Abstract
SINE-VNTR-Alus (SVA) are non-autonomous hominid specific retrotransposons that are associated with disease in humans. SVAs are evolutionarily young and presumably mobilized by the LINE-1 reverse transcriptase in trans. SVAs are currently active and may impact the host through a variety of mechanisms including insertional mutagenesis, exon shuffling, alternative splicing, and the generation of differentially methylated regions (DMR). Here we review SVA biology, including SVA insertions associated with known diseases. Further, we discuss a model describing the initial formation of SVA and the mechanisms by which SVA may impact the host.
Keywords: SVA, retrotransposon
Introduction
Most genomes are highly repetitive, with a large fraction of the DNA derived from transposons (Kazazian 2004; Belancio, Hedges et al. 2008). Some of these transposons, in particular retrotransposons, replicate and expand through an RNA intermediate by a “copy and paste” mechanism termed retrotransposition [1]. The non-LTR class of retrotransposons replicates by coupling reverse transcription and integration into DNA, a process termed target-primed reverse transcription (TPRT) [1, 2]. Long Interspersed Element-1 (L1) [3] is the most successful non-LTR retrotransposon in mammals [4–6] and is evolutionarily old as evidenced by its presence in C.albicans [7]. Human L1 is present in approximately 500,000 copies, comprising some 17% of the entire genome sequence [6]. An intact L1 encodes two proteins [8, 9], one of which, ORF2, is a reverse transcriptase [10], the enzyme responsible for the reverse-transcription of retrotransposon RNA to DNA.
Despite the cis preference [11] of L1 proteins for their own encoding RNA, a variety of other multi-copy sequences [12–14], in particular, non-autonomous retrotransposons such as SINEs [15, 16] and processed pseudogenes [17], amplify through an RNA intermediate by hijacking the L1 reverse transcriptase [18]. The factors that enable these RNAs to be preferential substrates for the L1 machinery are currently unknown.
Another interesting non-autonomous retrotransposon that likely uses the L1 machinery to enter the genome is the hominid specific SVA [19]. SINE VNTR Alu (SVA), as it was originally named [20], is a composite retrotransposon currently active in humans [21] and present in about 2700 copies [19] in the human genome reference sequence. SVAs were originally described as SINE-R elements [22], a retrotransposon containing 5’ GC-rich tandem repeats along with env (envelope) and LTR sequence from an endogenous retrovirus [23]. Since then, progress has been made, primarily through bioinformatics and sequence analysis, illuminating our understanding of SVA. Nevertheless, relatively little is known about SVA compared to L1 due to a lack of experimental data, especially an SVA retrotransposition cell culture assay. Here we review what is known about SVA biology, including what can be learned from the individual SVA domains, and examine general mechanisms by which SVA may impact the genome, and sometimes cause disease.
A repeat of repeats
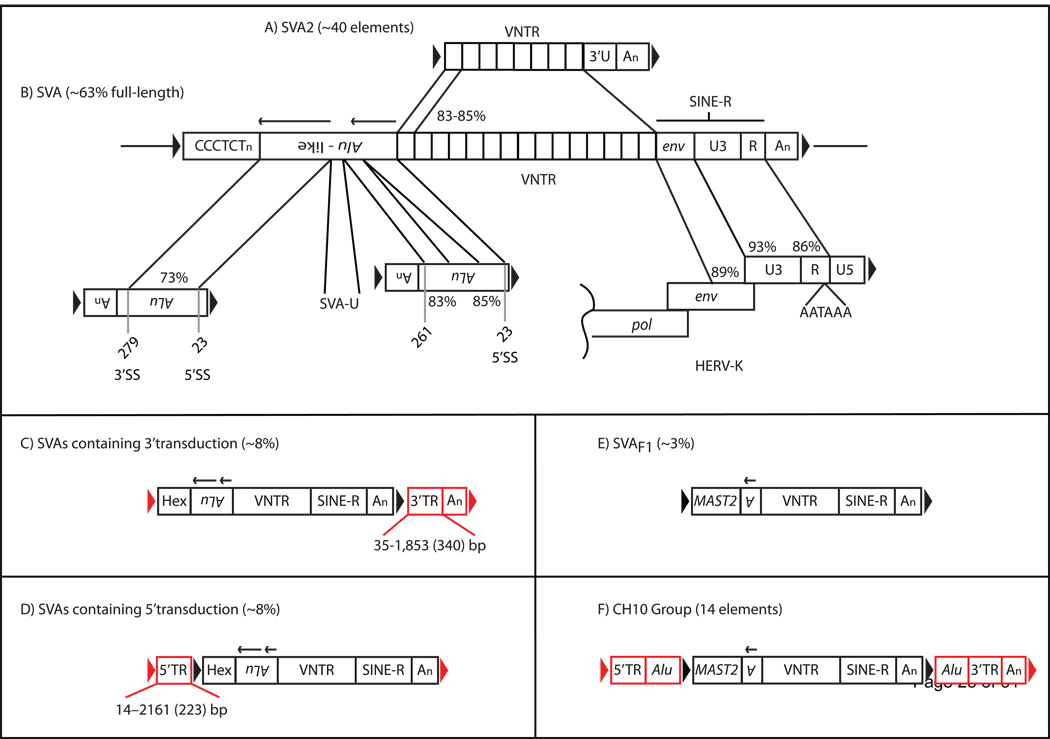
SVA is a composite non-coding retrotransposon [24, 25](Figure 1B) that in all likelihood relies on the L1 ORF2 reverse transcriptase for its mobilization [21], a presumption that has not yet been experimentally demonstrated. Each domain of SVA is derived from either a retrotransposon or a repeat sequence. A canonical SVA is on average ~2 kilobases (kb) but SVA insertions may range in size from 700–4000 basepairs (bp) [19, 26] in the human genome. Starting at its 5’ end, a canonical SVA (Figure 1B) consists of a hexameric CCCTCT repeat, followed by sequence sharing homology to two antisense Alu fragments, a variable number of GC-rich tandem repeats (VNTR), presumably derived from the SVA2 element [27–29] of the Rhesus macaque [30](Figure 1A), and sequence sharing identity to the env gene and right LTR of an ancient endogenous retrovirus, HERV-K10[22], followed by a canonical polyadenylation signal (polyA), AATAAA. SVA genomic insertions exhibit the classical hallmarks of L1 mediated retrotransposition and TPRT: 1) insertion at a consensus L1 endonuclease recognition motif 5′-TTTT/AA-3′ (where “/”denotes the cleavage site) [31], 2) a target-site duplication flanking the SVA insertion and ranging from 4-20bp in length, 3) a polyA tail of varying length, 4) the occurrence of 5’truncations, 5) internal rearrangements and inversions [21, 32, 33] and 6) 3’ transductions (Figure 1C)[21, 34–39]. However, one primary difference between L1 and SVA genomic insertions exists. Most SVAs are full-length, 63% and 42% in human and chimp, respectively [19]. While most (99.8%) L1 insertions are inactive due to 5’ truncations, inversions, and point mutations [6, 40]. Many SVA variants exist in hominid genomes, in addition to SVAs containing 3’transductions, SVAs may also contain 5’transductions (Figure 1D), upstream exons (Figure 1E) or both 5’ and 3’ transductions [26, 29](Figure 1F).
Figure 1.
The structure of a full-length SINE VNTR Alu (SVA) and SVA genomic variants. A) The SVA2 element. An SVA2 element consisting of a variable number of tandem GC-rich repeats (VNTR), followed by a unique 3’ sequence (3’U), followed by a polyA tail with the entire insertion flanked by a target-site duplication (black arrows) is shown. (B) A full-length SVA element consisting of in order from the 5’ end 1) CCCTCT hexameric repeats, 2) the Alu-like domain consisting of two antisense Alu fragments (black arrows above the Alu-domain indicate directionality of Alu sequences) and an intervening unique sequence, SVA-U, 3) a VNTR domain derived from the ancestral SVA2 element (A), 4) the SINE-R domain consisting of sequence sharing homology to the 3’end of the HERV-K10 env gene and U3, R, polyA signal (right LTR), terminating with a polyA tail (An) with the entire SVA insertion flanked by a target-site duplication. DNA sequence identities were obtained by pairwise BLAST alignments between the individual SVA domains and ancestral repeats (Alu, SINE-R, VNTR identity is between individual tandem repeats). Alignments consisted of using the following Repbase [27, 28] reference sequences: SVA2, SVA, Alu, HERV-K, and LTR5. The numbers below the antisense Alus correspond to nucleotide positions in AluRep and whether or not this position corresponds to a known splice site within Alu. Different SVA variants exist within the human genome, some contain additional 3’ sequence (C, red boxes), referred to as 3’ transductions (3’TR) or additional 5’ sequence (D, red boxes), referred to as 5’ transductions. A new target-site duplication (red arrows) flanks the SVA insertion and transduction. Transductions (C,D, red boxes) can be used to identify the source locus of a retrotransposon. Some 5’ transductions may be acquired via splicing of an upstream sequence into a downstream SVA element. This may result in novel SVA subfamilies such as SVAF1 (E) that acquired the MAST2 exon through splicing. SVA elements may also contain both 5’ and 3’ transductions (F), such as elements within the CH10 group. The number or percentage of SVAs containing a defining characteristic within the human genome reference sequence is in parentheses. Furthermore, the upper and lower limits in basepairs for reported 5’ [29] and 3’ [19] transductions is displayed below the transduction with the mean in parentheses.
SVA lifecycle and retrotransposition
The SVA RNA is likely a RNA polymerase (Pol) II transcript based upon several sequence features [19] and SVA expression analysis (Figure 2). SVAs contain multiple RNA Pol III terminators (TTTT) throughout the Alu-like and SINE-R domains. Also, the SVA RNA is ~2–3 kb, much longer than Pol III transcribed RNAs. SVA RNAs are presumably 5’ capped as indicated by the presence of guanine residues at the 5’end of ~1/3 SVA genomic insertions, similar to L1 insertions[41], and by the ability to amplify SVA RNA by 5’RACE[26], a technique reliant on a 5’cap structure. Furthermore, there is a small subset of SVAs [42], including the ARH insertion[43], that contains an alternative CCCTCT hexamer, ((CCCTCT)2 CCCGTCT)n, where the “G” might represent an example in which the 5’ cap was reverse-transcribed, and this nucleotide addition has expanded along with the hexamer.
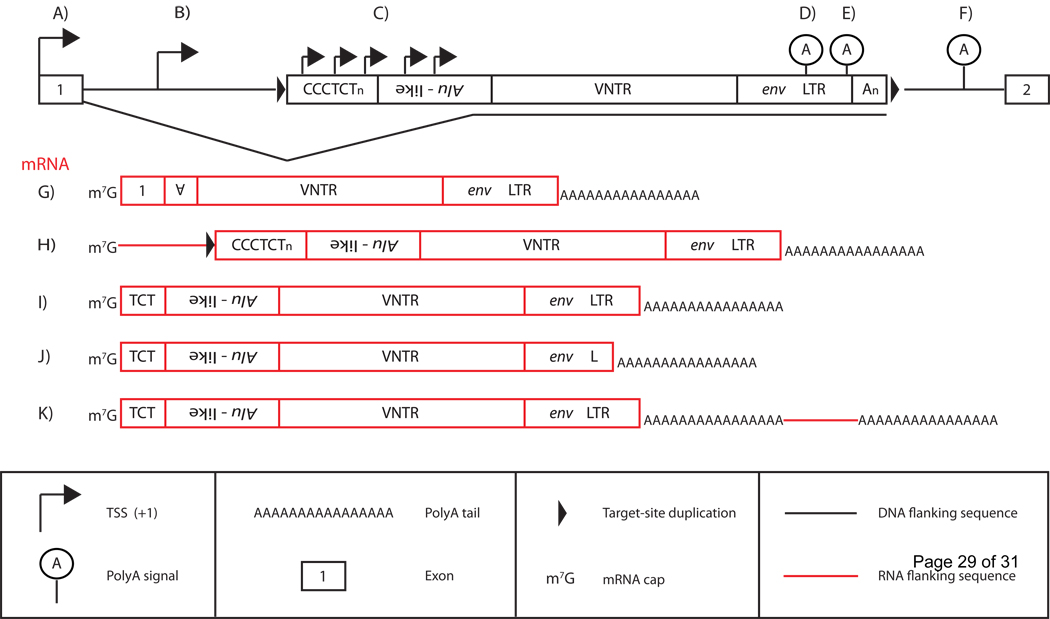
Figure 2.
SVA transcription and SVA mRNA structure. A full-length SVA, with individual domains labeled, in the genome residing in an intron (top; black line) of a gene with exons numbered 1 and 2 (top; black boxes) is displayed. Black bent arrowheads indicate different sites of SVA transcriptional initiation (A–C). The different SVA RNAs present in the human and chimpanzee transcriptome (red boxes; G–K) due to variable 5’ TSSs and 3’ polyA sites (D–F) are shown below. A) mRNA transcription may initiate at an upstream exon (black box labeled 1) that may subsequently splice into an SVA generating a “5’ truncated” SVA mRNA containing exonic sequence (G) terminating at the canonical SVA polyA signal (E). SVA-mediated exontrapping may enable SVA evolution as is the case for the SVAF1 subfamily. SVA transcription may initiate at an upstream TSS (B), presumably mediated by upstream promoter elements generating SVA mRNAs containing SVA 5’-flanking sequence (H; red line). Both (A,G) and (B,H) may result in retrotransposition of SVA 5’-flanking sequence, a process termed 5’-transduction. SVA RNAs may also initiate transcription internally (C) resulting in mRNAs resembling full-length genomic insertions (I), or terminate at internal non-canonical polyA sites in the SINE-R (D) resulting in 3’truncated SVA RNAs, or bypass the SVA polyA signal, terminating at a downstream polyA signal (F), resulting in an SVA mRNA containing 3’-flanking sequence (K; red line). Note that an SVA transcript may contain both 5’ and 3’ transductions (not shown).
The SVA contains a canonical polyadenylation signal, AATAAA (Figure 2E) at its 3’end. However, SVA transcription may occasionally bypass its own polyA signal, resulting in transcriptional readthrough, and terminate at a downstream polyA signal (Figure 2F)[21, 39]. This might result in retrotransposition of an SVA 3’ flank along with its sequence to another genomic location, a process termed 3’ transduction, with the potential for exon-shuffling [35]. Finally, SVAs have been identified that terminate at other internal non-canonical polyA sites within the SINE-R (Figure 2D), resulting in 3’ truncated SVA genomic insertions[19, 29].
The exact SVA transcriptional unit, along with its promoter and regulatory elements, is still undefined. Nonetheless, SVA RNAs are classified into three types of transcripts based upon the location of the 5’ transcriptional start site (TSS)[26]: 1) SVA sequences into which upstream exons are spliced, also referred to as SVA exon-trapping (Figure 2A and 2G), 2) SVAs that initiate transcription upstream of their genomic location (Figure 2B and H), and 3) SVAs that initiate transcription internally (Figure 2C, 2I, 2J, 2K). SVA mRNAs can be further subdivided on the basis of polyA signal selection: 1) internal polyA (Figure 2D), resulting in 3’ truncated SVAs (Figure 2J), 2) canonical polyA signal (Figure 2E), and 3) downstream polyA signal (Figure 2F), resulting in an SVA mRNA containing 3’flanking sequence (Figure 2K).
It is unclear which type of SVA transcription is the preferred mode of SVA mRNA expression. Still, SVA mRNAs are expressed in a variety of ways in cell lines and in vivo [26, 29, 44]. We know that SVAs initiating transcription upstream (Group 1 and 2) are retrotransposition competent because of the identification of SVA insertions containing retrotransposed 5’ flanking sequence [26, 29] due to 1) SVA exon-trapping or 2) upstream transcriptional initiation. SVAs retrotransposing 5’ flanking sequence, referred to as 5’ transductions, account for ~8% of total SVA insertions in the human genome [29]. SVA 5’ transduction as a result of upstream transcriptional initiation (Figure 2B) is much more common than 5’ transduction by exon-trapping (Figure 2A) as indicated by the number of distinct 5’ transduction groups relative to the number of SVA insertions identified that transduce upstream exons due to splicing. Furthermore, SVA elements are retrotransposition competent independent of polyA signal selection.
Therefore, SVAs are able to enter the transcriptome by three distinct routes 1) SVA mediated exon-trapping (Figure 2A and 2G), 2) upstream TSS mediated in most part by upstream promoters (Figure 2B and 2H), and 3) internal transcriptional start sites (TSS) (Figure 2C, 2I, 2J, 2K). Three different potential models exist for SVA transcription: 1) Similar to L1 and Alu, SVAs rely exclusively on their own promoter and/or regulatory elements, 2) SVAs themselves contain no regulatory elements and exclusively rely on external regulatory elements, or 3) SVAs contain some regulatory elements that may act synergistically with external promoters/external regulatory elements enabling SVA transcription. Experiments to localize any internal SVA promoter activity have led to ambiguous results (M.C. Seleme and H.H. Kazazian, unpublished data).
Whether or not the internal and upstream TSSs both rely on upstream promoter units is unclear. One possibility is that many SVAs depend on upstream promoters. This would suggest that many SVA master elements, retrotransposon loci that are the source of many genomic copies, in addition to the recently described CH10 SVA master element [26, 29], are present in different hominid genomes. If most of the human genome is transcribed [45, 46], with different transcripts having multiple different transcriptional start sites [47, 48] including transposon derived TSSs [49] and transcriptional readthrough is the primary mode of SVA transcription, then one might expect Alu genomic insertions to occasionally contain 5’ transductions. To the best of our knowledge, Alus containing 5’ transductions have not yet been identified. Moreover, if all SVAs are expressed due to upstream promoters, then SVA elements containing the CCCTCT hexamer would be 5’ truncated elements, presumably due to the inability of L1 ORF2 to reverse transcribe the CCCTCT repeat. Be that as it may, it is then difficult to reconcile how ORF2 is able to effectively reverse transcribe the VNTR of SVA.
Regarding SVA transcription model 3, SVAs may contain the inherent ability to mediate transcriptional initiation upstream of their genomic location due to an internal enhancer element. This enhancer element may be able to recruit transcription factors to upstream promoter elements, ultimately leading to transcription at or near the 5’ end of SVA. SVA elements contain many predicted transcription factor binding sites, such as SP1 binding sites in the VNTR and potential hormone response element (HRE) half-sites throughout the SVA domains [19, 50–52]. The HREs and the SP1 sites may cooperate, as in one study for Alu [50], to drive SVA transcription. It is particularly interesting that the SINE-R contains a glucocorticoid response element and an enhancer core element [22]. Both were originally described when HERV-Ks were cloned [53] and subsequently described in SINE-R elements [22]. Notably, HERV-K mRNA expression was up-regulated in a human breast cancer cell line treated with progesterone followed by estradiol treatment [54]. The lack of an internal promoter and the presence of enhancer elements in the SINE-R may account for the variation in SVA transcript structure and is consistent with the observation that active SVA elements may lack the CCCTCT hexamer and Alu-like domain [26, 29]. Ono et al [22] have postulated that the glucocorticoid response element and enhancer core may act as a steroid dependent enhancer element for SINE-R elements.
After transcription, the SVA RNA needs to 1) come in contact with the L1 ORF2 protein, and 2) out-compete the L1 RNA for the attention of ORF2 reverse transcriptase. L1 and Alu RNA competition for ORF2 presumably takes place at the ribosome [55, 56] or at the very least where Alu gets incorporated into the L1 ribonucleoprotein particle (RNP). Yet, the location in the cytoplasm and/or nucleus where SVA and other RNAs are incorporated by the L1 RNP and subsequently interact with ORF2 protein is unknown. The vast number, more than 2000 copies, of ribosomal processed pseudogenes [14] suggests that ORF2 competition may occur at the ribosome. To the contrary, since U6 snRNAs transit through the nucleolus, the presence of U6-L1 chimeric retrogenes [12] indicates that the nucleolus [57] may also be a site where the L1 RNP may acquire RNAs.
It has been hypothesized that the Alu-like domain localizes SVA RNA to the ribosome by annealing with Alu RNAs [58]. However, the identification of multiple retrotransposed SVAs lacking the majority of the Alu-like domain, due to SVA mediated alternative splicing, in particular SVA F1s, a human specific SVA subfamily distinguished by the presence of exon 1 from the MAST2 gene [26, 29, 59], argues against an SVA-Alu hybridization requirement for SVA retrotransposition. Future experiments describing which RNAs and their abundance in L1 cytoplasmic and nuclear RNP complexes will assist in resolving this question.
After incorporation into the L1 RNP, SVAs are reverse transcribed in the nucleus by ORF2 probably by a template choice mechanism [18]. The present lack of any described SVA-L1 chimeras and the dearth of known retrogene chimeras [18, 60], other than U6-L1 chimeras, disagrees with a template switching mechanism for ORF2 mediated SVA reverse transcription. However, template-switching by ORF2 between polyA tails [18] of L1 and SVA RNAs cannot be ruled out.
SVA elements that have been spliced into, resulting in loss of the CCCTCT hexamer and most of the Alu-like domain, followed by subsequent retrotransposition and those that are 3’ truncated have provided insight into requirements of SVA retrotransposition. These SVA insertions suggest much of the SVA is dispensable and unnecessary for successive rounds of retrotransposition. The VNTR is the core sequence of SVA, and the highly-structured nature of the tandem repeats probably plays a yet undefined functional role. RNAs that have increased mRNA stability are over represented as processed pseudogenes [61] and the VNTR alone or within the context of SVA may increase RNA stability. The SINE-R domain is probably responsible for SVA expansion and likely enables SVA expression. The variation in TSSs may be due to the looping of the SINE-R over the VNTR to the 5’ end of SVA. Longer VNTRs may lead to internal SVA TSSs, while shorter VNTRs may lead to transcriptional initiation further upstream into flanking DNA. Recruitment of transcription factors to the SINE-R and the interaction of these factors with SP-1 proteins may assist in the assembly of the transcription pre-initiation complex.
The only obvious functional requirement for SVA retrotransposition is the polyA tail. The polyA tail is indispensable for L1 [34] and Alu [16] retrotransposition in cell culture. Human L1s share no sequence homology with Alu and SVA other than polyA stretches. However, both Alu [62] and SVA are highly structured RNAs and the 3’UTR of many L1s contain GC-rich sequences that have been shown to be structured for Rat L1 [63]. Therefore, we propose in order to reconcile the lack of sequence homology between human L1, Alu, and SVA that RNA structure is fundamental for ORF2 substrate recognition and that RNA structure is the primary determinant of whether a RNA will be retrotransposed while the polyA tail is secondary. Work by Eickbush and colleagues has shown that the R2 retrotransposon RNA secondary structure in the 3’ UTR is required for TPRT and that the R2 protein from B. mori is able to carry out TPRT with the 3’UTR from D. melanogaster R2 element in vitro [64]. Furthermore R2 RNAs ending in fewer adenines are more preferential substrates for target-primed reverse transcription in vitro [65].
Two obvious caveats exist with this model; 1) the human L1 3’UTR is not required for retrotransposition in the cell culture retrotransposition assay [34] and 2) the presence of retropseudogenes derived from tRNAs that lack polyA tails [66] and the retroposed snRNAs [12, 18], like U6, that also lack a 3’ polyA stretch. The dispensability of the 3’UTR can be explained by human L1’s intense cis preference[11] for its own encoding RNA. The amplification of tailless tRNA retroelements and U6-L1 insertions can be explained by the fact that tRNAs and U6 RNAs are highly structured, and at least in the case of tRNA its ability to localize to the ribosome enhances its incorporation into the L1 RNP. Hence, successful competition for the reverse transcriptase of a non-LTR element is contingent upon a highly structured RNA or sequence containing 3’ non-LTR sequence, as in the case of tRNA derived SINEs [67] and snoRTE [68], which ultimately enables these elements to mimic retrotransposon RNA structure. Secondary to RNA structure, the ability of a RNA to localize to the ribosome determines its retrotranspositional success as indicated by Alus, tRNA-derived SINEs, and tailless tRNAs. Lastly, the polyA stretch of non-LTR elements is fundamentally important for retrotransposition, providing somewhat of a flat runaway for RT loading or enabling accessibility of RT for its template because the polyA lacks secondary structure. This is consistent with the longer polyA tails associated with active elements [69] and the length [70] and homogeneity [71] of the Alu polyA tails impacting their retrotransposition efficiency.
SVA Origins
Although retrotransposons derived from repeat sequences are not uncommon, the structure of SVA is unique to say the least [72]. Chimeric retrotransposed sequences are present in nature and are not rare [73, 74]. DNA recombination or retroelement insertion into a transcription unit may generate new sequences and increase their retrotransposition capability. Despite this fact, many retroelement chimeras are likely formed at the RNA level. U6-L1 chimeras are common in the human genome [12, 18] and in primates [75]. An example of a new hominid gene, PIPSL, formed from a retrotransposed chimeric transcript derived from an alternative splicing event involving adjacent genes was recently described [76]. Likewise, snoRTE [68], a chimeric retrotransposon, consisting of a 5′-H/ACA-snoRNA containing the 3’end of a BovB Plat RTE LINE, has been extremely successful, exceeding more than 40,000 copies in the platypus genome. Okada’s group has shown that the 3’ ends of tRNA-derived SINEs are derived from the 3’ ends of LINEs [77]. Furthermore, it has been proposed that LTR retrotransposons and retroviruses were derived from the fusion of a DNA transposon and a non-LTR retrotransposon [78].
SVAs are evolutionarily young which enables easier identification of the origins of their multiple domains (Alu-like, SINE-R). SVA evolutionary analysis provides insight into 1) how non-autonomous retrotransposons are created, 2) what sequence features might enable retrotransposition of a pseudogene and 3) how genomes evolve. In all regards, SVA is a successful pseudogene. SVA is currently more active than high-copy pseudogenes, such as processed ribosomal pseudogenes, as evidenced by seven published SVA insertions associated with disease [43, 79–84](Table 1) and no disease associated pseudogene insertions. Second, each mRNA pseudogene originates from primarily one source locus, while retrotransposed SVAs are derived from many loci, as indicated by the variation in 5’ [29] and 3’ transductions [39], indicating multiple SVA source loci.
Table 1.
SVA insertions and disease
| Gene | Insertion (kb) | HG19 | Full-length | Subfamily** | Associated disease | Genotype | Potential Mechanism | Progenitor | Note | SVA Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| HLA-A | 2 | No | Yes* | F1 | Leukemia | SVA/+ | Deletion | 3p21.31X | Founder Insertion (JPN) | AB291067, AB291066 |
| NF2 | 1.7 | Yes | Yes | D | Neurofibromatosis 2 | SVA/+ | Deletion | N/A | HG19 | |
| BTK | 0.25 | No | No | N/A | X-linked agammaglobulinemia (XLA) | SVA/Y | Exon skipping | N/A | Alu inserton at same site | Conley et al 2005 |
| α-spectrin | 0.63 | No | No | E | Heriditary elliptocytosis and pyropoikilocytosis | SVA/+ | Exon skipping | 3q25.1X | Inverted 3' transduction | dbRIPY |
| TAF1 | 2.6 | No | Yes | F | X-linked dystonia-parkinsonism (XDP) | SVA/Y | DNA methylation | N/A | Founder Insertion (PHI) | AB191243 |
| LDLRAP1 | 2.6 | No | Yes | E | Autosomal Recessive Hypercholesterolemia (ARH) | SVA/SVA | Reduced mRNA | N/A | Italian Ancestry | Wilhund et al 2002 |
| Fukutin | 3.1 | No | Yes | E | Fukuyama-type muscular dystrophy (FCMD) | SVA/SVA | Reduced mRNA | N/A | Founder Insertion (JPN) | AB185332 |
Published SVA insertions associated with disease. A full-length SVA insertion is defined as the presence of either the CCCTCT hexamer or MAST2 sequence.
Contains MAST2 sequence
SVA Subfamily Determined by Repeatmasker (http://www.repeatmasker.org) according Wang et al 2005 subfamily classification.
present in Human Genome (hg19) UCSC browser (http://genome.ucsc.edu/)
Work by Batzer and colleagues [30], as part of the Rhesus macaque genome consortium, documented the absence of SVAs from the Rhesus genome, but they noted that each SVA domain, CCCTCT hexamer, Alu-like, VNTR, and SINE-R was present independent of the other domains. They also reported that the VNTR was present ~40 times and contained a non-SVA sequence at its 3’ end followed by a polyA tail with the entire sequence flanked by a target-site duplication. These data suggest that the VNTR was retrotransposition-competent in the past. VNTRs similar to those described in Rhesus and referred to as SVA2 elements, had been briefly described by Repbase [27, 28] and more recently in a study characterizing SVA genomic insertions [29]. Whether or not the SVA2 (VNTR) can be classified as a retrotransposon or a relatively successful pseudogene is unclear because at least 15 different non-ribosomal processed pseudogenes have more than 30 copies in the human genome [85] as compared to the 40 SVA2 copies in Rhesus.
The lack of SVAs in old world monkeys [19, 30], suggests that SVAs are hominid specific retroelements [86]. Thus, SVA2 elements acquired the other current SVA domains sometime after the divergence of old world monkeys and hominids. Knowledge of the individual SVA domains has enabled us to model some of the events that likely occurred to create the present day SVA.
Alus are the most successful primate retrotransposons [87] with about one million copies in the human genome reference sequence [6]. Additionally, Alus are known to be frequently alternatively spliced when in the antisense orientation relative to the transcriptional unit [88–90]. SVA contains sequence with identity to two antisense Alus orientated head to tail [20, 21, 24](Figure 1B). The 5’ most Alu, of the Alu-like domain of SVARep is 255 bp long and aligns with 73% identity between nucleotides 279 and 23 relative to AluRep (Figure 1B)[42]. Following the first antisense Alu is a 31 nucleotide stretch termed SVA-U [25, 42]. The origin of this sequence is unclear [25]. Using BLAT and the genome sequences available on the UCSC browser website, this sequence could only be identified within SVA elements [42]. The second antisense Alu is shorter than the 5’Alu, spanning 93 nucleotides due to an internal deletion of 152 nt. The 5’ end of the second Alu spans nts 261-209 followed by the deletion, an insertion of 6 nt, followed by Alu nts 56-23 (Figure 1B). Overall, the entire Alu-domain consisting of the first Alu, SVA-U, and the second Alu, is 376 nts in length.
The Alu-like domain may have been formed through alternative splicing of two Alus [25, 42] and the unknown sequence, SVA-U. It is noteworthy that 3 out of 4 of the 5’ and 3’ positions of both Alu fragments correspond to known 5’ and 3’ splice sites identified in antisense Alus [88]. Alu alternative splicing has been well documented in the literature [88, 89]. Experimental analyses [89] have demonstrated that two primary 3’splice sites (SS) within Alu are utilized, one at position 279 and referred to as the proximal AG, and the second 3’SS at position 275 and referred to as the distal AG. The 5’ end of the first Alu aligns to position 279, while the 5’ end of the second Alu aligns to position 261. Multiple 5’ SS have been identified in antisense Alus with the second most common site in EST data being position 23 [88]. The 3’ end for both the Alus aligns to position 23. That the 5’ end of the second Alu does not correspond to a known Alu 3’SS can be explained by a deletion occurring between the junction of the 3’end of SVA-U and the 5’ end of the second Alu (Figure 3E).
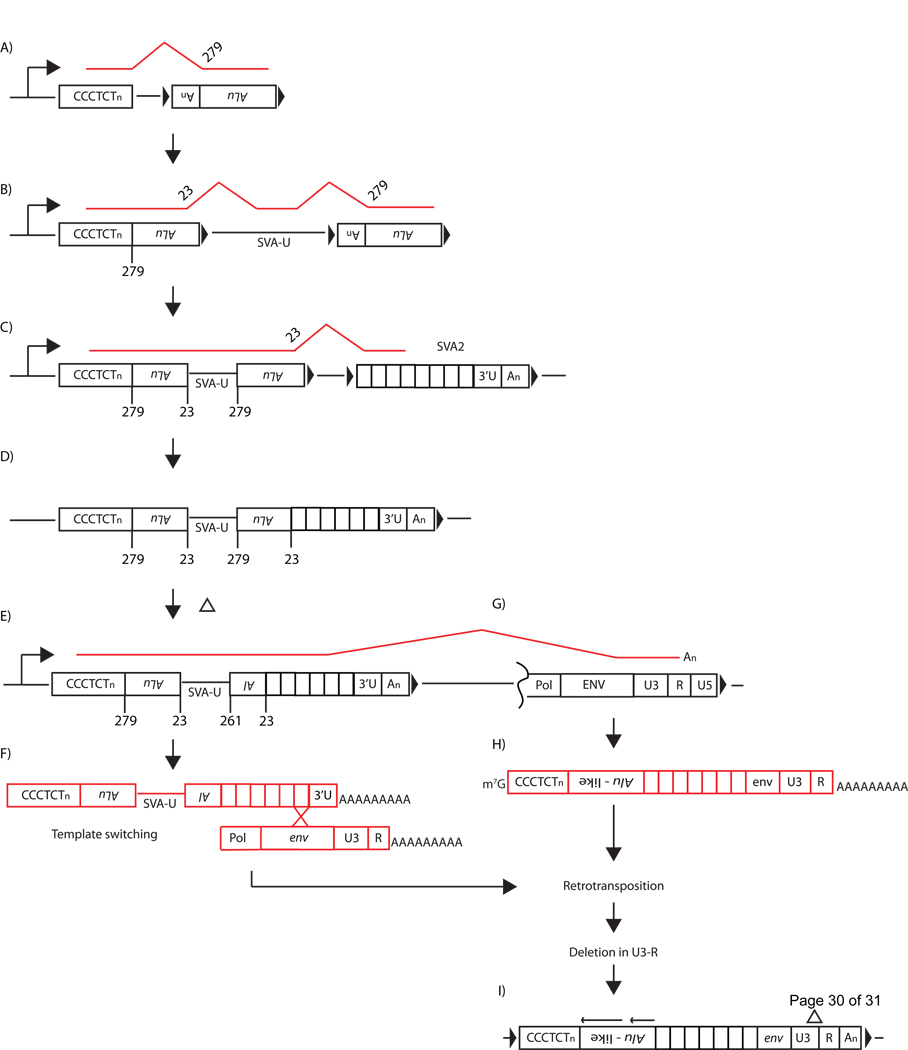
Figure 3.
The origin of SVA. A model outlining events that may have taken place to generate each junction and domain within a modern SVA is displayed. The events and junction formation are ordered from 5’ to 3’ for clarity (see text for further description). However whether the SVA domains were acquired by SVA2 independently or simultaneously is unknown. (A) An mRNA (red line) consisting of a CCCTCT hexameric repeat transcribed (black bent arrows) from a genomic location (black boxes) spliced into a downstream antisense Alu at the known 3’SS position 279 relative to AluRep creating the junction between the CCCTCT hexamer and the first Alu (B). Most likely in the same mRNA (B, red line), the first Alu was spliced out at position 23, a known Alu 5’SS, followed by subsequent splicing into a sequence of unknown origin, referred to as SVA-U. SVA-U presumably spliced into the second Alu presumably at a known Alu 3’SS between positions 273–281. Here we display Alu position 279 as the 3’SS utilized in the incorporation of the second Alu. The CCCTCT hexamer along with the Alu-like domain junctions are displayed (C) prior to deletions in the second Alu. The 3’end of the second Alu is a 5’SS, therefore the second Alu and VNTR junction was likely created by alternative splicing into a genomic SVA2 (C). During its evolution, two deletions in the second Alu from 1) the 3’SS to nt 262 and 2) nts 208-57 were generated (E). The SINE-R domain may have been acquired by potentially one of two RNA based mechanisms: 1) ORF2 reverse transcriptase template switching (F) or 2) alternative splicing (G) at an unidentifiable 3’SS in the env sequence of a HERV-K resulting in an mRNA resembling a modern SVA (H). Subsequently, the mRNA was retrotransposed resulting in present-day SVA (I). It is unclear whether the 367 nt deletion (H; black triangle) present within the LTR occurred before or after SVA assembled.
The observation that the terminal nucleotides of the Alu fragments correspond to known splice sites, led us to propose that the SVA-U sequence is also likely the remnant of an unidentifiable alternatively spliced sequence. This is consistent with the notion that the intersection of the first Alu and SVA-U is a splice junction, and that SVA-U was incorporated via alternative splicing. Briefly, an mRNA containing the CCCTCT hexamer presumably spliced into the first Alu at position 279 and out of that Alu at position 23 joining the hexamer and 5’ Alu segments (Figure 3A). Next, the first Alu spliced into downstream sequence, SVA-U, followed by splicing into the second Alu at an unidentifiable 3’SS (Figure 3B), followed by splicing out at position 23 (Figure 3C). Two deletions to the second Alu involving 1) the 3’SS to nt 262 and 2) nts 208-57 probably occurred after SVA domain acquisition by alternative splicing (Figure 3E). The abundance of Alus and satellite sequences in primate genomes suggests assembly of the Alu-like domain by mRNA splicing is possible.
Recently, we identified 5’ and 3’ SS within the VNTR region [26]. Similar to the first Alu and SVA-U junction, the intersection of the 3’ end of the second Alu and VNTR may represent a splice junction (Figure 3D). It is unclear which 3’ SS within the VNTR would have been utilized due to the repetitive nature of the tandem repeats. However, due to the GC-richness and asymmetry of the tandem repeats, multiple pyrimidine stretches are positioned 5’ of the canonical CAG trinucleotide splice acceptor.
The 3’ end of SVA is referred to as the SINE-R, where R indicates retroviral origin [22]. This sequence shares homology to the env gene and right LTR of a HERV-K10. The env sequence is 81 nucleotides long sharing about 88% identity with HERV-KRep. 3’ of the env sequence is the right LTR, consisting of the U3, R, and polyA signal derived from a HERV-K. This LTR harbors a 367 nt deletion of nucleotides 331–697 relative to LTR5Rep [22]. The 5’ portion of the LTR shares 90% identity while the 3’ portion is 87% identical to LTR5Rep. Similar sequence identity and the 367 nt deletion is observed when SVARep is compared to the HERV-K10 in Genbank (#M14123.1).
The right LTR does not contain U5 sequence and terminates at the HERV-K polyA signal. This suggests that the SINE-R was not incorporated into SVA through a DNA based mechanism, but through a RNA based mechanism. The mechanism must be able to account for the loss of the unique 3’ SVA2 sequence [27, 28], SVA2 polyA signal and polyA tail (Figure 1A). An attractive mechanism for the acquisition of the SINE-R sequence is template-switching [18] between the HERV-K and VNTR mRNA during reverse transcription (Figure 3F). A less appealing possibility is alternative splicing of the VNTR into the env sequence of the HERV-K (Figure 3G). HERVs [91, 92], and more specifically HERV-K sequences [93], are known to be spliced into mRNA and could account for the loss of the SVA2 3’ end. However, no predicted splice site in HERV-K corresponds to the nucleotide intersection of the VNTR and env sequences. Additionally, it is unclear whether the deletion in the LTR occurred before or after SVA incorporation.
To date, no reports of SVA intermediate structures have been identified in genomic DNA sequence. It is unclear whether SVA intermediates are rare or no intermediates may have ever existed [42]. The likelihood that 6 individual events, acquisition of sequence followed by retrotransposition of the 1) CCCTCT hexamer, 2) first Alu, 3) SVA-U, 4) second Alu, 5) VNTR, and 6) the SINE-R, occurred independently and sequentially may be more probable, however the lack SVA intermediates is evidence against this model. Acquisition of each SVA domain simultaneously may be less probable, yet it only needed to occur once. It may be difficult to completely understand how SVA evolved; still, the model proposed is consistent with the data and the literature.
Genomic Impact of SVA
SVA has the ability to influence a genomic locus at the DNA, RNA, and epigenetic levels (Figure 4). Retrotransposons are insertional mutagens [94] and may result in disease in humans (reviewed in [95]). SVA insertions, similar to L1, have been associated with deletion of genomic DNA: 1) a 14kb deletion including the entire HLA-A gene [80], and 2) 2 different cases of neurofibromatosis 2 [82], where in one case the breakpoint exists within the SVA and in the second case the DNA breakpoint is within 400 bp of the same SVA (Table 1). The deletions may be due to non-allelic recombination (NAHR) which has been documented for L1 [96] and Alu [87] associated deletions (Figure 4D). As described earlier, SVA is a repeat of repeats, therefore any of the individual repeat domains are potentially capable of misaligning with another genomic locus containing a similar SVA or repeat, resulting in NAHR. The variation in copy number both in the CCCTCT hexamer and VNTR hints that mispairing (Figure 4D) between SVAs does occur, similar to mini-satellites [97], microsatellites, and tandem repeats [98], leading to NAHR and the expansion and contraction of these SVA domains. Ostertag et al [21] noted the relatedness between neighboring SVA VNTRs and speculated that this sequence identity was likely due to NAHR. The CCCTCT hexamer and VNTR likely evolve by additional mechanisms: 1) DNA slippage during replication [99], 2) slippage during transcription, 3) slippage during reverse transcription [29], 4) and gene conversion [100, 101]. It has been demonstrated experimentally in yeast that expansion and contraction of repeats may allow quantitative and reversible functional adaptive changes [102]. It is unknown currently if the evolution of the CCCTCT hexamer or VNTR is functionally important or rather a consequence of maintaining direct DNA repeats.
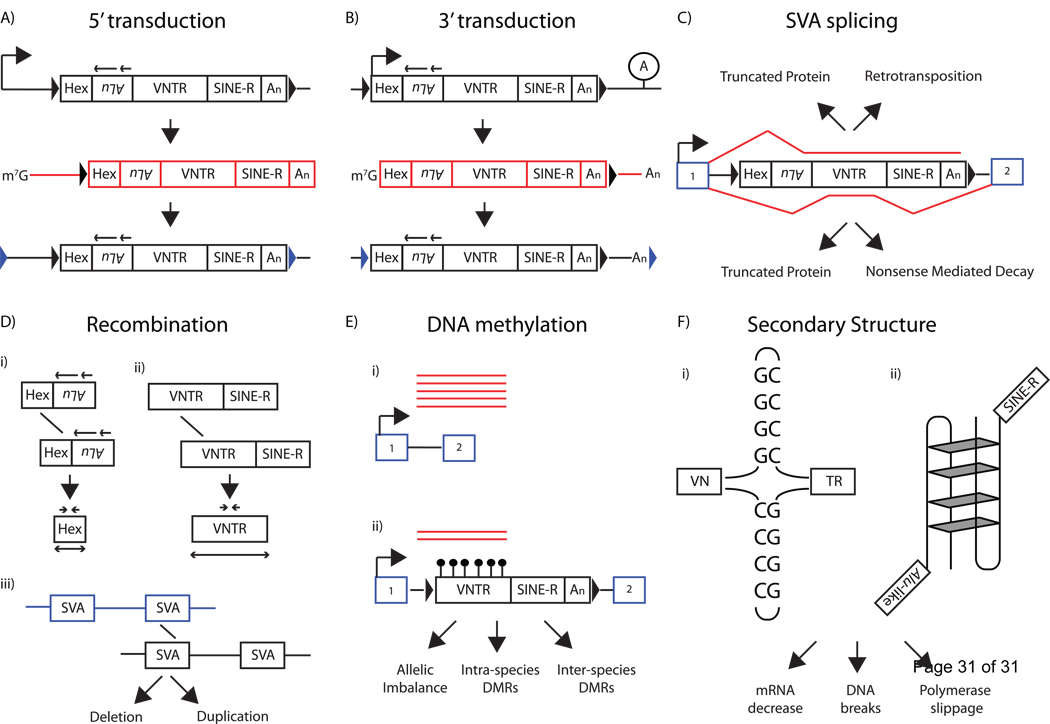
Figure 4.
Genomic Impact of SVA. A) SVAs are known to contain upstream transcriptional start sites (top, black bent arrow). This upstream transcriptional initiation will lead to an SVA mRNA (middle, red boxes) containing 5’ flanking sequence and sometimes result in retrotransposition of this sequence to a new genomic location (bottom), a process termed 5’ transduction. 5’ transduced sequence can be identified by the location of the target-site duplication positioned (blue arrowheads). B) SVAs are known to bypass their polyA signal and terminate mRNA transcription at a downstream polyA signal (lollipop A). This transcriptional readthrough will result in an SVA mRNA (middle, red boxes) containing 3’ flanking sequence and sometimes result in retrotransposition of this sequence to a new genomic location, a process termed 3’ transduction. 3’ transduced sequence can be identified by the location of the target-site duplication (blue arrowheads). The same SVA element is able to transduce 5’ and 3’ flanking sequences (not shown). C) SVA alternative splicing reduces host gene expression. SVA splicing may result in exon trapping (top, red line) that may result in truncated proteins or retrotransposition of upstream exons. SVA exonization (bottom, red line) may introduce nonsense codons into the mRNA leading to truncated proteins or nonsense-mediated decay. D) SVA elements may result in non-allelic homologous recombination within the CCCTCT hexamer (i) or the VNTR (ii), or (iii) between different SVA elements leading to deletion of genomic DNA. E) SVA VNTRs are known to be densely methylated (bottom, black lollipops) which may result in reduced mRNA expression (red lines) leading to allelic imbalance and inter- and intra- species-specific differentially methylated regions (DMR). F) The SVA VNTR is composed of GC-rich tandem repeats. Imperfect palindromes within individual tandem repeats may lead to the formation of cruciform structures in DNA (i). The VNTR may also result in more complex DNA structure, such as G-quadruplexes (ii). Structures formed due to the GC-richness of the VNTR may lead to decreases in mRNA, DNA breaks during replication, and DNA polymerase slippage, resulting in VNTR deletions. However, the RNA secondary structure of the VNTR is presumed to be functionally important in SVA retrotransposition.
The VNTR length has increased over evolutionary time [19]. The younger SVAs of subfamilies E, F, and F1 elements tend to have longer VNTRs relative to those of the older subfamilies, B, C, and D. There is a linear relationship between VNTR size and subfamily age if the oldest subfamily, SVAA is excluded [19]. The VNTR size of SVA orthologues differs between species, as indicated by the size variation of the duplicated SVARHOT1 insertions [26]. SVARHOT1 insertions are a group of three SVAs where the original SVA retrotransposed following an SVA- mediated alternative splicing event. A fusion mRNA containing six exons and SVA sequence retrotransposed to CH13 sometime after our last common ancestor (LCA) with the orangutan. This SVA containing the RHOT1 exons inserted into a larger copy number variant which subsequently duplicated twice, once since our LCA with gorilla to CH18, and once since our LCA with chimp to CH21 [26]. It is noteworthy, that the 3 human SVARHOT1 insertions also differ in VNTR size [26], consistent with a DNA-based mechanism for VNTR evolution.
Two SVA insertions have also been associated with exon-skipping (Table 1). An inherited SVA insertion into the Bruton’s tyrosine kinase gene (BTK) interrupted an exon leading to exon skipping identifiable in the patient’s cDNA [81]. The SVA insertion was not de novo in the patient as it was inherited from grandmother to mother to patient. This insertion disrupted the 5’ SS of BTK exon 9 resulting in loss of protein and X-linked agammaglobulinemia (XLA). This SVA insertion was 253 bp long, contained only SINE-R sequence, with a 92-bp polyA tail, and was flanked by a 16-bp target site duplication. Oddly, an Alu insertion has been identified in a different patient that occurred at the exact same site, to the nucleotide, also resulting in XLA [103].
An additional example of an SVA insertion disrupting an exon led our lab to become interested in SVA (Table 1). A report describing a family with hereditary elliptocytosis and pyropoikilocytosis was associated with a truncated α-spectrin protein [83](OMIM #182860). Further analysis revealed a 632 bp insertion interrupting exon 5 of α-spectrin. The insertion contained a polyA tail with the entire insertion flanked by a target-site duplication, but this sequence shared no homology to known retrotransposons. At the time that the insertion was first described, it was unknown that L1s [34–38] and SVAs [19, 21] were able to retrotranspose sequences 3’ of their genomic location (Figure 4B). It turned out that the unidentifiable retrotransposon insertion in α-spectrin was a severely truncated SVA insertion that lacked SVA sequence. The insertion sequence represented a secondary SVA 3’ transduction event that was inverted and contained a 22bp deletion at the site of inversion[21].
Three examples have been described where an SVA insertion has been associated with loss of mRNA expression (Table 1). The first, is a 2.6 kb SVA insertion into intron 32 of the TAF1 gene that has been hypothesized to cause X-linked dystonia-parkinsonism (XDP)[79] in individuals originating from the Philippine island of Panay [104] (OMIM #313650, #314250). Reduced TAF1 mRNA expression in the caudate nucleus of XDP patients was associated with hyper-DNA methylation of the SVA as indicated by HpaIII/MspI restriction analyses [79]. SVA DNA is known to be methylated [24, 105] and this methylation presumably occurs in the GC-rich VNTR (Figure 4E). One study that performed a genome-wide screen analysis for DNA methylation sites, described SVA sequence as comprising 70% of their library and they noted that the SVA VNTR was completely methylated in adult tissues [24]. Due to SVA insertional polymorphism in humans [19, 106, 107] and across species [108], SVA represents differentially methylated regions (DMR). The functional significance of SVA DNA methylation still needs to be demonstrated but it may lead to a decrease in gene expression (Figure 3E).
The last two examples of SVA insertions associated with disease resulted in almost complete loss of gene expression in the patients (Table 1). The first is a patient that had autosomal recessive hypercholestrolemia (ARH) and was homoyzygous for a full-length SVA insertion into the first intron of the LDLRAP gene [43]. This individual had no mRNA expression as indicated by Northern blot. Strangely, this SVA insertion was not detected in any other individuals, and whether consanguinity was present in this family is unknown. The homozygosity of this patient, and the inability to detect it in other individuals, suggest that the SVA insertion is relatively old or that there might be loss of heterozygosity at this locus in this individual.
Finally, another ancient SVA insertion has been described that results in Fukuyama-type muscular dystrophy (FCMD) (OMIM# 607440, #253800)[84]. FCMD is one of the most common autosomal recessive disorders in Japan. Patients are homozyogous for a full-length SVA insertion in the 3’UTR of the fukutin gene. Unexpectedly, both patients and carriers display little to no expression of the fukutin gene. The mechanism by which SVA mediates loss of mRNA expression in the ARH patient and FCMD patients is currently unknown. Unlike the TAF1 insertion, both the ARH and FCMD insertions are in the sense orientation relative to the disease gene.
Recently, it was shown that SVAs contain many functional 5’ and 3’SS on the sense strand of the element,[26]. SVA-mediated alternative splicing defined as exon-trapping (Figure 4C; top) or SVA exonization (Figure 4C; bottom) may result in a decrease of mRNA output from a gene. Both SVA exon-trapping and SVA exonization may lead to nonsense-mediated decay (Figure 4C). An alternative mechanism that may explain the loss of fukutin mRNA is that the SVA insertion may have resulted in elongation of the 3’ UTR resulting in NMD[109]. In summary, SVA insertions may result in exon-skipping, generate a novel DMR, or decrease mRNA output, potentially due to SVA mediated alternative splicing.
SVAs may also create genetic instability through other mechanisms, primarily related to the GC-richness of the SVA VNTR and its potential to form stable structures (Figure 4F). The presence of imperfect palindromes within the VNTR, GGGGGGTCAGCCCCCC, may potentially generate cruciform structures [23] that may present problems during DNA replication resulting in VNTR deletions, resulting in variation in VNTR copy number. VNTRs may also have the capability to form G-quadruplexes [110]. Using a G-quadruplex prediction website [111], seven G-quadruplexes are predicted in SVARep, with five having modest scores [42]. Interestingly, a structured GC element has been previously characterized in the Rat L1 3’UTR [63]. The secondary structure of the VNTR may lead to a reduction in mRNA output or DNA breaks during DNA replication (Figure 4F). Nevertheless, the VNTR makes it difficult to PCR amplify and sequence SVA DNA as observed by the numerous gaps in the chimpanzee and orangutan reference genome draft sequences corresponding to SVA VNTRs.
SVA retrotransposons and cancer
The precise role human retrotransposons play in cancer is unknown. The negative impact of retrotransposons in cancer may or may not rely on whether these elements are actively retrotransposing. For example, LINE-1 is likely more deleterious as an insertional mutagen relative to SVA in cancer. However, how “active” are retrotransposons in human cancer is still of great debate and interest.
Here, we have described multiple mechanisms by which an SVA may alter gene expression. Of particular interest is the ability of SVA-mediated alternative splicing to result in either 1) the production of a dominant negative product or 2) an overall decrease in mRNA output from a specific gene. Furthermore, it is well established that DNA methylation patterns are disrupted in cancer resulting in inappropriate silencing or activation of genes. Therefore, it is worthwhile to investigate the DNA methylation state of specific SVAs and the role of this methylation or lack thereof on local gene expression in cancer.
Notably, as described in the SVA lifecycle section, SVA elements contain a HRE within the SINE-R. Since the HERV-K from which the SINE-R is derived is inducible upon addition of progesterone followed by estrogen, it is likely that SVA mRNA expression may also be induced upon addition of hormone. Nevertheless, SVA contains HREs that are likely functional, and these HREs in polymorphic SVAs may be oncogenic or contribute to cancer progression.
Human Variation and Evolution
SVA can be divided into six families, A thru F, based upon point mutation and indels within the SINE-R [19] or in the case of the F1 subfamily, presence of the MAST2 first exon [26, 29, 59]. Similar to L1 [112], phylogenetic analyses suggests one dominant retrotransposing SVA family at a time [19]. SVAs from the D and human-specific subfamilies E, F, F1 are polymorphic in humans [19, 29, 106]. A recent study identified 14 SVA insertions present in the HuRef genome not shared with HGWD [107]. Previous studies estimate that ~40% of SVAs [106] are polymorphic, in particular 37% of E and 27% F SVA elements [19]. The personal genome era combined with high-throughput DNA sequencing technologies will enable a better estimate of SVA polymorphism levels.
SVAs residing in genes are potentially disruptive in either orientation. About 1/3 of all SVAs in the human genome reside in genic regions [19], with about 20% of those SVAs being the same orientation as a gene [26]. The depletion of SVAs on the coding strand suggests selection against insertions on the sense strand of a gene. A similar pattern is observed for chimp SVAs. This under-representation may be due to SVA-mediated alternative splicing or SVA induced DNA methylation. Considering SVA’s ability to transduce genomic sequence along with its ability to mediate alternative splicing and the high degree of SVA polymorphism, SVA is capable of generating considerable inter-individual variation in gene expression at loci in which it resides.
It has been estimated that ~80% of SVA insertions occurred after the human-chimp split ~6 mya [106]. A more recent study comparing draft genomes identified 800 SVA insertions as human-specific and about 400 SVAs as chimp-specific [108], while the chimpanzee genome project estimated about 1000 lineage specific SVA insertions [113]. A higher coverage chimpanzee genome draft sequence along with an exhaustive genotyping approach will provide better insight into the number of SVA elements fixed and polymorphic between the two species. Furthermore, the authors of the chimpanzee genome analysis speculated that SVAs may generate species-specific differences due to multiple CpGs and potential transcription factor binding sites.
SVAs evolved from repeats and are currently evolving in humans, as indicated by the acquisition of MAST2 sequence via splicing forming the SVAF1 subfamily (Figure 1E) and the many transduction groups identified recently. The F1 subfamily comprises at least 32% of all SVAFs [29]. SVAF1s have further evolved by acquiring 5’ and 3’ Alu transductions forming a group that contains at least 13 elements in the HGWD and a non-reference insertion associated with disease derived from an SVA master element locus on chromosome 10 [26, 29]. How the MAST2 sequence or the Alus enhance SVA retrotransposition is unknown. On the other hand, it is clear that SVAs may acquire additional sequence such as novel TSSs through retrotransposition of upstream exons as indicated by the expression of SVATPTE in chimp testes from transcription initiated in the first exon transduced by SVA [26]. Alternatively, genes may pick up SVA sequence as in the case of LEPR [114]. Here, an SVA is expressed as the C-terminal coding exon of the leptin receptor isoform RNA. SVA sequence incorporation into the transcriptome is probably common, however the multiple nonsense codons in each SVA reading frame presumably prohibit SVA sequence from being translated into protein. SVA may also create new gene families, as described for the retrotransposon-mediated duplication of the AMAC gene due to SVA 3’ transduction [39]. In this instance, three AMAC copies were duplicated by SVA 3’ transductions and at least in humans maintain intact ORFs.
Concluding Remarks
In summary, SVA is alive and well and its activity impacts the human genome by the mechanisms reviewed here along with other unknown mechanisms. It is of critical importance to develop a robust SVA cell culture retrotransposition assay to further study SVA. The lack of a bona fide progenitor element to a de novo SVA insertion has impeded development of the assay. On a more fundamental level, it is necessary to understand not only what enables SVA retrotransposition, but what enables its transcription. How active SVA is in humans is presently unknown, but the current DNA sequencing technologies will provide unprecedented opportunities for retrotransposon research. Good estimates of L1, Alu, and SVA retrotransposon activity in humans will exist within the next couple years. Progress on characterizing SVA biology has been slow and difficult at times; however more recent progress has revealed exciting findings and new directions. As other genomes are finished, such as the gorilla and orangutan, along with individual humans, the impact of SVA on individual variation and disease will slowly be revealed.
Acknowledgements
D.C.H. is funded by a T32GM008216 from the N.I.H. Research in the Kazazian laboratory is funded by R01s awarded to H.H.K. from the N.I.H. We thank the guest editors for the invitation to submit this review. We also thank Dr. John Goodier for critical reading of this manuscript and helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 2.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skowronski J, Singer MF. The abundant LINE-1 family of repeated DNA sequences in mammals: genes and pseudogenes. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):457–464. doi: 10.1101/sqb.1986.051.01.055. [DOI] [PubMed] [Google Scholar]
- 4.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 5.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin TJ, Ormandy JE, Poulter RT. L1-like non-LTR retrotransposons in the yeast Candida albicans. Curr Genet. 2001;39:83–91. doi: 10.1007/s002940000181. [DOI] [PubMed] [Google Scholar]
- 8.Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, et al. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 10.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 11.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3' terminus of l1. Genomics. 2002;80:402–406. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- 13.Pavlicek A, Paces J, Elleder D, Hejnar J. Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 2002;12:391–399. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Harrison P, Gerstein M. Identification and Analysis of Over 2000 Ribosomal Protein Pseudogenes in the Human Genome. Genome Research. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3' sequence. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 16.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 17.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–611. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, et al. SVA Elements: A Hominid-specific Retroposon Family. Journal of Molecular Biology. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 20.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, et al. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. Journal of Biological Chemistry. 1994;269:8466–8476. [PubMed] [Google Scholar]
- 21.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA Elements Are Nonautonomous Retrotransposons that Cause Disease in Humans. The American Journal of Human Genetics. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono M, Kawakami M, Takezawa T. A novel human nonviral retroposon derived from an endogenous retrovirus. Nucleic Acids Res. 1987;15:8725–8737. doi: 10.1093/nar/15.21.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Z, Hsieh S, Bentley D, Campbell R, Volanakis J. A variable number of tandem repeats locus within the human complement C2 gene is associated with a retroposon derived from a human endogenous retrovirus. J Exp Med. 1992;175:1783–1787. doi: 10.1084/jem.175.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, et al. A Genome-Wide Screen for Normally Methylated Human CpG Islands That Can Identify Novel Imprinted Genes. Genome Research. 2002;12:543–554. doi: 10.1101/gr.224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strichman-Almashanu LZ. A novel class of CPG islands-methylated in normal tissues [microform] 2000 [Google Scholar]
- 26.Hancks D, Ewing A, Chen JE, Tokunaga K, Kazazian H. Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009 doi: 10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurka J. Repbase Update: a database and an electronic journal of repetitive elements. Trends in Genetics. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 28.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and Genome Research. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 29.Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, et al. 5'-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Research. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han K, Konkel MK, Xing J, Wang H, Lee J, Meyer TJ, et al. Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- 31.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 32.Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0052. research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 35.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 36.Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 37.Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3'-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 38.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proceedings of the National Academy of Sciences. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 2004;14:2253–2260. doi: 10.1101/gr.2745804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hancks DC, Kazazian HH., Jr Unpublished data and observations. In. [Google Scholar]
- 43.Wilund KR, Yi M, Campagna F, Arca M, Zuliani G, Fellin R, et al. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum Mol Genet. 2002;11:3019–3030. doi: 10.1093/hmg/11.24.3019. [DOI] [PubMed] [Google Scholar]
- 44.Georgiou I, Noutsopoulos D, Dimitriadou E, Markopoulos G, Apergi A, Lazaros L, et al. Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum Mol Genet. 2009;18:1221–1228. doi: 10.1093/hmg/ddp022. [DOI] [PubMed] [Google Scholar]
- 45.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 47.Trinklein ND, Karaöz U, Wu J, Halees A, Force Aldred S, Collins PJ, et al. Integrated analysis of experimental data sets reveals many novel promoters in 1% of the human genome. Genome Research. 2007;17:720–731. doi: 10.1101/gr.5716607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, et al. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Research. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 50.Piedrafita FJ, Molander RB, Vansant G, Orlova EA, Pfahl M, Reynolds WF. An Alu element in the myeloperoxidase promoter contains a composite SP1-thyroid hormone-retinoic acid response element. J Biol Chem. 1996;271:14412–14420. doi: 10.1074/jbc.271.24.14412. [DOI] [PubMed] [Google Scholar]
- 51.Vansant G, Reynolds WF. The consensus sequence of a major Alu subfamily contains a functional retinoic acid response element. Proc Natl Acad Sci U S A. 1995;92:8229–8233. doi: 10.1073/pnas.92.18.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norris J, Fan D, Aleman C, Marks JR, Futreal PA, Wiseman RW, et al. Identification of a New Subclass of Alu DNA Repeats Which Can Function as Estrogen Receptor-dependent Transcriptional Enhancers. Journal of Biological Chemistry. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- 53.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol. 1987;61:2059–2062. doi: 10.1128/jvi.61.6.2059-2062.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boeke JD. LINEs and Alus--the polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 56.Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, et al. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buzdin A, Gogvadze E, Lebrun MH. Chimeric retrogenes suggest a role for the nucleolus in LINE amplification. FEBS Lett. 2007;581:2877–2882. doi: 10.1016/j.febslet.2007.05.034. [DOI] [PubMed] [Google Scholar]
- 58.Mills RE, Bennett EA, Iskow RC. Devine SE. Which transposable elements are active in the human genome? 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Bantysh OB, Buzdin AA. Novel Family of Human Transposable Elements Formed Due to Fusion of the First Exon of Gene MAST2 with Retrotransposon SVA. Biochemistry (Moscow) 2009;74:1393–1399. doi: 10.1134/s0006297909120153. [DOI] [PubMed] [Google Scholar]
- 60.Buzdin A, Gogvadze E, Kovalskaya E, Volchkov P, Ustyugova S, Illarionova A, et al. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 2003;31:4385–4390. doi: 10.1093/nar/gkg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pavlicek A, Gentles AJ, Paces J, Paces V, Jurka J. Retroposition of processed pseudogenes: the impact of RNA stability and translational control. Trends Genet. 2006;22:69–73. doi: 10.1016/j.tig.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weichenrieder O, Wild K, Strub K, Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- 63.Usdin K, Furano AV. The structure of the guanine-rich polypurine:polypyrimidine sequence at the right end of the rat L1 (LINE) element. Journal of Biological Chemistry. 1989;264:15681–15687. [PubMed] [Google Scholar]
- 64.Mathews DH, Banerjee AR, Luan DD, Eickbush TH, Turner DH. Secondary structure model of the RNA recognized by the reverse transcriptase from the R2 retrotransposable element. RNA. 1997;3:1–16. [PMC free article] [PubMed] [Google Scholar]
- 65.Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmitz, Churakov G, Zischler H, Brosius A Novel Class of Mammalian-Specific Tailless Retropseudogenes. Genome Research. 2004;14:1911–1915. doi: 10.1101/gr.2720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okada N, Hamada M, Ogiwara I, Ohshima K. SINEs and LINEs share common 3' sequences: a review. Gene. 1997;205:229–243. doi: 10.1016/s0378-1119(97)00409-5. [DOI] [PubMed] [Google Scholar]
- 68.Schmitz J, Zemann A, Churakov G, Kuhl H, Grutzner F, Reinhardt R, et al. Retroposed SNOfall--a mammalian-wide comparison of platypus snoRNAs. Genome Res. 2008;18:1005–1010. doi: 10.1101/gr.7177908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roy-Engel AM, Salem AH, Oyeniran OO, Deininger L, Hedges DJ, Kilroy GE, et al. Active Alu element "A-tails": size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dewannieux M, Heidmann T. Role of poly(A) tail length in Alu retrotransposition. Genomics. 2005;86:378–381. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 71.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: What causes Alu elements to die? Genome Research. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malik HS, Eickbush TH. The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol Biol Evol. 1998;15:1123–1134. doi: 10.1093/oxfordjournals.molbev.a026020. [DOI] [PubMed] [Google Scholar]
- 73.Gogvadze E, Barbisan C, Lebrun MH, Buzdin A. Tripartite chimeric pseudogene from the genome of rice blast fungus Magnaporthe grisea suggests double template jumps during long interspersed nuclear element (LINE) reverse transcription. BMC Genomics. 2007;8:360. doi: 10.1186/1471-2164-8-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buzdin AA. Retroelements and formation of chimeric retrogenes. Cell Mol Life Sci. 2004;61:2046–2059. doi: 10.1007/s00018-004-4041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hasnaoui M, Doucet AJ, Meziane O, Gilbert N. Ancient repeat sequence derived from U6 snRNA in primate genomes. Gene. 2009;448:139–144. doi: 10.1016/j.gene.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 76.Babushok DV, Ohshima K, Ostertag EM, Chen X, Wang Y, Mandal PK, et al. A novel testis ubiquitin-binding protein gene arose by exon shuffling in hominoids. Genome Res. 2007;17:1129–1138. doi: 10.1101/gr.6252107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ohshima K, Hamada M, Terai Y, Okada N. The 3' ends of tRNA-derived short interspersed repetitive elements are derived from the 3' ends of long interspersed repetitive elements. Mol Cell Biol. 1996;16:3756–3764. doi: 10.1128/mcb.16.7.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 79.Makino S, Kaji R, Ando S, Tomizawa M, Yasuno K, Goto S, et al. Reduced Neuron-Specific Expression of the TAF1 Gene Is Associated with X-Linked Dystonia-Parkinsonism. The American Journal of Human Genetics. 2007;80:393–406. doi: 10.1086/512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takasu M, Hayashi R, Maruya E, Ota M, Imura K, Kougo K, et al. Deletion of entire HLA-A gene accompanied by an insertion of a retrotransposon. Tissue Antigens. 2007;70:144–150. doi: 10.1111/j.1399-0039.2007.00870.x. [DOI] [PubMed] [Google Scholar]
- 81.Rohrer J, Minegishi Y, Richter D, Eguiguren J, Conley ME. Unusual mutations in Btk: an insertion, a duplication, an inversion, and four large deletions. Clin Immunol. 1999;90:28–37. doi: 10.1006/clim.1998.4629. [DOI] [PubMed] [Google Scholar]
- 82.Legoix P, Sarkissian HD, Cazes L, Giraud S, Sor F, Rouleau GA, et al. Molecular Characterization of Germline NF2 Gene Rearrangements. Genomics. 2000;65:62–66. doi: 10.1006/geno.2000.6139. [DOI] [PubMed] [Google Scholar]
- 83.Hassoun H, Coetzer TL, Vassiliadis JN, Sahr KE, Maalouf GJ, Saad ST, et al. A novel mobile element inserted in the alpha spectrin gene: spectrin dayton. A truncated alpha spectrin associated with hereditary elliptocytosis. J Clin Invest. 1994;94:643–648. doi: 10.1172/JCI117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- 85.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of Years of Evolution Preserved: A Comprehensive Catalog of the Processed Pseudogenes in the Human Genome. Genome Research. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim HS, Wadekar RV, Takenaka O, Hyun BH, Crow TJ. Phylogenetic analysis of a retroposon family in african great apes. J Mol Evol. 1999;49:699–702. doi: 10.1007/pl00000083. [DOI] [PubMed] [Google Scholar]
- 87.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 88.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3' splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 90.Makalowski W, Mitchell GA, Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends in Genetics. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 91.Goodchild NL, Freeman JD, Mager DL. Spliced HERV-H endogenous retroviral sequences in human genomic DNA: evidence for amplification via retrotransposition. Virology. 1995;206:164–173. doi: 10.1016/s0042-6822(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 92.Kowalski PE, Freeman JD, Mager DL. Intergenic splicing between a HERV-H endogenous retrovirus and two adjacent human genes. Genomics. 1999;57:371–379. doi: 10.1006/geno.1999.5787. [DOI] [PubMed] [Google Scholar]
- 93.van de Lagemaat LN, Medstrand P, Mager DL. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 2006;7:R86. doi: 10.1186/gb-2006-7-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 95.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 96.Han K, Lee J, Meyer TJ, Remedios P, Goodwin L, Batzer MA. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci U S A. 2008;105:19366–19371. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jeffreys AJ, Neil DL, Neumann R. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J. 1998;17:4147–4157. doi: 10.1093/emboj/17.14.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 99.Usdin K. The biological effects of simple tandem repeats: Lessons from the repeat expansion diseases. Genome Research. 2008;18:1011–1019. doi: 10.1101/gr.070409.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roy AM, Carroll ML, Nguyen SV, Salem AH, Oldridge M, Wilkie AO, et al. Potential gene conversion and source genes for recently integrated Alu elements. Genome Res. 2000;10:1485–1495. doi: 10.1101/gr.152300. [DOI] [PubMed] [Google Scholar]
- 101.Chen J-M, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 102.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Conley ME, Partain JD, Norland SM, Shurtleff SA, Kazazian HH., Jr Two independent retrotransposon insertions at the same site within the coding region of BTK. Hum Mutat. 2005;25:324–325. doi: 10.1002/humu.9321. [DOI] [PubMed] [Google Scholar]
- 104.Deng H, Le WD, Jankovic J. Genetic study of an American family with DYT3 dystonia (lubag) Neurosci Lett. 2008;448:180–183. doi: 10.1016/j.neulet.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 105.Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, et al. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448:151–167. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, et al. Mobile elements create structural variation: Analysis of a complete human genome. Genome Research. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, et al. Recently Mobilized Transposons in the Human and Chimpanzee Genomes. The American Journal of Human Genetics. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mendell JT, Dietz HC. When the Message Goes Awry: Disease-Producing Mutations that Influence mRNA Content and Performance. Cell. 2001;107:411–414. doi: 10.1016/s0092-8674(01)00583-9. [DOI] [PubMed] [Google Scholar]
- 110.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends in Cell Biology. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 111.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucl Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Consortium TCSaA. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 114.Damert A, Lower J, Lower R. Leptin Receptor Isoform 219.1: An Example of Protein Evolution by LINE-1-Mediated Human-Specific Retrotransposition of a Coding SVA Element. Mol Biol Evol. 2004;21:647–651. doi: 10.1093/molbev/msh056. [DOI] [PubMed] [Google Scholar]