in a humidified incubator. Cells were used for experiments when they were in exponential growth.
Confocal Microscopy
MCF-7c3 cells were plated in 35-mm glass-bottomed tissue-culture dishes (MatTek Corp., Ashland, Massachusetts) at 2×105 cells per dish. Prior to addition of Pc 4, the cultures were treated for 1 h to block mitochondrial function, either by heating at 65°C or by the addition of 10 μM CCCP, a mitochondrial uncoupler, which abolishes the mitochondrial membrane potential. After treatment, the medium was removed and the cultures were incubated with 200 nM Pc 4 in complete medium for 1 h to allow cell uptake of the photosensitizer. This time had been previously shown to allow maximum uptake. Then, the complete medium was removed and replaced with serum-free RPMI (1 mL). Imaging was carried out with a 63× numerical aperture (NA) 1.4 oil immersion planapochromat objective on a Zeiss LSM 510 confocal microscope (Thornwood, New York) in the Case Comprehensive Cancer Center Confocal Microscopy Core Facility. A He∕Ne laser supplied the 633-nm excitation light to excite Pc 4, and a 650-nm long-pass filter was used to collect emission. Cells without pretreatment but with Pc 4 were included as controls.
Monitoring Mitochondrial Membrane Potential
Cells were grown as above for 48 h, then loaded with 100 nM TMRM for 30 min at 37°C. The medium was removed, and the cells were kept in 10 nM TMRM in calcium buffer (130 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 1 mM MgCl2, 25 mM Hepes, and 0.1% BSA). Images of TMRM fluorescence were collected using 561-nm excitation light from a solid state diode laser, and a 580 to 650-nm bandpass filter and carried out with a 100× NA 1.4 oil immersion planapochromat objective. After recording the first image, 10 μM CCCP was added to the cells in calcium buffer containing TMRM (10 nM). Control cells received 10 nM TMRM in calcium buffer, but without CCCP. Imaging was performed at 5-s intervals over a period of 1 min or until depolarization was observed. For evaluation of the effect of heat, we used flow cytometry and showed that 100% of the cells were dead after 1 h at 65°C.
Flow Cytometry
Cells were plated in 60-mm tissue culture dishes at 6×105 cells per dish and allowed to grow for 24 h. Albumin-bound fatty acids were prepared by stirring fatty acid sodium salts at 37°C in a molar ratio of 2:1 fatty acid to BSA. (Ref. 30). The solution was filtered through a 0.22-μm filter. Cells were serum-starved in RPMI containing 0.5% fatty acid-free BSA for 12 h, then incubated with BSA conjugated to palmitate (0.1 mM) for up to 8 h. The final concentration of BSA was always adjusted to 0.5%. After incubation with BSA-fatty acid, the medium was removed and the cells were incubated with Pc 4 (200 nM) or NAO (200 nM) in RPMI for 60 or 30 min, respectively, times previously shown to allow maximum uptake, and then harvested by trypsinization. Flow cytometric analysis was performed in the Case Comprehensive Cancer Center Flow Cytometry Core Facility, using a BD™ LSR II Flow Cytometer (San Jose, California). NAO was excited by a laser (488±5 nm), and fluorescence emission was collected with a 525±5-nm bandpass filter. Pc 4 was excited by a UV laser (355 nm), and fluorescence emission was collected with a 650-nm long-pass filter. Autofluorescence of control cells was subtracted. Cells not exposed to BSA-palmitate were used as controls.
Measurements of CL Level
Lipid extraction and separation by TLC were used to confirm the decrease of CL content of the variously treated MCF-7c3 cells. Cells were treated as described for flow cytometry. After trypsinization and counting of the cells, identical numbers of control and palmitate-treated cells were centrifuged and resuspended into 1.5 mL of milliQ water saturated with NaCl. Phospholipids were extracted by vortexing with 3 mL of chloroform:methanol [2:1, v∕v]. After centrifugation, the recovered organic phase was dried with MgSO4, filtered using cotton, and evaporated under nitrogen. Each dried extract was dissolved in chloroform (25 μL) and applied to a silica gel TLC plate. The mobile phase consisted of methanol∕chloroform∕acetic acid∕water39 (3∕0.52∕0.36∕0.12). TLC was visualized by iodine vapors to detect total lipid.34 Commercial CL served as a standard. Densitometric analysis of the bands was accomplished with the Image Quantum TL v 2005 software (Amersham Biosciences Corp., Sunnyvale, California).
Pc 4-Mediated Photo-oxidation of CL in Liposomes
Liposomes (0.2 mM) were prepared as already described but containing 80% DMPC and 20% CL and loaded with 6 μM Pc 4. Aliquots of 1 mL were transferred into 35-mm tissue culture dishes and exposed to 100 mW∕cm2 red light produced by a light-emitting diode array (EFOS, Mississauga, Ontario, Canada, λmax 670 to 675 nm) at room temperature for 40 min. Control samples were kept in the dark. Lipids were extracted from the liposome solution with 2 mL chloroform:methanol [2:1]. CL was separated by TLC as described above. The CL spot was scraped into a 0.6-mL Eppendorf tube and extracted with 100 μL chloroform:methanol [1:1]. The chloroform:methanol solution of CL was analyzed by Thermo Finnigan LCQ Advantage spectrometer (Waltham, Massachusetts) using negative-ion electrospray ionization-mass spectrometry (ESI-MS) with direct infusion.
Results and Discussion
Spectroscopic Studies
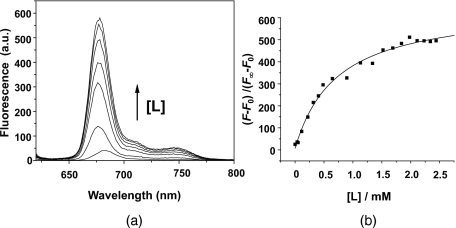
Figure 1a shows the changes in fluorescence spectrum of Pc 4 on titration with lipid. The addition of liposomes results in an increase in the fluorescence intensity at a fixed Pc 4 concentration until a maximum fluorescence intensity is reached, at which point Pc 4 is maximally monomerized, and a 1 to 2-nm blue shift in the peak wavelength has occurred, denoting incorporation of Pc 4 into the liposomes. The blue shift is typical of the change in shape of spectra for other phthalocyanines upon addition of lipid.37 Similar changes were found for all lipid compositions studied (Table 1). The identical shape of the emission spectra in PBS and liposomes and the nonsignificant wavelength shift indicates that the monomer fluorescent specie is the same in both conditions.
Figure 1.
Increase in Pc 4 fluorescence intensity with increasing liposome concentration. In this example, the liposomes contained 45% DMPC, 45% DMPE, 10% CL and [Pc 4]=6 μM. (a) Fluorescence spectra of Pc 4 in PBS with no lipid or increasing lipid concentration [L]=0.102; 0.412; 0.630; 0.889; 1.120 and 1.5510 mM, and (b) titration curve for no lipid up to [L]=2.5 mM.
Table 1.
UV absorbance and fluorescence data for Pc 4 in liposomes of different compositions.
| Liposome Composition (%) | Maxima | ||||
|---|---|---|---|---|---|
| DMPC | DMPE | CL | Cholesterol | λabs (nm) | λfl (nm) |
| 50 | 50 | — | — | 671 | 675 |
| 45 | 45 | 10 | — | 671 | 676 |
| 40 | 40 | 20 | — | 672 | 677 |
| 35 | 35 | — | 30 | 672 | 676 |
| 27.5 | 27.5 | — | 45 | 674 | 676 |
Determination of Binding Constants
The major polar lipid components found in mitochondria from liver, heart, and kidney are CL, phosphatidyl choline, and phosphatidyl ethanolamine; these are present23, 40 in a molar ratio of 1:4:4. We chose to work with liposomes of similar lipid composition to mitochondria, using DMPC and DMPE in a ratio of 1:1 and adding CL (or not), but keeping the total lipid concentration constant, to determine whether Pc 4 displayed any preferential binding to CL-containing liposomes.
On the basis of fluorescence spectra, it is possible to compare the degree of incorporation of Pc 4 as monomer into liposomes with different phospholipid compositions. The association constants were calculated by hand according to Eq. 1; they were also calculated by plotting (F−F0)∕(F∞−F0) versus [L] and fitting to Eq. 1 using Origin software (OriginLab Corporation, Northampton, Massachusetts). Only minor differences in Ka values were obtained using the two calculation methods. The averages of Ka values for Pc 4 in 0, 10, and 20% CL were 2.1×103 (n=2), 1.7×103 (n=1), and 2.4×103 (n=2) M−1, respectively (Table 2). Therefore, no trend in binding affinity versus CL content was observed, which suggests that Pc 4 does not show preferential binding to CL in the liposomal system. Since our earlier studies using confocal microscopy showed that Pc 4 did not localize to the plasma membrane,10, 11 liposomes containing DMPC, DMPE, and cholesterol were also studied. Cholesterol is a major lipid of the plasma membrane of human cells. With liposomes containing 30 or 45% cholesterol, the average of Ka values for Pc 4 were 1.3×103 (n=2) and 2.1×103 (n=2) M−1, respectively. The similarity of these constants to the prior results (Table 2) suggests that Pc 4 has similar affinities to different lipids in our system. Furthermore, the mean and standard deviation of all of the results is 1.9±0.7 M−1 (n=9). Since all but one of the individual values fall within the standard deviation, it appears that there is no significant influence of lipid composition on the affinity of Pc 4 for liposomes. Thus, another factor besides CL is likely to be responsible for the preferential localization of Pc 4 in the mitochondrial and ER membranes.
Table 2.
Binding constants for Pc 4 in liposomes of different compositions.
| Liposome Composition | Ka (M−1×10−3) | |||||
|---|---|---|---|---|---|---|
| DMPC | DMPE | CL | CHOL | First Experiment | Second Experiment | Average |
| 50 | 50 | — | — | 2.28 | 1.95 | 2.1 |
| 45 | 45 | 10 | — | 1.67 | — | 1.7 |
| 40 | 40 | 20 | — | 3.33 | 1.38 | 2.4 |
| 40 | 40 | — | 30 | 1.27 | 1.26 | 1.3 |
| 27.5 | 27.5 | — | 45 | 2.06 | 2.20 | 2.1 |
Fluorescence-Quenching Experiments
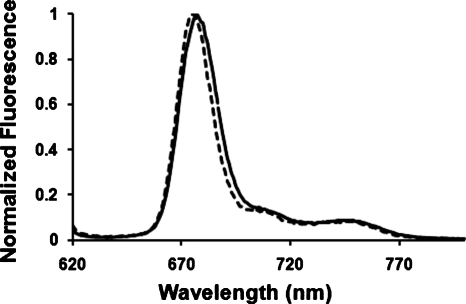
Since there is a blue shift of 1–2 nm in the emission of Pc 4 when CL is introduced to the liposomes (Fig. 2 and Table 1), fluorescence quenching was studied to explore the possibility of a dissimilar distribution of Pc 4 in liposomes of different composition. The quenching of the fluorescence of Pc 4 was measured in DMPC liposomes with and without CL using AQS− as quencher, which doesn’t penetrate the lipid bilayer. The fluorescence quenching data, as analyzed by the Stern-Volmer equation [Eq. 2], gave quenching constants Kq (mean±standard deviation) of (4.4±0.9)×109 M−1 (n=3) and (4.3±0.7)×109 M−1 (n=3) in liposomes with and without CL, respectively. Since the kq values are essentially identical, these data suggest that the presence of CL in the lipid bilayer does not affect the distribution of Pc 4 molecules in the liposome. The kq values are in agreement with the values found37 for other phthalocyanines in liposomes (4.6×109, 9.6×109, and 7.8×109 M−1 for three different zinc phthalocyanines).
Figure 2.
Fluorescence spectra of [Pc 4]=6 μM in (dashed line) liposomes at 0% CL and [L]=2.23 mM and (solid line) liposomes at 20% CL and [L]=1.33 mM.
Role of Mitochondrial Membrane Potential in the Preferential Localization of Pc 4 in Mitochondria
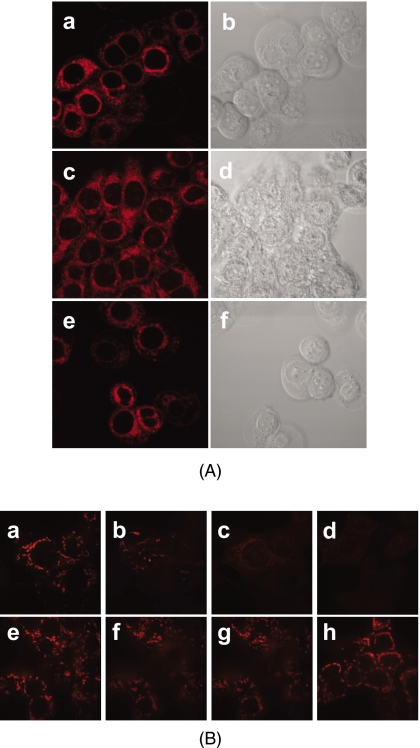
Aside from the presence of CL, another factor that could account for the preferential localization of Pc 4 in mitochondria is its cationic character. Hydrophobic compounds with delocalized positive charges can be driven across the inner membrane lipid bilayer by the mitochondrial electrochemical potential established by the proton-pumping mechanism of respiratory complexes.41 Using V8.0 Batch pKa Prediction Software, the calculated pKa of Pc 4 was approximately 9.6. Although the positive charge is localized, rather than delocalized as in mitochondrion-targeting dyes such as rhodamine-123 and JC-1, we considered the possibility that the preferential uptake of Pc 4 required a strong mitochondrial membrane potential.41 Two methods were used to block mitochondrial function and∕or uncouple oxidative phosphorylation: heating cells and introducing an uncoupler, CCCP. As shown in Fig. 3A the distribution of Pc 4 (200 nM) in individual cells treated with heat or CCCP was similar to that in the control cells, with Pc 4 bound to intracellular membranes and excluded from the nucleus; the distributions were similar to those previously published.10, 11, 12, 13, 14 Although these images have not been quantified, individual cells appear to have similar levels of Pc 4 fluorescence irrespective of the loss of some cells from the CCCP-treated population. The same pattern was found for a lower concentration (50 nM) of Pc 4 (data not shown).
Figure 3.
Influence of mitochondrial membrane potential on the binding of Pc 4 to mitochondria. (A) Confocal images of intracellular distribution/accumulation of Pc 4. MCF-7c3 cells were plated at 2×105 per 35-mm dish, allowed to attach overnight (16 to 18 h), and then treated for 1 h in one of three ways: (a) and (b) untreated cells, (c) and (d) cells heated at 65°C, and (e) and (f) cells incubated with CCCP (10 μM). After that, the medium was removed and the cultures were incubated 1 h with Pc 4 (200 nM) in a 37°C incubator. All cells were washed twice in PBS and overlaid with phenol red-free Hank’s balanced salt solution prior to imaging. (B) Mitochondrial membrane potential visualized by confocal microscopy. Cells were grown as already described, then loaded with 100 nM TMRM for 30 min at 37°C. The medium was removed, and the cells were kept in 10 nM TMRM in calcium buffer. After recording the first image, 10 μM CCCP was added to the cells in calcium buffer containing TMRM (10 nM). Control cells received 10 nM TMRM in calcium buffer but without CCCP. Imaging was performed at 5-s intervals over a period of 1 min or until depolarization was observed. Upper panel from left to right: (a) cells before adding CCCP, 10 μM; (b) and (c) 48 s and 1 min after adding CCCP, respectively; (d) 1 min after adding CCCP in a different field. Lower panel from left to right: control cells (e) 0, (f) 1, and (g) 2 min, after the start of imaging; and (h) 8 min after the start of imaging but in a different field.
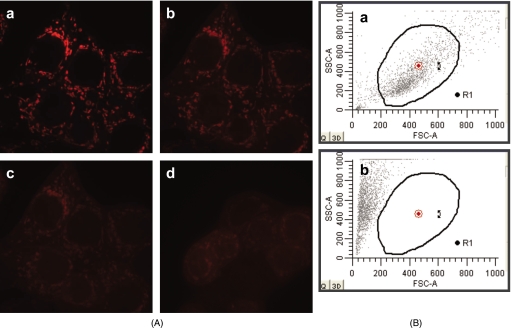
We performed experiments to monitor mitochondrial membrane potential using TMRM. Polarized mitochondria take up the cationic dye TMRM, which is released after depolarization. Using confocal microscopy, we evaluated the mitochondrial level of TMRM, as a measure of mitochondrial membrane potential. Figure 3B shows the loss of the mitochondrial membrane potential after adding CCCP (10 μM), the same concentration used in the experiments to evaluate Pc 4 localization [Fig. 3A]. Even 200 nM CCCP was able to completely depolarize the mitochondrial membranes [Fig. 4A]. Control cells loaded with vehicle alone did not release TMRM. For evaluation of the effect of heat, we used flow cytometry and showed that 100% of the cells were dead after 1 h at 65°C [Fig. 4B]. From these experiments, we concluded that the mitochondrial membrane potential appears not to be indispensable for Pc 4 uptake into mitochondria.
Figure 4.
(A) Mitochondrial membrane potential visualized by confocal microscopy. Cells were grown as described in methods for 48 h, then loaded with 100 nM TMRM for 30 min at 37°C. The medium was removed, and the cells were kept in 10 nM TMRM in calcium buffer. After recording the first image, 200 nM CCCP was added to the cells in calcium buffer containing TMRM (10 nM). Imaging was performed at 5-s intervals until depolarization was observed: (a) cells before adding CCCP; (b) and (c) 48 s and 1 min, respectively, after adding CCCP; and (d) 1 min after adding CCCP in a different field. (B) Flow cytometric evaluation of the effect of heat on cells. Graphs of side scatter (SSC) versus forward scatter (FSC) of (a) control cells kept at 37°C and (b) cells after 1 h at 65°C. Dead cells have lower FSC and higher SSC than living cells.
Role of CL Level in the Preferential Localization of Pc 4 in Mitochondria
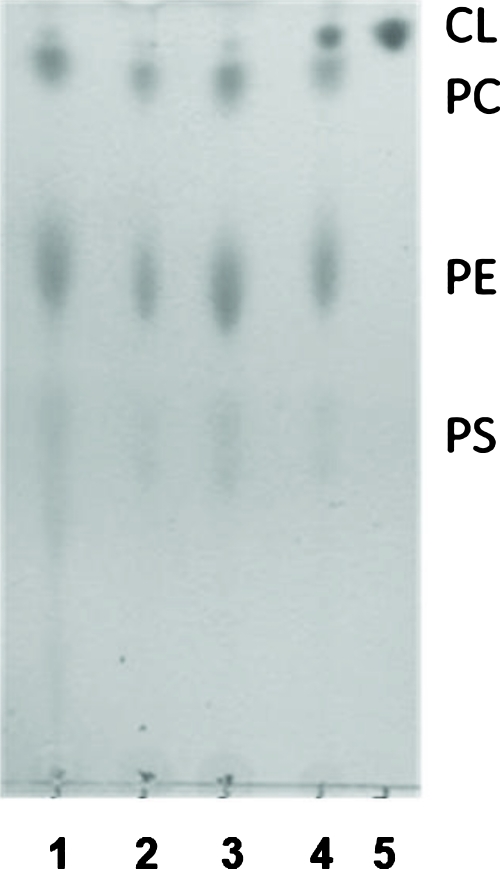
CL is a critical phospholipid of the mitochondrial inner membrane. We therefore asked whether reducing the CL content would affect the ability of Pc 4 to bind to the cells. Two laboratories have reported reduced mitochondrial levels of CL on exposure of rat neonatal cardiomyocytes or human breast cancer MDA-MB-231 cells to palmitate.30, 42 Because both studies found that the loss of CL on addition of this saturated fatty acid is followed by apoptosis, the MTT assay was used to identify a concentration where the CL level was reduced but the cells remained viable. With concentrations of palmitate ⩽0.5 mM, no toxicity was observed in MCF-7c3 cells for incubation times up to 6 h, but toxicity was found at longer times (data not shown). Using this condition, we extracted phospholipids from control and palmitate-treated cells, separated the phospholipids by TLC, and quantified the amount of CL using ImageQuant software based on the iodine staining.34 As shown in the TLC plate of Fig. 5, CL was well separated from the other phospholipids. Duplicate samples and duplicate controls were studied in each of three experiments. Using the phosphatidyl choline spot of each lane to normalize the amount of CL, we find that under the chosen conditions, palmitate-treated cells had 38±16% (n=6) of the CL content of untreated cells.
Figure 5.
Separation of cellular lipids by TLC. MCF-7c3 cells were treated as described in Sec. 2. Phospholipids were extracted with chloroform/methanol (2:1, v/v) from cells suspended in saturated saline solution. After that, the organic phase was recovered and dried under nitrogen. Each dried extract was dissolved in 25 μL chloroform and applied to a silica TLC plate. The mobile phase consisted of methanol/chloroform/acetic acid/water (3∕0.52∕0.36∕0.12). TLC was developed by iodine vapors to detect total lipid. Commercial CL was analyzed similarly as a standard. Lanes are 1, control cells; 2 and 3, palmitate-treated cells; 4, control cells plus addition of CL standard; and 5, CL standard. CL, cardiolipin; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine.
Cells treated with palmitate for 6 h to reduce their CL content and control cells were then evaluated for the uptake of Pc 4 and NAO by flow cytometry. When the control cells were exposed to a range of NAO concentrations from 20 to 300 nM, a linear increase in fluorescence was observed in the cells, without evidence of saturation (data not shown). For study of uptake of NAO into palmitate-treated cells, 200 nM NAO was used, a concentration that was within the linear range of the measurements. For 200 nM NAO, uptake into palmitate-treated cells was not significantly different (i.e., 93±23%; n=6) from that into control cells. The data agree with previous findings43, 44 that NAO cannot be used to quantify CL. For Pc 4 (200 nM), the mean channel fluorescence in palmitate-treated cells was found to be 94.9±5.4% (n=6) of the values for nontreated cells, indicating that there was no significant difference in the uptake of Pc 4 into cells as a function of palmitate treatment.
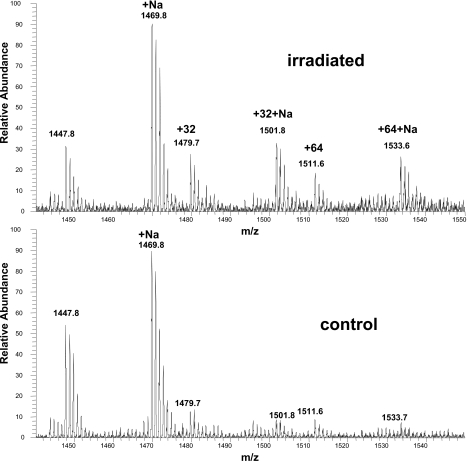
Although no specific binding between Pc 4 and CL was found, oxidation of CL was demonstrated to occur in the liposome system using ESI-MS and ESI-MS∕MS. For these experiments, higher doses of Pc 4 and light were used than are typically applied in our cellular studies. Note, however, that the actual photosensitizer concentration at the critical sites is not known either in cells or within the liposomes, so doses were chosen to enable identification of products of CL oxidation. The mass spectrum of CL isolated from the Pc 4-containing liposomes after photoirradiation showed two major oxidized species with mass increments of +32 and +64 from the unmodified tetralinoleoyl CL (Fig. 6). The MS∕MS spectra suggest that the +32 and +64 are mono- and dihydroperoxidized CL species, respectively. These could be produced directly by PDT-generated singlet oxygen, or they might result from radical chain reactions initiated and propagated within the CL pool.28, 29
Figure 6.
ESI-MS spectra of CL extracted from liposomes. Liposomes with Pc 4 were prepared as described in Sec. 2. Control liposomes were kept in the dark in the presence of Pc 4, while experimental liposomes were irradiated with 100 mW∕cm2 red light at room temperature for 40 min. The peak at m∕z 1447.8 is [M-H+]− of tetralinoleoyl CL.
Conclusion
In contrast to our proposal based on the earlier FRET studies, Pc 4 does not display preferential binding to CL, suggesting that other factor(s) must influence the preferential localization of Pc 4 to the mitochondrial and ER membranes. The binding constant of Pc 4 seems to be unaffected by the lipid composition, as shown by the similar binding constants obtained in cholesterol-containing membranes. Cholesterol is a major component of plasma membranes,45 but Pc 4 does not localize to that membrane in cells.10, 11 In these experiments, the highly hydrophobic Pc 4 macrocycle binds lipid in preference to aggregating in aqueous solution, but there appears to be no preference for any particular lipid.
The colocalization of Pc 4 and CL within the mitochondrial membranes and the oxidation of CL when Pc 4-loaded liposomes are photoirradiated remains consistent with CL being one target of Pc 4-PDT. A loss of molecular interaction between Cyt-c and CL due to the lipid peroxidation was reported by Petrosillo et al.46 to induce release of Cyt-c from submitochondrial particles to initiate apoptosis. Thus, the singlet oxygen or other radical species produced when Pc 4-loaded mitochondria are photoirradiated may oxidize CL, triggering apoptosis, even if CL is not the only target for Pc 4-PDT.
Disclosures
Two of the authors, MEK and NLO, are inventors on patents concerning Pc 4-PDT and are associated with Fluence Therapeutics, Inc., a company developing photodynamic therapy with the photosensitizer Pc 4.
Acknowledgments
This research was supported by grants from the U.S. National Cancer Institute, Department of Health and Human Services (DHHS), Grants R01 CA106491 and P30 CA43703, and by the National Institutes of Health, T35 Short-Term Research Training Grant “Research Training in Heart, Lung, Blood, & Sleep Disorders,” Grant No. HL082544.
References
- Dougherty T. J., Gomer C. J., Henderson B. W., Jori G., Kessel D., Korbelik M., Moan J., and Peng Q., “Photodynamic therapy,” J. Natl. Cancer Inst. 90(12), 889–905 (1998). 10.1093/jnci/90.12.889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleinick N. L., Morris R. L., and Belichenko I., “The role of apoptosis in response to photodynamic therapy: what, where, why, and how,” Photochem. Photobiol. Sci. 1(1), 1–21 (2002). 10.1039/b108586g [DOI] [PubMed] [Google Scholar]
- MacDonald I. and Dougherty T. J., “Basic principles of photodynamic therapy,” J. Porphyr. Phthalocyanines 5, 105–129 (2001). 10.1002/jpp.328 [DOI] [Google Scholar]
- Weishaupt K. R., Gomer C. J., and Dougherty T. J., “Identification of singlet oxygen as the cytotoxic agent in photoinactivation of a murine tumor,” Cancer Res. 36(7, Pt. 1), 2326–2329 (1976). [PubMed] [Google Scholar]
- Buytaert E., Dewaele M., and Agostinis P., “Molecular effectors of multiple cell death pathways initiated by photodynamic therapy,” Biochim. Biophys. Acta 1776(1), 86–107 (2007). [DOI] [PubMed] [Google Scholar]
- Suwa K., Kimura T., and Schaap A. P., “Reaction of singlet oxygen with cholesterol in liposomal membranes. Effect of membrane fluidity on the photo-oxidation of cholesterol,” Photochem. Photobiol. 28(4–5), 469–473 (1978). 10.1111/j.1751-1097.1978.tb06951.x [DOI] [Google Scholar]
- Korytowski W. and Girotti A. W., “Singlet oxygen adducts of cholesterol: photogeneration and reductive turnover in membrane systems,” Photochem. Photobiol. 70(4), 484–489 (1999). 10.1111/j.1751-1097.1999.tb08242.x [DOI] [PubMed] [Google Scholar]
- Oleinick N. L., Antunez A. R., Clay M. E., Rihter B. D., and Kenney M. E., “New phthalocyanine photosensitizers for photodynamic therapy,” Photochem. Photobiol. 57(2), 242–247 (1993). 10.1111/j.1751-1097.1993.tb02282.x [DOI] [PubMed] [Google Scholar]
- Miller J. D., Baron E. D., Scull H., Hsia A., Berlin J. C., McCormick T., Colussi V., Kenney M. E., Cooper K. D., and Oleinick N. L., “Photodynamic therapy with the phthalocyanine photosensitizer Pc 4: the Case experience with preclinical mechanistic and early clinical-translational studies,” Toxicol. Appl. Pharmacol. 224(3), 290–299 (2007). 10.1016/j.taap.2007.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi N. S., Wang H. W., Nieminen A. L., Oleinick N. L., and Izatt J. A., “Quantitative analysis of Pc 4 localization in mouse lymphoma (LY-R) cells via double-label confocal fluorescence microscopy,” Photochem. Photobiol. 71(5), 634–639 (2000). [DOI] [PubMed] [Google Scholar]
- Lam M., Oleinick N. L., and Nieminen A. L., “Photodynamic therapy-induced apoptosis in epidermoid carcinoma cells. Reactive oxygen species and mitochondrial inner membrane permeabilization,” J. Biol. Chem. 276(50), 47379–47386 (2001). 10.1074/jbc.M107678200 [DOI] [PubMed] [Google Scholar]
- Morris R. L., Azizuddin K., Lam M., Berlin J., Nieminen A. L., Kenney M. E., Samia A. C., Burda C., and Oleinick N. L., “Fluorescence resonance energy transfer reveals a binding site of a photosensitizer for photodynamic therapy,” Cancer Res. 63(17), 5194–5197 (2003). [PubMed] [Google Scholar]
- Usuda J., Chiu S. M., Murphy E. S., Lam M., Nieminen A. L., and Oleinick N. L., “Domain-dependent photodamage to Bcl-2. A membrane anchorage region is needed to form the target of phthalocyanine photosensitization,” J. Biol. Chem. 278(3), 2021–2029 (2003). 10.1074/jbc.M205219200 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. E., Zhang P., Azizuddin K., Delos Santos G. B., Chiu S., Xue L., Berlin J. C., Peng X., Wu H., Lam M., Nieminen A.-L., Kenney M. E., and Oleinick N. L., “Structural factors and mechanisms underlying the improved photodynamic cell killing with silicon phthalocyanine photosensitizers directed to lysosomes versus mitochondria,” Photochem. Photobiol. 85(5), 1189–1200 (2009). 10.1111/j.1751-1097.2009.00558.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel D., Luo Y., Deng Y., and Chang C. K., “The role of subcellular localization in initiation of apoptosis by photodynamic therapy,” Photochem. Photobiol. 65(3), 422–426 (1997). 10.1111/j.1751-1097.1997.tb08581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel D. and Luo Y., “Mitochondrial photodamage and PDT-induced apoptosis,” J. Photochem. Photobiol., B 42(2), 89–95 (1998). 10.1016/S1011-1344(97)00127-9 [DOI] [PubMed] [Google Scholar]
- Xue L. Y., Chiu S. M., and Oleinick N. L., “Photochemical destruction of the Bcl-2 oncoprotein during photodynamic therapy with the phthalocyanine photosensitizer Pc 4,” Oncogene 20(26), 3420–3427 (2001). 10.1038/sj.onc.1204441 [DOI] [PubMed] [Google Scholar]
- Usuda J., Chiu S. M., Azizuddin K., Xue L. Y., Lam M., Nieminen A. L., and Oleinick N. L., “Promotion of photodynamic therapy-induced apoptosis by the mitochondrial protein Smac/DIABLO: dependence on Bax,” Photochem. Photobiol. 76(2), 217–223 (2002). [DOI] [PubMed] [Google Scholar]
- Chiu S. M., and Oleinick N. L., “Dissociation of mitochondrial depolarization from cytochrome c release during apoptosis induced by photodynamic therapy,” Br. J. Cancer 84(8), 1099–1106 (2001). 10.1054/bjoc.2000.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan V. E., Bayır H. A., Belikova N. A., Kapralov O., Tyurina Y. Y., Tyurin V. A., Jiang J., Stoyanovsky D. A., Wipf P., Kochanek P. M., Greenberger J. S., Pitt B., Shvedova A. A., and Borisenko G., “Cytochrome c/cardiolipin relations in mitochondria: a kiss of death,” Free Radic Biol. Med. 46(11), 1439–1453 (2009). 10.1016/j.freeradbiomed.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M., Rua D., and Greenberg M. L., “The biosynthesis and functional role of cardiolipin,” Prog. Lipid Res. 39(3), 257–288 (2000). 10.1016/S0163-7827(00)00005-9 [DOI] [PubMed] [Google Scholar]
- McMillin J. B. and Dowhan W., “Cardiolipin and apoptosis,” Biochim. Biophys. Acta 1585(2–3), 97–107 (2002). [DOI] [PubMed] [Google Scholar]
- Malisan F. and Testi R., “Mitochondrial lipids as apoptosis regulators,” Curr. Top. Med. Chem. 10(16), 1573–1580 (2003). [DOI] [PubMed] [Google Scholar]
- Lesnefsky E. J., Slabe T. J., Stoll M. S., Minkler P. E., and Hoppel C. L., “Myocardial ischemia selectively depletes cardiolipin in rabbit heart subsarcolemmal mitochondria,” Am. J. Physiol. 280(6), 2770–2778 (2001). [DOI] [PubMed] [Google Scholar]
- Moan J. and Berg K., “The photodegradation of porphyrins in cells can be used to estimate the lifetime of singlet oxygen,” Photochem. Photobiol. 53(4), 549–553 (1991). 10.1111/j.1751-1097.1991.tb03669.x [DOI] [PubMed] [Google Scholar]
- Kim J., Rodriguez M. E., Guo M., Kenney M. E., Oleinick N. L., and Anderson V. E., “Oxidative modification of cytochrome-c by singlet oxygen,” Free Radic Biol. Med. 44(9), 1700–1711 (2008). 10.1016/j.freeradbiomed.2007.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Rodriguez M. E., Oleinick N. L., and Anderson V. E., “Photo-oxidation of cardiolipin and cytochrome c by bilayer-embedded Pc 4” Free Radic Biol. Med. 49(5), 718–725 (2010). 10.1016/j.freeradbiomed.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotti A. W., “Photosensitized oxidation of membrane lipids: reaction pathways, cytotoxic effects, and cytoprotective mechanisms,” J. Photochem. Photobiol., B 63(1–3), 103–113 (2001). 10.1016/S1011-1344(01)00207-X [DOI] [PubMed] [Google Scholar]
- Girotti A. W., “Translocation as a means of disseminating lipid hydroperoxide-induced oxidative damage and effector action,” Free Radic Biol. Med. 44(6), 956–968 (2008). 10.1016/j.freeradbiomed.2007.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander D. B., Sparagna G. C., Amoscato A. A., McMillin J. B., and Dowhan W., “Decreased cardiolipin synthesis corresponds with cytochrome c release in palmitate-induced cardiomyocyte apoptosis,” J. Biol. Chem. 276(41), 38061–38067 (2001). 10.1074/jbc.M103689200 [DOI] [PubMed] [Google Scholar]
- Wilson B. C., Olivo M., and Singh G., “Subcellular localization of photofrin and aminolevulinic acid and photodynamic cross-resistance in vitro in radiation-induced fibrosarcoma cells sensitive or resistant to photofrin-mediated photodynamic therapy,” Photochem. Photobiol. 65(1), 166–176 (1997). 10.1111/j.1751-1097.1997.tb01894.x [DOI] [PubMed] [Google Scholar]
- Kriska T., Korytowski W., and Girotti A. W., “Role of mitochondrial cardiolipin peroxidation in apoptotic photokilling of 5-aminolevulinate-treated tumor cells,” Arch. Biochem. Biophys. 433(2), 435–446 (2005). 10.1016/j.abb.2004.09.025 [DOI] [PubMed] [Google Scholar]
- Verma S., Watt G. M., Mai Z., and Hasan T., “Strategies for enhanced photodynamic therapy effects,” Photochem. Photobiol. 83, 996–1005 (2007). 10.1111/j.1751-1097.2007.00166.x [DOI] [PubMed] [Google Scholar]
- Palumbo G. and Zullo F., “The use of iodine staining for the quantitative analysis of lipids separated by thin layer chromatography,” Lipids 22(3), 201–205 (1987). 10.1007/BF02537303 [DOI] [PubMed] [Google Scholar]
- Li Y.-S. and Kenney M. E., “Methods of syntheses of phthalocyanine compounds,” U.S. Patent No. 5763602 (1998).
- Laroche C., Simonin H., Beney L., and Gervais P., “Phase transitions as a function of osmotic pressure in Saccharomyces cerevisiae whole cells, membrane extracts and phospholipid mixtures,” Biochim. Biophys. Acta 1669(1), 8–16 (2005). 10.1016/j.bbamem.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. E., Awruch J., and Dicelio L. E., “Photophysical properties of zinc(II) phthalocianinates incorporated into liposomes,” J. Porphyr. Phthalocyanines 6, 122–129 (2002). 10.1142/S1088424602000166 [DOI] [Google Scholar]
- Fujitsuka M., Ito O., and Konami H., “Photoexcited state properties of silicon phthalocyanine monomer, dimer, and trimer,” Bull. Chem. Soc. Jpn. 74, 1823–1829 (2001). 10.1246/bcsj.74.1823 [DOI] [Google Scholar]
- Ventrella A., Catucci L., Mascolo G., Corcelli A., and Agostiano A., “Isolation and characterization of lipids strictly associated to PSII complexes: focus on cardiolipin structural and functional role,” Biochim. Biophys. Acta 1768(6), 1620–1627 (2007). 10.1016/j.bbamem.2007.03.024 [DOI] [PubMed] [Google Scholar]
- Fleischer S., Rouser G., Fleischer B., Casu A., and Kritchevsky G., “Lipid composition of mitochondria from bovine heart, liver, and kidney,” J. Lipid Res. 8(3), 170–180 (1967). [PubMed] [Google Scholar]
- Rosania G. R., “Supertargeted chemistry: identifying relationships between molecular structures and their sub-cellular distribution,” Curr. Top. Med. Chem. 3(6), 659–685 (2003). 10.2174/1568026033452410 [DOI] [PubMed] [Google Scholar]
- Hardy S., El-Assad W., Przybytkowski E., Joly E., Prentki M., and Langelier Y., “Saturated fatty acid-induced apoptosis in MDA-MB-231 breast cancer cells. A role for cardiolipin,” J. Biol. Chem. 278(34), 31861–31870 (2003). 10.1074/jbc.M300190200 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. E., Azizuddin K., Zhang P., Chiu S. M., Lam M., Kenney M. E., Burda C., and Oleinick N. L., “Targeting of mitochondria by 10-N-alkyl acridine orange analogues: role of alkyl chain length in determining cellular uptake and localization,” Mitochondrion 8(3), 237–246 (2008). 10.1016/j.mito.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson J., Duchen M. R., and Heales S. J., “Intracellular distribution of the fluorescent dye nonyl acridine orange responds to the mitochondrial membrane potential: implications for assays of cardiolipin and mitochondrial mass,” J. Neurochem. 82(2), 224–233 (2002). 10.1046/j.1471-4159.2002.00945.x [DOI] [PubMed] [Google Scholar]
- Zager R. A., “Plasma membrane cholesterol: a critical determinant of cellular energetics and tubular resistance to attack,” Kidney Int. 58(1), 193–205 (2000). 10.1046/j.1523-1755.2000.00154.x [DOI] [PubMed] [Google Scholar]
- Petrosillo G., Ruggiero F. M., and Paradies G., “Role of reactive oxygen species and cardiolipin in the release of cytochrome c from mitochondria,” FASEB J. 17(15), 2202–2208 (2003). 10.1096/fj.03-0012com [DOI] [PubMed] [Google Scholar]