We report the use of hairpin-shaped and circular oligodeoxynucleotides as high-affinity ligands in the binding of homopyrimidine DNA and RNA strands. The complexation involves the use of two connected binding domains to bind a target strand by triple-helix formation. Previous work in our2a–h and others' 3a–d laboratories has shown that homopurine target sequences can be bound between connected pyrimidine domains using the pyr·pur·pyr triplex4–7 motif; however, pyrimidine-rich sequences cannot be complexed in this fashion, thus limiting the number of accessible targets. We now describe an approach in which oligonucleotides are directed to homopyrimidine sequences by triple-helix formation, resulting in high binding affinity. In this strategy (Figure 1), the single-stranded DNA and RNA sequences are bound utilizing a purine-rich ligand in a variation of the pur·pur·pyr motif.8,9 The new strategy expands the range of possible triplex binding sites in single-stranded RNA and DNA to include either purine-rich or pyrimidine-rich targets.
Figure 1.
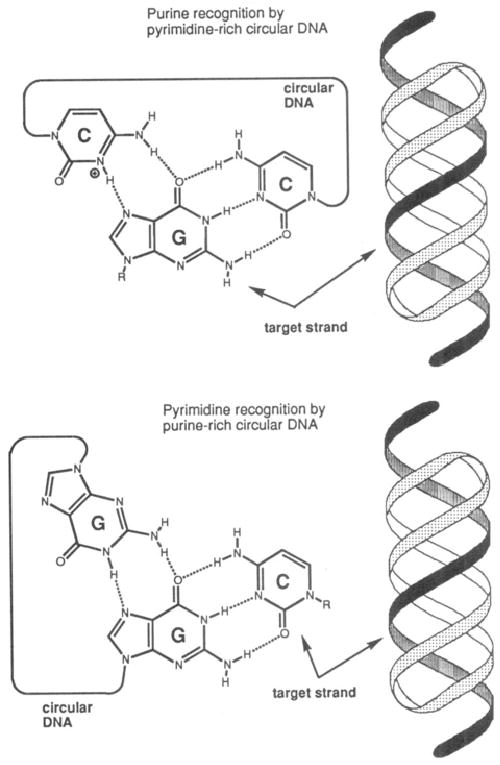
Two strategies for the high-affinity binding of single-stranded sequences by triplex formation. Top: Previously reported strategy2a,c,h for binding purine (A,G)-rich sequences. Bottom: Strategy, described herein, for binding pyrimidine (C,T,U)-rich sequences. Not shown are similar T-A-T(U) triad structures for the binding of A (top) or T,U (bottom). Hairpin-like structures can also be used as ligands in both cases if only one loop connects the binding domains (see Table 1).
Oligodeoxynucleotide ligands 1–3 (see sequences in Table 1) were constructed for the binding of dodecanucleotide sequences containing 12 pyrimidines. Ligands similar to compound 1 were recently reported in a study of H-DNA properties,10 but the binding of RNA was not explored and the affinities with either DNA or RNA were not characterized. Also synthesized for comparison was the linear 12-mer Watson–Crick DNA complement of the pyrimidine target. Oligodeoxynucleotide 1 and circle 2 are designed to bind the 12-mer sequence dCTCCTCCCTCCT (or rCUCCUCCCUCCU); one 12-nucleotide domain is complementary to the target in antiparallel Watson–Crick sense, and the opposing 12-nucleotide domain is complementary to the first domain in antiparallel reverse Hoogsteen sense9 (see Figure 1). All compounds were constructed with pentanucleotide bridging loops of sequence -CACAC-.2c,h
Table 1.
Melting Transition Temperatures (Tm (°C)) and Free Energies (−ΔG° (kcal/mol)) for Complexes of Linear and Circular Purine-Rich DNAs with Complementary Pyrimidine DNA and RNA Single Strands at pH 7.0 (Lines Indicate Watson–Crick Complementarity, Dots Hoogsteen Complementarity, and Arrows 5′ to 3′ Directionalitya
| ligand | complex | Tm(°C)a,b | -ΔG°37b (kcal/mol) |
-ΔG°60b (kcal/mol) |
|---|---|---|---|---|
| DNA target | ||||
| linear 12mer | 56.9 | 14.2 | 8.4 | |
| 1 |  |
68.4 | 16.9 | 11.6 |
| 2 | 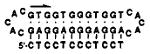 |
71.0 | 17.3 | 12.0 |
| 3 |  |
61.9 | 12.4 | 9.4 |
| RNA target | ||||
| linear 12mer | 58.9 | 15.6 | 8.3 | |
| 1 | 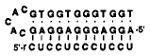 |
65.3 | 14.5 | 10.4 |
| 2 |  |
69.2 | 15.4 | 11.2 |
| 3 |  |
64.3 | 15.3 | 10.3 |
Conditions: 10 mM Mg2+, 100 mM Na+, 10 mM Na·PIPES (pH 7.0), at 4.0 μM total DNA concentration.
Uncertainties in Tm values and in free energies are estimated at ±1.0 °C and ±15%, respectively.
Melting studies of ligands 1 and 2 alone (data not shown) indicate that both compounds have substantial helical structure in solution. Both ligands show weakly cooperative melting transitions, with the same Tm of 57 °C and a hypochromicity of 14% at pH 7.0. Whether this is the result of a single-stranded helix or a Hoogsteen-type duplex structure remains to be determined.
The circular oligomer (2) was constructed to test the effects of covalent closure on binding. Previous studies in the binding of purine targets2c,g have shown that circular compounds can have a substantial binding advantage over linear ones, probably due to the entropic benefit of preorganization.11 In addition, circular oligomers offer the practical advantage of being exceptionally stable in biological media.2e In the current study, the circular oligomer was constructed using a previously described ligation method,2c,g,h which involves the nonenzymatic template-directed closure of end-phosphorylated precursors. The cyclization of 5′-pdGAGGAGCACACGTGGTGGGTGGTCACACAGGAGG to give 2 was carried out using the 12-nucleotide DNA complement dCTCCTCCCTCCT as a template for the ligation. The esterification reaction was carried out in an aqueous buffer containing imidazole hydrochloride, NiCl2, and BrCN. The circularity of the product was confirmed by partial S1 nuclease digestion, which resulted initially in a single band migrating as a linear 34-mer by denaturing gel electrophoresis.
The binding properties of compounds 1–3 were characterized by thermal denaturation and gel titration studies with DNA and RNA target strands monitored at 260 nm. Results of the binding studies with single-stranded DNA (Table 1) show that compounds 1 and 2 do in fact bind the 12-mer homopyrimidine target, and with considerably higher affinity than does a Watson–Crick 12-mer complement. Specifically, oligomer 1 binds dCTCCTCCCTCCT with a Tm of 68.4 °C (conditions: pH 7.0 (10 mM Na·PIPES buffer), 100 mM Na+, 10 mM Mg2+) and a free energy estimated at −16.9 kcal/mol at 37 °C.14 This is in contrast to a simple 12-mer Watson–Crick complement, which binds with a Tm of 56.9 °C and a free energy of −14.2 kcal/mol under identical conditions. The closed circular oligomer 2 binds the target with the highest thermal stability, with a Tm of 71.0 °C and a free energy of −17.3 kcal/mol. This represents an advantage of 2 orders of magnitude in association constant over simple Watson–Crick recognition. A similar advantage is seen in free energies calculated at 60 °C (Table 1). The stoichiometry of the complex of 2 was measured by titration of the target with the ligand, monitored by denaturing gel electrophoresis (data not shown). This confirmed 1:1 stoichiometry as predicted for the expected triple-helical complex. The single-mismatched ligand 3 was also hybridized to the pyrimidine complement. Results show (Table 1) that it binds the DNA target with significantly lower affinity; this confirms the importance of the Hoogsteen strand in increasing the affinity of binding, even though it presumably does not come in direct contact with the target (Figure 1).
Importantly, oligomers 1 and 2 also bind single-stranded RNA with high affinity (Table 1, bottom). Hairpin-shaped compound 1 binds rCUCCUCCCUCCU with a Tm of 65.3 °C, for an advantage of 5.4 °C over the Watson–Crick complement, and circular compound 2 binds with the highest thermal stability, with a Tm advantage of 10.3 °C over simple Watson–Crick binding. Interestingly, the estimated free energies at 37 °C do not reflect this difference; however, more accurate values calculated for 60 °C14 do mirror the advantage seen in the melting temperatures. Somewhat surprisingly, mismatched ligand 3 binds the RNA target as strongly as the fully complementary hairpin; it seems that with RNA as the target the third-strand interaction is less favorable (compare the second Table 1 entries for DNA and RNA), and it may be that a single T–G Hoogsteen mismatch is not significantly destabilizing in this context. In any case, there is a clear binding advantage for hairpin and circular ligands over the simple Watson–Crick complement. It is interesting that both DNA and RNA strands can be strongly bound using the same DNA ligands; studies have recently shown that, in the other (pyr·pur·pyr) triplex motif, stabilities can be highly dependent on DNA versus RNA structure.2h,15 A comprehensive study of these effects has not yet been reported for the pur·pur·pyr motif, although early studies have indicated that in third-strand triplex binding the backbone can have a significant effect.15c,16
As is generally true for pur·pur·pyr-motif triplexes,8,9 the circle complex 2·dCTCCTCCCTCCT is not significantly pH dependent, giving the same Tm, within experimental error, at pH 5.5 as that measured at pH 7.0. The higher affinities of the complexes of 1 and 2 relative to the Watson–Crick complement demonstrate positive contributions from the Hoogsteen domains, and comparison of circular 2 to hairpin-shaped 1 shows the benefit of circularity. While the high-affinity ligand 2 presumably binds directly to the target using only Watson–Crick interactions (Figure 1), it likely uses the Hoogsteen domain to rigidify the ligand, thus gaining an entropic benefit in complexation.
The results presented here clearly demonstrate that high-affinity binding of DNA or RNA sequences containing pyrimidines is readily achievable using this new strategy. Thus, two types of strong circle·single strand triple-helical complexes have now been characterized in the binding of DNA and RNA (Figure 1): the pyr·pur·pyr type (used in targeting purine sites)2,3 and the pur·pur·pyr type (for pyrimidine sites, ref 10 and this work). The present result should very significantly increase the number of sequences which can be targeted by triplex formation.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health and the Office of Naval Research for support of this work.
Footnotes
Supplementary Material Available: Details of synthesis, characterization, and experimental methods (7 pages). This material is contained in many libraries on microfiche, immediately follows this article in the microfilm version of the journal, and can be ordered from the ACS; see any current masthead page for ordering information.
References
- 1.Camille and Henry Dreyfus Teacher-Scholar (1993–1995); Alfred P. Sloan Fellow (1994).
- 2.(a) Kool ET. J Am Chem Soc. 1991;113:6265–6266. [Google Scholar]; (b) Prakash G, Kool ET. J Chem Soc, Chem Commun. 1991:161–1162. doi: 10.1039/C39910001161. 1994, 646. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Prakash G, Kool ET. J Am Chem Soc. 1992;114:3523–3528. doi: 10.1021/ja00035a056. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) D'Souza DJ, Kool ET. J Biomol Struct Dyn. 1992;10:141–152. doi: 10.1080/07391102.1992.10508634. [DOI] [PubMed] [Google Scholar]; (e) Rumney S, Kool ET. Angew Chem. 1992;104:1686–1689. [Google Scholar]; Angew Chem, Int Ed Engl. 1992;31:1617–1619. doi: 10.1002/anie.199216171. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Perkins TA, Goodman JL, Kool ET. J Chem Soc, Chem Commun. 1993:215–217. [Google Scholar]; (g) D'Souza DJ, Kool ET. Bioorg Med Chem Lett. 1994;4:965–970. doi: 10.1016/S0960-894X(01)80664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Wang S, Kool ET. Nucleic Acids Res. 1994;22:2326–2333. doi: 10.1093/nar/22.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Giovannangeli C, Montenay-Garestier T, Rougée M, Chassignol M, Thuong NT, Hélène C. J Am Chem Soc. 1991;113:7775–7776. [Google Scholar]; (b) Salunkhe M, Wu T, Letsinger RL. J Am Chem Soc. 1992;114:8768–8772. [Google Scholar]; (c) Giovannangeli C, Thuong NT, Hélène C. Proc Natl Acad Sci USA. 1993;90:10013–10017. doi: 10.1073/pnas.90.21.10013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gryaznov SM, Lloyd DH. Nucleic Acids Res. 1993;21:5909–5915. doi: 10.1093/nar/21.25.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Hudson RHE, Damha MJ. Nucleic Acids Symp Ser. 1993;29:97–99. [PubMed] [Google Scholar]
- 4.Felsenfeld G, Davies DR, Rich A. J Am Chem Soc. 1957;79:2023–2024. [Google Scholar]
- 5.Moser HE, Dervan PB. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 6.Le Doan T, Perrouault L, Praseuth D, Habhoub N, Decout JL, Thuong NT, Lhomme J, Hélène C. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajagopal P, Feigon J. Nature. 1989;339:637–640. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- 8.(a) Marck C, Thiele D. Nucleic Acids Res. 1978;5:1017–1028. doi: 10.1093/nar/5.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Broitman SL, Im DD, Fresco JR. Proc Natl Acad Sci USA. 1987;84:5120–5124. doi: 10.1073/pnas.84.15.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Letai AG, Palladino MA, Fromm E, Rizzo V, Fresco JR. Biochemistry. 1988;27:9108–9112. doi: 10.1021/bi00426a007. [DOI] [PubMed] [Google Scholar]; (d) Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 9.(a) Beal PA, Dervan PB. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]; (b) Chen FM. Biochemistry. 1991;30:4472–4479. doi: 10.1021/bi00232a014. [DOI] [PubMed] [Google Scholar]; (c) Pilch DS, Levenson C, Shafer RH. Biochemistry. 1991;30:6081–6087. doi: 10.1021/bi00239a001. [DOI] [PubMed] [Google Scholar]; (d) Durland RH, Kessler DJ, Gunnell S, Duvic M, Pettitt BM, Hogan ME. Biochemistry. 1991;30:9246–9255. doi: 10.1021/bi00102a017. [DOI] [PubMed] [Google Scholar]; (e) Beal PA, Dervan PB. Nucleic Acids Res. 1992;20:2773–2776. doi: 10.1093/nar/20.11.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Radhakrishnan I, Patel DJ. J Am Chem Soc. 1993;115:1615–1617. [Google Scholar]; (g) Cherny DI, Malkov VA, Volodin AA, Frank-Kamenetskii MD. J Mol Biol. 1993;230:379–383. doi: 10.1006/jmbi.1993.1154. [DOI] [PubMed] [Google Scholar]; (h) Clarenc JP, Lebleu B, Léonetti JP. Nucleosides Nucleotides. 1994;13:799–809. [Google Scholar]
- 10.Samadashwily GM, Dayn A, Mirkin SM. EMBO J. 1993;12:4975–4983. doi: 10.1002/j.1460-2075.1993.tb06191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cram D. J CHEMTECH. 1987;17:120–125. [Google Scholar]
- 12.Beaucage SL, Caruthers MH. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 13.Horn T, Urdea M. Tetrahedron Lett. 1986;27:4705–4708. [Google Scholar]
- 14.Free energies were determined by curve fitting, see: Petersheim M, Turner DH. Biochemistry. 1982;22:256–263. doi: 10.1021/bi00271a004. Values at 60 °C are more accurate than those at 37 °C because of smaller extrapolation from the Tm values.
- 15.(a) Roberts RW, Crothers DM. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]; (b) Han H, Dervan PB. Proc Natl Acad Sci USA. 1993;90:3806–3810. doi: 10.1073/pnas.90.9.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Escudé C, François JC, Sun J, Ott G, Sprinzl M, Garestier T, Hélène C. Nucleic Acids Res. 1993;21:5547–5553. doi: 10.1093/nar/21.24.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skoog JU, Maher LJ. Nucleic Acids Res. 1993;21:2131–2138. doi: 10.1093/nar/21.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


