Abstract
We developed a fast method for denoising pyrosequencing for community 16S rRNA analysis. We observe a 2–4 fold reduction in the number of observed OTUs (operational taxonomic units) comparing denoised with non-denoised data. ~50,000 sequences can be denoised on a laptop within an hour, two orders of magnitude faster than published techniques. We demonstrate the effects of denoising on alpha and beta diversity of large 16S rRNA datasets.
Keywords: next generation DNA sequencing, ribosomal RNA, community sequence analysis, microbial ecology, denoising
Pyrosequencing1 has revolutionized microbial community analysis by allowing the simultaneous assessment of hundreds of microbial communities in multiplex with sufficient depth to resolve meaningful biological patterns2. These techniques have been used to gain striking new insight into microbial processes on scales ranging from continents3 to within an individual’s body4.
Although powerful new analysis tools such as GAST5, Mothur6, and QIIME7 greatly streamline the process of interpreting microbial community information obtained by pyrosequencing, especially similarities and differences among communities, substantial questions remain about the suitability of pyrosequencing to address questions concerning alpha diversity, the amount of diversity within each individual community and non-phylogenetic beta-diversity measures (phylogenetic beta-diversity measures such as UniFrac, which measure similarities between different communities, are relatively robust to these issues8). In particular, noise introduced during pyrosequencing and the PCR amplification stage can inflate estimates of the number of OTUs (chosen at the 97% identity level) in a given habitat by orders of magnitude9, 10. The current state-of-the-art is to reduce noise by clustering the flowgrams (patterns of intensities in each read) before conversion to sequences to eliminate issues due to homopolymer read errors10, yet this approach is exceedingly computationally expensive and beyond the reach of most individual investigators who do not have access to large-scale computing facilities.
Methods
Inability to accurately determine which sequences are present in a sample, and hence the abundances of rare taxa, greatly inhibits our ability to infer important ecological parameters such as rank-abundance curves, yet ironically the portion of the rank-abundance curve that can be inferred, i.e. of the common taxa, provides a solution to the conundrum of the expense of denoising. Empirical rank-abundance curves, especially from human-associated samples, tend to be dominated by a relatively small number of abundant taxa. Given this feature of actual microbial communities, performing all-on-all comparisons for clustering is exceedingly inefficient: instead, a subset of reads suffices to identify the common OTUs, which can then be iteratively removed by recruitment to an existing cluster. Consequently, we can rapidly determine the OTUs that are most likely to be abundant, concentrate initially on comparing reads to the small number of abundant OTUs (removing matches from the analysis), and then cluster only the leftover reads representing more divergent sequences.
We can thus reduce the total number of sequence comparisons using empirical features of the abundance distribution of real datasets as follows. First, we devised a fast pre-filter, removing reads that are strict prefixes of other reads, and compute an initial sequence distribution. We then sort the prefix clusters in descending order of abundance, and use this initial distribution to cluster similar reads, comparing each additional unclustered read to the most abundant clusters first because we expect the abundant clusters to yield a larger number of erroneous near-matching reads due to their numerical dominance alone. For a more detailed description of the algorithm, see Supplementary Methods. A similar method of pre-clustering on the sequence level and subsequent sequence clustering along the abundance distribution has been proposed recently11.
The method introduced here is a major improvement over previous flowgram-based denoising routines10 in terms of compute resources, yet retains the advantage that singletons are not discarded entirely, allowing exploration of the rare biosphere12. Previously, a mid-size 24-core cluster was needed to analyze a small dataset of around 40,000 sequences in around 10 hours. Our method allows the same dataset to be denoised in less than an hour on a single laptop computer (Table S1). We can also denoise full 454 runs with 500,000 sequences on a mid-size cluster in 1 day. We can thus address questions in community ecology that were previously intractable.
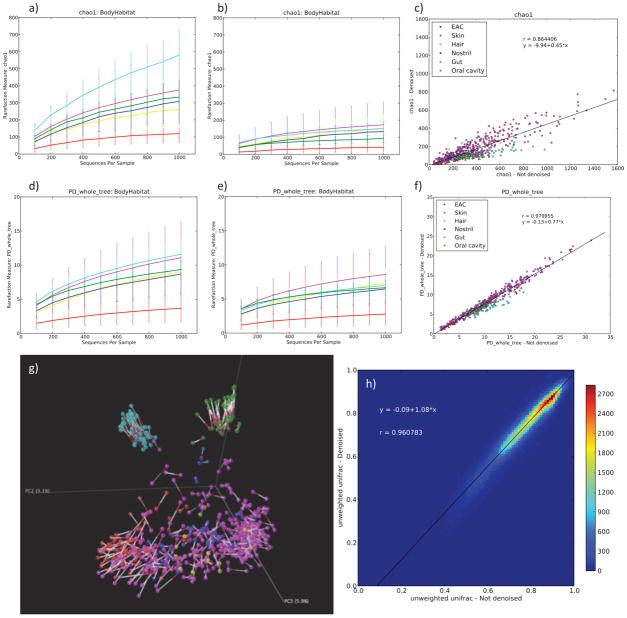
Applying these new methods to the most comprehensive survey of human-associated body habitats yet performed4, we find that denoising produces a substantial decrease in the diversity both at the OTU level and in terms of the phylogenetic diversity (the total branch length associated with each sample on a phylogenetic tree14). However, the results from the non-denoised (but filtered) and denoised data are highly correlated (r2 = 0.97, P <10−300 for phylogenetic diversity), suggesting that relative results concerning diversity within each sample are robust to the types of errors introduced by pyrosequencing (Fig. 1a–f). Interestingly, in spite of this high correlation, denoising changes the relative order of OTU richness of individual body habitats. Although the gut exhibits the highest OTU richness without denoising, it falls back into the middle ranks after denoising. This holds true for both Chao1 estimates and the phylogenetic diversity (Fig. 1a,d and 1b,e). The drastic reduction after denoising might be an effect of the sequence composition of the dominant OTUs in the gut (see Supplementary Methods for a more detailed discussion).
Figure 1.
Comparisons of non-denoised data (a–c) to denoised data (d–f) for alpha diversity for the Body Habitat study, and comparisons of beta diversity (g–h). Rarefaction plots of the “Body Habitat” study4 show a 3 to 4 fold decrease in the Chao1 estimate when comparing non-denoised (a) to denoised (b) data. Interestingly, denoising changes the relative order of OTU richness of individual body habitats: the gut exhibits the highest OTU richness without denoising, but falls back into the middle ranks after denoising. This holds true for both Chao1 estimates and phylogenetic diversity (PD). c) Scatter plots of alpha diversity metrics per sample show a high correlation overall, but a significant deviation from the average for gut and the oral cavity. (EAC = external auditory canal). g) Procrustes analysis of denoised and filtered unweighted UniFrac principal coordinates analysis (PCoA). Bars connect identical samples in the plot with the red side of the bar pointing towards the denoised data. There is no qualitative difference between denoised and filtered in the overall clustering, yet on a smaller scale we observe that the denoised samples are oriented more to the center than the filtered ones. This shows that denoising removes some of the artificial distance between samples introduced by false OTUs. h) Unweighted UniFrac distances for all pairs of samples for the denoised and filtered data set are highly correlated (r2=0.96). From the regression, it is clear that for similar samples noise has a greater effect than it has for dissimilar samples. The color bar gives the number of pairwise comparisons at a particular point.
Similarly, when clustering the samples using UniFrac, the non-denoised and denoised reads produce very similar patterns (Fig. 1g–h), reinforcing the point that errors introduced into each sample by noise or chimeras have little effect on beta diversity because they inflate the distances among all samples rather than introducing artifactual similarities between specific pairs of samples15.
We conclude that the availability of these new methods will make more accurate assessments of alpha diversity available to a wide range of researchers (especially in conjunction with improved chimera-checking methods such as ChimeraSlayer, http://microbiomeutil.sourceforge.net/), and will greatly improve our understanding of microbial communities in habitats with scales ranging from global to extremely personal. The efficiency of the new techniques and the fact that they can change conclusions about the relative diversity in different habitats suggests that they should be applied routinely in all pyrosequencing studies where estimates of diversity within each sample are the goal.
Supplementary Material
Acknowledgments
We thank Peter Turnbaugh for providing us with an excellent Mock community for testing and Chris Quince for unpublished insights into how PyroNoise works.
J.R. was supported in part by a postdoctoral scholarship from the DAAD. This work was supported in part by grants from the NIH and NASA, and by HHMI.
Footnotes
Availability
The program is available for download at http://www.microbio.me/denoiser/
References
- 1.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costello EK, et al. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 2009 doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huse SM, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors lead to artificial inflation of diversity estimates. Environ Microbiol. 2009 doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 10.Quince C, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 11.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61:1–10. [Google Scholar]
- 15.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.