Introduction
Having completed a PhD in molecular and cellular biology in mammalian reproduction at the University of Sydney, my first postdoctoral appointment began early in 1965 in the laboratory of Sir Hans Krebs at Oxford University where I had the pleasure of working with Dr Eric Newsholme on the regulation of gluconeogenesis and glycolysis during fetal development. My introduction to Pyruvate Carboxylase (PC) came early in 1966 when Michael Scrutton arrived back in Oxford to submit his D.Phil. thesis after spending a remarkably productive time working on PC in the laboratory of its discoverer, Professor Merton Utter, at Western Reserve University Medical School, Cleveland, Ohio. Not surprisingly, Michael bubbled with infectious enthusiasm about PC, and presented an outstanding seminar to the Department.
In late 1966 I moved to Professor Utter’s group and had an invaluable and enjoyable experience broadening my skills and filling some of the many gaps in my knowledge, particularly in biophysical techniques. The co-discoverer of PC, Professor Bruce Keech, returned to Cleveland in 1968 on sabbatical leave from the University of Adelaide, Australia, and we soon established an effective and agreeable collaboration. This collaboration was renewed in late 1969 when I arrived in Adelaide, where I secured a Faculty position in 1970, and continued until Bruce retired in 1983.
The occurrence and biological roles of Pyruvate Carboxylase
PC was discovered in the context of defining the pathway of gluconeogenesis in liver and kidney (1,2), but was soon recognised to play important roles also in lipogenesis (3), in glyceroneogenesis in liver and adipose tissue (4) as well as in the functions of brain (5) and pancreatic islets (5, 6). The central anaplerotic role of this mitochondrial enzyme in various mammalian tissues has been summarised in recent reviews (8, 9).
PC is also found in many, though not all, prokaryotes, and has been shown to play essential roles in several important pathogens (10, 11). PC is found in most fungi, generally in the cytoplasm, and in many invertebrates where its functions are equally significant.
The reaction for carboxylating pyruvate
PC catalyses the ATP-dependent formation of oxaloacetate from pyruvate, and in most species this enzyme is stimulated allosterically by acetyl-CoA. The overall reaction [Eq. I] is composed of two partial reactions [Eq. II & III] catalysed at separate sub-sites located on different subunits. The major effect of the allosteric activator is to enhance the rate of carboxylation of the biotin prosthetic group in the first partial reaction.
| (Eq. I) |
| (Eq. II) |
| (Eq. III) |
The structure of PC
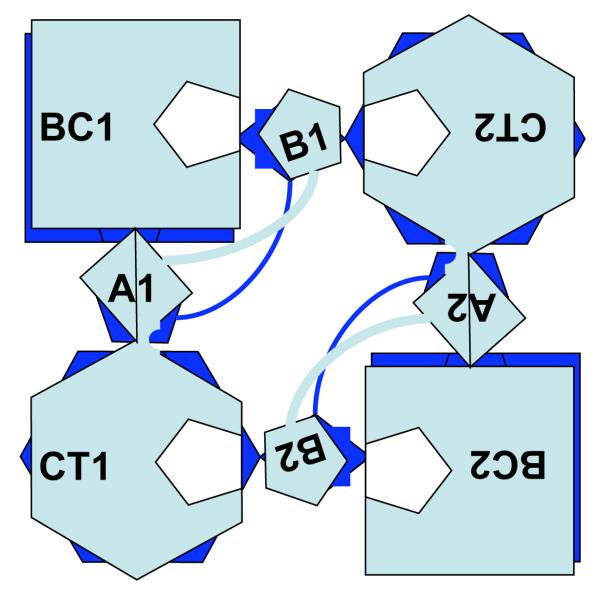
In mammals and most microbes PC is a homotetramer composed of identical subunits of 120-130 kDa, and is active only in its tetrameric form. Analysis of the first complete gene and protein sequence of PC in S. cerevisiae (12) revealed 3 regions with strong sequence identity to component subunits of other biotin-dependent enzymes. An N-terminal domain that includes the Biotin Carboxylation (BC) sub-site shares ~45% identity with the ATP-binding subunit of E. coli acetyl-CoA carboxylase. A centrally located domain accommodating the Carboxyltransferase (CT) sub-site shares 32% identity with the pyruvate-binding 5S subunit of methyl-malonyl-CoA transcarboxylase in Propionibacterium shermanii. The biotin prosthetic group in PC is attached covalently to the epsilon-amino group of a specific lysine residue in a highly-conserved motif ~35 residues from the enzyme’s C-terminus in a domain that shares a high degree of identity with the Biotin Carboxyl Carrier Protein (BCCP) subunit of E. coli acetyl-CoA carboxylase and the 1.3S biotinylated subunit of transcarboxylase in P. shermanii. High resolution structures of PC from Rhizobium etli by St. Maurice et al (13) and Staphylococcus aureus by Xiang & Tong (14) showed the subunits are arranged in rhombohedral geometry within the quaternary structure of the tetramer as a dimer of dimers. The monomers within each dimeric pair are arranged anti-parallel so that the BC sub-site of one is opposite and ~75 Angstroms from the CT sub-site of the other [see Fig. 1], but there is minimal direct contact between them. Within the dimeric pair of subunits 1 and 2 the BCCP domain of subunit 1 is located between the BC sub-site of subunit 1 and the CT sub-site of subunit 2, and vice versa. Subunits 3 and 4 combine to form another dimeric pair arranged orthogonal to and on another plane to the dimer of subunits 1 and 2. The BCCP domains of each dimer are located on opposite sides of the tetramer, as revealed by electron microscopy of 1:1 complexes of PC and avidin (15).
Figure 1. Schematic representation of the arrangement of the individual monomers making up the R.etli PC tetramer.
Light grey coloured shapes represent the uppermost dimer pair composed of subunits 1 and 2. Black shapes below represent the other dimer pair composed of subunits 3 and 4.
BC1 & BC2, Biotin carboxylase domains (residues 1-465) of subunits 1 & 2 respectively
A1 & A2, Allosteric domains (residues 471-489 + 1002-1073) of subunits 1 & 2 respectively
CT1 & CT2, Carboxyltransferase domains (residues 515-1000) of subunits 1 & 2 respectively
B1 & B2, Biotin carboxyl carrier domains (residues 1084-1154) of subunits 1 & 2 respectively
White pentagonal shapes within BC1 and CT2, and within BC2 and CT1 indicate notional positions of catalytic sub-sites served by B1 and B2 respectively.
The reaction mechanism
In the first partial reaction (Eqn II) bicarbonate is activated by ATP to form a carboxyphosphate intermediate (16) that then directly, or more likely indirectly, transfers CO2 onto the 1′-nitrogen of the covalently attached biotin in the BCCP domain. The biotin prosthetic group then serves to transport this carboxyl group from the BC sub-site to the opposing CT sub-site where pyruvate accepts it and is converted to oxaloacetate. The separation of the partial reactions was inferred from kinetic studies (17) and by analogy to the situation for acetyl-CoA carboxylase in E. coli (18) and methyl-malonyl-CoA transcarboxylase in P. shermanii (19). Both are biotin-dependent enzymes composed of three classes of subunits that are all required for catalysis of the overall reaction. Proof of this separation of sub-sites came with the determination of PC structures from R. etli (13) with ATP bound in the BC sub-site and from S. aureus (14) with pyruvate bound in the CT sub-site. The crucial roles in substrate binding and catalysis of particular amino acid side chains in the BC and CT sub-sites are becoming clearer as site-directed mutagenesis studies have been focused by the recent structural advances (9, 13, 20, 21).
Activation by acetyl-CoA
It has long been known that acetyl-CoA activates PC in vitro (22) and in vivo (23, 24) and that its major effect is exerted on the first partial reaction (9, 22). However, apart from the demonstration that its action was allosteric (25, 26), its binding site proved to be elusive until revealed recently when an analog of acetyl-CoA, viz. ethyl-CoA, was shown to be bound in a novel fold composed of residues located in the polypeptide chain between the BC and CT domains and also between the CT and BCCP domains (13). Whilst the conformational change induced by ethyl-CoA on the two subunits R. etli PC to which it was bound appeared to explain its activating effect, this does not seem to be the case with a weaker activator, Coenzyme A bound to S. aureus PC (21). Some microbial PCs, eg. Pseudomonas citronellolis (27) and Azotobacter vinelandii (28), have 4 protomers composed of two polypeptide chains, a biotinylated 75kDa (α)subunit and a 52 kDa (β)subunit arranged in an (αβ)4 structure. These bacterial PCs do not respond to the addition of acetyl-CoA, presumably because they lack the highly conserved residues involved in binding the nucleotide portion of acetyl-CoA that are located between the BC domain [= β-subunit] and CT domain [= α-subunit].
Regulation of expression and activity of PC
A single gene encoding PC resides on chromosome 11q13.4 in humans and chromosome 1q43 in rat. Both are composed of 19 coding exons arranged similarly (29). The hormones and other factors regulating gluconeogenesis, lipogenesis, glycerogenesis, neurotransmitter secretion and insulin secretion in specific tissues are all clearly different, and accordingly PC exhibits tissue-specific regulation (9, 29). In rat, alternative transcription from two promoters is responsible for producing five different mature transcripts expressed in a tissue-specific manner with all containing the same coding region but differing in their 5′ non-coding sequences (30).
Fasting and diabetes have been shown to induce 2-3-fold increases in hepatic PC activity in several species, most probably mediated via the effects of increased glucocorticoid secretion, and augmented by indirect actions of glucagon. Conversely, insulin has been shown to down-regulate PC expression in diabetic rats [reviewed in (29)].
Expression of PC has also been shown to be increased 2-5-fold at the onset of obesity in Zucker fatty rats (fa/fa) (31). The lipogenic role of PC has been further demonstrated by evidence of a functional PPARγ response element in the murine PC gene both in vitro and in vivo (32).
Unlike acetyl-CoA carboxylase, which is regulated by phosphorylation (33), PC activity does not appear to be controlled this way even under conditions when pyruvate dehydrogenase was phosphorylated (34).
PC deficiency
Given the range of tissues and major metabolic functions in which PC participates, it′s hardly surprising that a deficiency of PC in humans usually leads to severe clinical symptoms. PC deficiency (OMIM, 266150) is a rare autosomal recessive disease with three almost distinct phenotypes (35). Type A is characterised by hypoglycemia accompanied by mild to moderate lactic acidemia and sometimes elevated ketone body levels. Apart from the acute symptoms these patients also suffer from psychomotor retardation, though they rarely die at a young age. Type B, having no detectable PC protein in any tissues, is the most severe form which leads to death generally within three months from lactic acidemia accompanied by hyperammonemia, citrullinemia and hyperlysinemia. Type C has a benign phenotype associated with episodes of lactic acidemia and no psychomotor disorders.
Concluding remarks
Fifty years after the discovery of PC, and with only a little over 2000 papers listed by PubMed in response to a search for“pyruvate carboxylase”, it is not surprising that there remain many questions to be answered about this complex enzyme and its diverse functions. This quest will be challenging but also potentially very rewarding for an improved understanding and treatment of diseases involving PC activity.
Acknowledgements
Work in the author’s laboratory has been supported by the Australian Research Council and by the National Institutes of Health grant GM070455. The author apologies for any inadvertent oversight in the selection of relevant work due to space limitations.
REFERENCES
- 1.Utter MF, Keech DB. Pyruvate Carboxylase. I Nature of the reaction. J. Biol. Chem. 1963;238:2603–2608. [PubMed] [Google Scholar]
- 2.Keech DB, Utter MF. Pyruvate Carboxylase. II. Properties. J. Biol. Chem. 1963;238:2609–2614. [PubMed] [Google Scholar]
- 3.Ballard FJ, Hanson RW. The citrate cleavage pathway and lipogenesis in rat adipose tissue: replenishment of oxaloacetate. J Lipid Res. 1967;8:73–9. [PubMed] [Google Scholar]
- 4.Reshef L, Hanson RW, Ballard FJ. Glyceride-glycerol synthesis from pyruvate. Adaptive changes in phosphoenolpyruvate carboxykinase and pyruvate carboxylase in adipose tissue and liver. J. Biol. Chem. 1969;244:1994–2001. [PubMed] [Google Scholar]
- 5.Salganicoff L, Koeppe RE. Subcellular distribution ot pyruvate carboxylase, diphosphopyridine nucleotide and triphosphopyridine nucleotide isocitrate dehydrogenases, and malate enzyme in rat brain. J. Biol. Chem. 1968;243:3416–20. [PubMed] [Google Scholar]
- 6.Curi R, Carpinelli AR, Malaisse WJ. Hexose metabolism in pancreatic islets: pyruvate carboxylase activity. Biochimie. 1991;73:583–6. doi: 10.1016/0300-9084(91)90026-w. [DOI] [PubMed] [Google Scholar]
- 7.MacDonald MJ, Kaysen JH, Moran SM, Pomije CE. Pyruvate dehydrogenase and pyruvate carboxylase. Sites of pretranslational regulation by glucose of glucose-induced insulin release in pancreatic islets. J. Biol. Chem. 1991;266:22392–7. [PubMed] [Google Scholar]
- 8.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277:30409–12. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 9.Jitrapakdee S, St. Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem. J. 2008;413:369–87. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velayudhan J, Kelly DJ. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology. 2002;148:685–94. doi: 10.1099/00221287-148-3-685. [DOI] [PubMed] [Google Scholar]
- 11.Schär J, Stoll R, Schauer K, Loeffler DI, Eylert E, Joseph B, Eisenreich W, Fuchs TM, Goebel W. Pyruvate carboxylase plays a crucial role in carbon metabolism of extra- and intracellularly replicating Listeria monocytogenes. J. Bacteriol. 2010 doi: 10.1128/JB.01132-09. [Jan 22., Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim F, Morris CP, Occhiodoro F, Wallace JC. Sequence and domain structure of yeast pyruvate carboxylase. J. Biol. Chem. 1988;263:11493–7. [PubMed] [Google Scholar]
- 13.St. Maurice M, Reinhardt L, Surinya KH, Attwood PV, Wallace JC, Cleland WW, Rayment I. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science. 2007;317:1076–9. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- 14.Xiang S, Tong L. Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat. Struct. Mol. Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- 15.Johannssen W, Attwood PV, Wallace JC, Keech DB. Localisation of the active site of pyruvate carboxylase by electron microscopic examination of avidin-enzyme complexes. Eur. J. Biochem. 1983;133:201–6. doi: 10.1111/j.1432-1033.1983.tb07448.x. [DOI] [PubMed] [Google Scholar]
- 16.Attwood PV, Wallace JC. Chemical and catalytic mechanisms of carboxyl transfer reactions in biotin-dependent enzymes. Acc. Chem. Res. 2002;35:113–20. doi: 10.1021/ar000049+. [DOI] [PubMed] [Google Scholar]
- 17.Barden RE, Fung CH, Utter MF, Scrutton MC. Pyruvate carboxylase from chicken liver. Steady state kinetic studies indicate a“two-site” ping-pong mechanism. J. Biol. Chem. 1972;247:1323–33. [PubMed] [Google Scholar]
- 18.Alberts AW, Nervi AM, Vagelos PR. Acetyl CoA carboxylase, II. Demonstration of biotin-protein and biotin carboxylase subunits. Proc. Natl. Acad. Sci. U S A. 1969;63:1319–26. doi: 10.1073/pnas.63.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Northrop DB. Transcarboxylase. VI. Kinetic analysis of the reaction mechanism. J. Biol. Chem. 1969;244:5808–5819. [PubMed] [Google Scholar]
- 20.Zeczycki TN, St. Maurice M, Jitrapakdee S, Wallace JC, Attwood PV, Cleland WW. Insight into the carboxyl transferase domain mechanism of pyruvate carboxylase from Rhizobium etli. Biochemistry. 2009;48:4305–4313. doi: 10.1021/bi9003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu LP, Xiang S, Lasso G, Gil D, Valle M, Tong L. A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 2009;17:823–32. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scrutton MC, Keech DB, Utter MF. Pyruvate carboxylase IV. Partial reactions and the locus of activation by acetyl—CoA. J. Biol. Chem. 1965;240:574–81. [PubMed] [Google Scholar]
- 23.Barritt GJ, Zander GL, Utter MF. In: Gluconeogenesis. Hanson RW, Mehlman MA, editors. John Wiley & Sons; New York: 1976. pp. 3–46. [Google Scholar]
- 24.Williams DL, Spray GH, Hems R, Williamson DH. Metabolic effects of propionate in normal and vitamin B 12 -deficient rats. Biochem. J. 1971;124:501–7. doi: 10.1042/bj1240501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashman LK, Keech DB, Wallace JC, Nielsen J. Sheep kidney pyruvate carboxylase. Studies on its activation by acetyl coenzyme A and characteristics of its acetyl coenzyme A independent reaction. J. Biol. Chem. 1972;247:5818–24. [PubMed] [Google Scholar]
- 26.Ashman LK, Wallace JC, Keech DB. Densitization of pyruvate carboxylase against acetyl-CoA stimulation by chemical modification. Biochem Biophys Res Commun. 1973;51:924–31. doi: 10.1016/0006-291x(73)90015-6. [DOI] [PubMed] [Google Scholar]
- 27.Goss JA, Cohen ND, Utter MF. Characterization of the subunit structure of pyruvate carboxylase from Pseudomonas citronellolis. J. Biol. Chem. 1981;256:11819–25. [PubMed] [Google Scholar]
- 28.Scrutton MC, Taylor BL. Isolation and characterization of pyruvate carboxylase from Azotobacter vinelandii OP. Arch. Biochem. Biophys. 1974;164:641–54. doi: 10.1016/0003-9861(74)90076-9. [DOI] [PubMed] [Google Scholar]
- 29.Jitrapakdee S, Wallace JC. Structure, function and regulation of pyruvate carboxylase. Biochem. J. 1999;340:1–16. doi: 10.1042/bj3400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jitrapakdee S, Walker ME, Wallace JC. Identification of novel alternatively spliced pyruvate carboxylase mRNAs with divergent 5′-untranslated regions which are expressed in a tissue-specific manner. Biochem. Biophys. Res. Commun. 1996;223:695–700. doi: 10.1006/bbrc.1996.0958. [DOI] [PubMed] [Google Scholar]
- 31.Jitrapakdee S, Gong Q, MacDonald MJ, Wallace JC. Regulation of rat pyruvate carboxylase gene expression by alternate promoters during development, in genetically obese rats and in insulin-secreting cells. Multiple transcripts with 5′-end heterogeneity modulate translation. J. Biol. Chem. 1998;273:34422–8. doi: 10.1074/jbc.273.51.34422. [DOI] [PubMed] [Google Scholar]
- 32.Jitrapakdee S, Slawik M, Medina-Gomez G, Campbell M, Wallace JC, Sethi JK, O′Rahilly S, Vidal-Puig AJ. The peroxisome proliferator-activated receptor-gamma regulates murine pyruvate carboxylase gene expression in vivo and in vitro. J. Biol. Chem. 2005;280:27466–76. doi: 10.1074/jbc.M503836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brownsey RW, Boone AN, Elliott JE, Kulpa JE, Lee WM. Regulation of acetyl-CoA carboxylase. Biochem. Soc. Trans. 2006;34:223–7. doi: 10.1042/BST20060223. [DOI] [PubMed] [Google Scholar]
- 34.Leiter AB, Weinberg M, Isohashi F, Utter MF. Relationshiop between phosphorylation and activity of pyruvate dehydrogenase in rat liver mitochondria and the absence of such a relationship for pyruvate carboxylase. J. Biol. Chem. 1978;253:2716–23. [PubMed] [Google Scholar]
- 35.Robinson BH. Lactic acidemia and mitochondrial disease. Mol. Genet. Metab. 2006;89:3–13. doi: 10.1016/j.ymgme.2006.05.015. [DOI] [PubMed] [Google Scholar]