Abstract
We report the properties of hydrophobic isosteres of pyrimidines and purines in synthetic DNA duplexes. Phenyl nucleosides 1 and 2 are nonpolar isosteres of the natural thymidine nucleoside, and indole nucleoside 3 is an analog of the complementary purine 2-aminodeoxyadenosine. The nucleosides were incorporated into synthetic oligodeoxynucleotides and were paired against each other and against the natural bases. Thermal denaturation experiments were used to measure the stabilities of the duplexes at neutral pH. It is found that the hydrophobic base analogs are nonselective in pairing with the four natural bases but selective for pairing with each other rather than with the natural bases. For example, compound 2 selectively pairs with itself rather than with A, T, G, or C; the magnitude of this selectivity is found to be 6.5–9.3 °C in Tm or 1.5–1.8 kcal/mol in free energy (25 °C). All possible hydrophobic pairing combinations of 1, 2, and 3 were examined. Results show that the pairing affinity depends on the nature of the pairs and on position in the duplex. The highest affinity pairs are found to be the 1–1 and 2–2 self-pairs and the 1–2 heteropair. The best stabilization occurs when the pairs are placed at the ends of duplexes rather than internally; the internal pairs may be destabilized by imperfect steric mimicry which leads to non-ideal duplex structure. In some cases the hydrophobic pairs are significantly stabilizing to the DNA duplex; for example, when situated at the end of a duplex, the 1–1 pair is more stabilizing than a T–A pair. When situated internally, the affinity of the 1–1 pair is the same as, or slightly better than, the analogous T–T mismatch pair, which is known to have two hydrogen bonds. The studies raise the possibility that hydrogen bonds may not always be required for the formation of stable duplex DNA-like structure. In addition, the results point out the importance of solvation and desolvation in natural base pairing, and lend new support to the idea that hydrogen bonds in DNA may be more important for specificity of pairing than for affinity. Finally, the study raises the possibility of using these or related base pairs to expand the genetic code beyond the natural A–T and G–C pairs.
Introduction
The factors contributing to the thermodynamic stability of the DNA double helix have been the source of scientific inquiry since the original discovery of this structure.1 Early attention centered on the specific hydrogen bonds formed between the bases as a source both of affinity between the strands and specificity of pairing. Over time the importance and complexity of noncovalent bonding in biological systems has become increasingly recognized, and many studies have focused on the interactions which stabilize nucleic acid structure as well as other important biomolecular associations such as protein folding and protein–nucleic acid recognition.2,3
It is now well established that hydrogen bonds between molecules in aqueous solution are relatively weak, due to the high dielectric constant of the medium and to hydrogen bonding competition with the solvent itselfe4 In nucleic acids, studies in RNA duplexes5 and hairpins6 place the value of individual hydrogen bonds in water at ~0.4–1.3 kcal, although the value is considerably context dependent.6
It is also clear that base pairing in nucleic acids is the result of more than just hydrogen bonding interactions. Base pairing stability is dependent on nearest neighbor interactions,7 and it is now evident that base stacking–the interaction between the flat aromatic π systems of the bases–is a very important factor in stabilizing secondary structure.8 One experimental value, measured for RNA, estimates that base stacking and hydrogen bonding each contribute about 1 kcal of free energy to the stability of a base paire9 Very few such studies have been carried out in DNA.10 While studies of simple model systems have addressed what are the important contributing factors in aqueous π–π stacking,11–13 there are few experimental studies which examine these contributions in the context of the DNA structure.14 Theoretical studies have examined several factors, including solvophobic effects,15 van der Waals’ forces,16 and electrostatic effects,15–17 as potentially attractive forces involved in this interaction. The understanding of the contributing factors in DNA base pairing is important from at least two standpoints. First, it is such a basic part of biological function that its importance is almost implicit. Second, such knowledge can aid in the design of new biologically functional structures which may be useful in biochemical study and potentially as therapeutic agents as well.18
In order to generate new data to test the relative importance of hydrogen bonding and base stacking in the structure, stability, and functions of DNA, we have designed nonpolar isosteric analogs19 (1–3) of the natural DNA nucleosides. We have chosen substituted benzenes as close steric analogs of pyrimidines, and indoles as purine analogs. In this paper we describe the incorporation of these new nucleosides into synthetic oligodeoxynucleotides, and we measure their base pairing properties when placed opposite the natural DNA bases as well as each other. Some of the nonnatural pairs are found to be highly stabilizing to DNA duplexes. In addition, it is found that the hydrophobic nucleosides are selective in pairing with each other rather than with the natural nucleosides. The findings stress the importance of solvation and desolvation in DNA base pairing.
Experimental Section
General Synthetic Methods
1H and 13C NMR spectra were obtained on a 300 MHz instrument and are reported on the parts per million scale; J values are reported in hertz. Mass spectral analyses were performed by the Midwest Center for Mass Spectrometry with partial support by the National Science Foundation, Biology Division (Grant No. DIR9017262), or by the University of California–Riverside Mass Spectrometry Facility. Pyridine was dried by distillation from barium oxide. Methylene chloride and triethylamine were distilled from calcium hydride. 4,4′-Dimethoxytrityl chloride and 2-cyanoethyl N,N- diisopropylchlorophosphoramidite were purchased from Aldrich. Long chain alkyl amino controlled pore glass (LCAA CPG) was purchased from CPG Inc. (Fairfield, NJ). All other solvents and chemicals were purchased from Aldrich, Fisher, J. T. Baker, or Sigma and were used without further purification. The derivatization of the controlled pore glass and the calculation of the loading were performed according to the published procedure.20
1′,2′-Dideoxy-1′-(2,4-difluorotolyl)-5′-O-(4,4′-dimethoxytrityl)-β-D-ribofuranose (1b)
Nucleoside 1a (280 mg, 1.15 mmol) (synthesis described previously)19 was co-evaporated twice with dry pyridine and then dissolved in 5 mL of dry pyridine. To this was added triethylamine (120 mg, 1.15 mmol) and 4,4′-dimethoxytrityl chloride (544 mg, 1.60 mmol). The mixture was stirred at room temperature under an atmosphere of nitrogen for 4 h. Methanol (1 mL) was added to quench any remaining 4,4′-dimethoxytrityl chloride and the reaction mixture was stirred for an additional 30 min. The mixture was then poured into saturated sodium bicarbonate and extracted with methylene chloride. The organic layers were washed with brine and dried over anhydrous sodium sulfate. The solution was filtered, concentrated, and purified by silica gel column chromatography, eluting with ethyl acetate–hexanes (20:80 plus 5% triethylamine). The product was obtained as a yellow foam in 52% yield (330 mg, 0.60 mmol): 1H NMR (CDCl3, ppm) 7.47 (1H, d, J = 8, ArH (trityl)), 7.37–7.21 (8H, m, ArH (trityl)), 6.85 (5H, d, J = 6, ArH (trityl)), 6.74 (1H, t, ArH), 5.32 (1H, t, J = 9, H1′), 4.43 (1H, br s, H3′), 4.21 (1H, br s, H4′), 3.80 (6H, s, OCH3), 3.41–3.23 (2H, m, H5′, 5″), 2.80–2.71 (1H, m, H2′), 2.25 (3H, s, CH3), 1.98–1.94 (1H, m, 2″); 13C (CDCl3, ppm) 14.0, 42.2, 55.2, 64.6, 73.8, 74.8, 84.6, 103.1 (t), 113.2, 120.8 (d), 125.7 (d), 126.8, 127.9, 128.1, 128.9, 129.0, 129.1, 130.0, 135.9, 144.8, 158.1, 159.5 (dd); HRMS (FAB+, 3-NBA matrix) mass calcd for C33H32F2O5 546.2218, found 546.2200.
1′,2′-Dideoxy-5′-O-(4,4′-dimethoxytrityl)-1′-(1,2,4-trimethylphenyl)-β-D-ribofuranose (2b)
The nucleoside 2a (described previously)19 was tritylated in a manner similar to that described above. The product was obtained in 56% yield as an off-white foam after purification by silica gel chromatography eluting with ethyl acetate–hexanes (40:60 plus 5% triethylamine): 1H (CDCl3, ppm) 7.48 (2H, d, J = 8, ArH (trityl)), 7.37 (6H, d, J = 8, ArH (trityl)), 7.32, (1H, s, H6), 7.25 (1H, d, J = 8, ArH (trityl)), 6.93 (1H, s, H3), 6.86 (4H, d, J = 8, ArH (trityl)), 5.33 (1H, t, J = 6, H1′), 4.47 (1H, br s, H3′), 4.27 (1H, br s, H4′), 3.82 (6H, s, OCH3), 3.44–3.26 (2H, m, H5′, 5″), 2.78–2.69 (1H, m, H2′), 2.27 (3H, s, CH3), 2.24 (3H, s, CH3), 2.23 (3H, s, CH3), 1.98–1.89 (1H, br s, H2″); 13C (CDCl3, ppm) 18.4, 19.0, 19.2, 42.0, 54.9, 64.7, 74.9, 76.8, 84.1, 112.9, 125.6, 131.2, 131.5, 134.0, 134.9, 138.1, 144.7, 158.3; HRMS (FAB+, 3-NBA matrix) mass calcd for C35H37O5 537.2641, found 537.2648.
1′,2′-Dideoxy-5′-O-(4,4′-dimethoxytrityl)-1′-(4,6-dimethylindoIyl)-β-D-ribofuranose (3b)
The nucleoside 3a (synthesis described previously)19 was tritylated using the method described above. The product was obtained in 79% yield after purification by silica gel chromatography eluting with ethyl acetate–hexanes (20:80 plus 5% triethylamine): 1H NMR (CDCl3, ppm) 7.46–7.26 (9H, m, ArH (trityl)), 7.17 (1H, d, J = 4, H2), 7.14 (1H, s, H7), 6.83 (1H, s, H5). 6.80 (4H, s, ArH (trityl)), 6.49 (1H, d, J = 4, H3), 6.40 (1H, t, J = 6, H1′), 4.62 (1H, br s, H3′). 4.10 (1H, br s, H4′), 3.34–3.29 (2H, m, H5′, 5″), 2.69–2.58 (1H, m, H2′), 2.51 (3H, s, CH3), 2.42 (3H, s, CH3), 2.44–2.37 (1H, m, H2″); 13C (CDCl3, ppm) 18.3, 21.5, 39.6, 54.9, 63.9, 72.8, 84.4.86.2, 101.1, 107.2, 112.9, 121.9, 122.4, 126.6, 127.6, 127.9, 129.8, 131.7, 135.5, 144.4, 158.2; HRMS (FAB+, 3-NBA matrix) calcd for C36H37NO5 563.2672, found 563.2658.
1′,2′-Dideoxy-1′-(2,4-difluorotolyl)-5′-O-(4,4′-dimethoxytrityl)-β-D-ribofuranose Cyanoethyl N,N-Diisopropylphosphoramidite(1C)
The tritylated compound lb (284 mg, 0.52 mmol) was dissolved in 4 mL of dry methylene chloride and to this was added diisopropylethylamine (0.36 mL, 2.1 mmol) and 2-cyanoethyl N,N-diisopropylchlorophosphoramidite (0.17 mL, 0.78 mmol). The reaction mixture was stirred for 2 h under an atmosphere of nitrogen. Ethyl acetate was added (10 mL) and the solution washed with saturated sodium bicarbonate and brine. The organic layer was dried over anhydrous sodium sulfate. The product was purified by silica gel column chromatography, eluting with ethyl acetate–hexanes (50:50 plus 5% triethylamine). The product was obtained as an off-white foam in 72% yield (320 mg, 0.43 mmol) as a mixture of diastereomers: 1H NMR (CDCl3, ppm) 7.51 (1H, d, ArH), 7.43–7.22 (9H, m, ArH (trityl)), 6.88–6.82 (4H, m, ArH (trityl)), 6.74 (1H, t, J = 10, ArH), 5.43 (1H, t, J = 6, H1′), 4.57 (1H, br s, H3′), 4.38 (1H, br s, H4′), 3.45–3.66 (3H, m, –OCH2, –NCH–), 3.38–3.17 (2H, m, H5′, 5″), 2.79–2.70 (1H, m, H2′), 2.25 (3H, s, CH3), 1.14–1.02 (12 H, m, –CH(CH3)2; 31P (CDCl3, ppm) 148.6, 149.1; HRMS (FAB+, 3-NBA matrix) calcd for C42H49F2N2O6Na 769.3194, found 769.3209.
1′,2′-Dideoxy-5′-O-(4,4′-dimethoxytrityl)-1′-(1,2,4-trimethylphenyl)-β-D-ribofuranose Cyanoethyl N,N-Diisopropylphosphoramidite (2c)
The phosphoramidite of compound 2b was obtained in a manner similar to that described above. The product 2c was obtained as a mixture of diastereomers in 88% yield as an off-white foam after purification by silica gel chromatography, eluting with ethyl acetate–hexanes (35:65 plus 5% triethylamine): 1H NMR (CDCl3, ppm) 7.35–7.23 (10H, m, ArH + H6), 6.94 (1H, s, H3), 6.87–6.82 (4H, m, ArH (trityl)), 5.42 (1H, t, J = 6, H1′), 4.71–4.60 (1H, m, H3′), 4.38 (1H, br s, H4′), 3.82 (6H, s, OCH3), 3.63–3.53 (2H, m, OCH2–), 3.57–3.18 (4H, m, H5′, 5″), 2.79–2.68 (1H, m, H2′), 2.48–2.39 (4H, m, –CH2CN), 2.29 (6H, s, CH3), 2.25 (3H, s, CH3), 2.06–1.97 (1H, m, H2″), 1.18–1.02 (12H, m, CH3); 31P (CDCl3, ppm) 148.4, 148.9; HRMS (FAB+, 3-NBA matrix) (M + H)+ calcd for C44H55N2O6P 739.3876, found 739.3870.
1′,2′-Dideoxy-5′-O-(4,4′-dimethoxytrityl)-1′-(4,6-dimethylindolyl)-β-D-ribofuranose Cyanoethyl N,N-Diisopropylphosphoramidite(3c)
The synthesis of 3c was performed in a manner similar to compounds 1C and 2c above. The product was obtained as two separate diastereomers in 83% total yield as an off-white foam after purification by column chromatography, eluting with ethyl acetate–hexanes (35:65 plus 5% triethylamine): 1H NMR (CDCl3, ppm) 7.47–7.25 (9H, m, ArH (trityl)), 7.24 (1H, d, J = 4, H2), 7.18 (1H, s, H7), 6.80 (4H, br s, ArH (trityl)), 6.78 (1H, br s, H5), 6.50 (1H, d, J = 4, H3), 6.40 (1H, t, J = 6, H1′), 4.74 (1H, br s, H3′), 4.27 (1H, br s, H4′), 3.80 (3H, s, OCH3), 3.76–3.63 (4H, m, –OCH2, –NCH–), 3.41–3.25 (2H, m, H5′), 2.71–2.55 (1H, m, H2′), 2.52 (3H, s, CH3), 2.49 (2H, t, J = 6, –CH2CN (2.64, t, J = 6, other diastereomer)), 2.41 (3H, s, CH3), 1.21 (12H, t, J = 8, –CH(CH3)2 (1.12, d, J = 8, and 1.21, d, J = 8, other diastereomer)); 31P (CDCl3, ppm) 149.0, 149.4; HRMS (FAE+, 3-NBA matrix) calcd for C45H54N3O6P 763.3750, found 763.3748.
1′-(2,4-Difluorotolyl)-5′-O-(4,4′-dimethoxytrityl)-3′-O-succinyl-β-D-ribofuranose (4)
Compound lb was dissolved in dry pyridine and to this was added 4-(dimethylamino)pyridine (15 mg, 0.12 mmol) and succinic anhydride (24 mg, 0.24 mmol). The reaction was allowed to stir overnight under an atmosphere of nitrogen. The solvent was evaporated and the residual pyridine removed by co-evaporation with toluene. The residue was dissolved in methylene chloride and washed with 10% citric acid and water then dried over anhydrous sodium sulfate. The product was purified by silica gel column chromatography, eluting initially with CH2Cl2, then CH2C12–MeOH (95:5). The product was obtained in 79% yield (140 mg, 0.21 mmol); 1H NMR (CDCl3, ppm) 7.46, (2H, d, J =9, ArH (trityl)), 7.41–7.20 (8H, m, ArH (trityl) + H6), 6.84 (4H, d, J = 9, ArH (trityl)), 6.72 (1H, t, J =9, H3), 5.50 (1H, t, J = 9, H1′), 5.37 (1H, s, H3′), 4.43 (1H, br s, H4′), 3.80 (6H, s, OCH3), 3.33–3.26 (2H, m, H5′, 5″), 2.47 (4H, br s, –COCH2CH2– CO–), 2.26 (3H, s, CH3), 2.01–1.93 (1H, m, H2″) (H2′ signal obscured by (CH3).
1′-(2,4-Difluorotolyl)-5′-O-(4,4′-dimethoxytrityl)-3′-(p-nitrophenylsuccinyl)-β-D-ribofuranose Controlled Pore Glass (CPG)
Succinic ester (4) (130 mg, 0.29 mmol) was dissolved in anhydrous dioxane containing several drops of pyridine. To this was added p-nitrophenol (30 mg, 0.21 mmol) and dicyclohexylcarbodiimide. The reaction was stirred for 2.5 h. The reaction was filtered and to the supernatant was added long chain alkyl amino controlled pore glass (LCAA CPG) (1.0 g) and several drops of triethylamine. The reaction was allowed to stand overnight and was then filtered and the CPG was washed with DMF, methanol, and ether and then air-dried. Any remaining free amines on the support were capped by treating the CPG with acetic anhydride (0.2 mL) and (dimethylamino)pyridine (10 mg) in pyridine for 30 min. The support was then filtered and washed with methanol and ether, then air and vacuum dried.
Oligonucleotide Synthesis
DNA oligonucleotides were synthesized on an Applied Biosystems 392 synthesizer using standard β-cyanoethyl phosphoramidite chemistry.21 Nonnatural nucleoside phosphoramidites were incorporated using the standard ABI coupling cycle; stepwise coupling yields for the nonnatural residues were all greater than 95% as determined by trityl cation monitoring. Oligomers containing nucleoside 1 at the 3′ end were prepared using a controlled pore glass derivatized with that tritylated nucleoside (see above). All oligonucleotides were deprotected in concentrated NH4OH (55 °C, 12 h), purified by preparative 20% denaturing polyacrylamide gel electrophoresis, isolated by the crush and soak method, and quantitated by absorbance at 260 nm. Molar extinction coefficients were calculated by the nearest neighbor method.22 Values for oligonucleotides containing nonnatural residues were obtained in the following way: The extinction coefficient for each of the new nucleosides was measured at 260 nm. The molar extinction coefficients for 1–3 were found to be 1200, 851, and 6362, respectively. The individual extinction coefficients for all the bases in a given oligomer were summed and compared to the sum from the corresponding sequence in which the nonnatural residues were replaced by T. The ratio of the two was used to scale downward the molar extinction coefficient of the T-containing sequence derived from nearest neighbor parameters. Since in most cases the content of nonnatural residues in the sequences is low, this estimation method is unlikely to generate large errors in concentration. Oligodeoxynucleotides were obtained after purification as the sodium salt. Intact incorporation of residues 1–3 was confirmed by synthesis of short oligomers of sequence T–X–T (where X = 1, 2, or 3); the aromatic region of the proton NMR shows the presence of the intact phenyl or indole structures with the expected integration relative to the C-6 protons of the thymines. Analysis by enzymatic digestion with snake venom phosphodiesterase and bacterial alkaline phosphatase was not possible, because the nonnatural residues inhibit the phosphodiesterase cleavage. The intact structure of the indole nucleoside (3) in a short oligonucleotide was separately confirmed by negative ion FAB-MS, which gave a strong parent ion at mlz 868 (M + H).
Thermal Denaturation Studies
Solutions for the thermal denaturation studies contained a one-to-one ratio of two complementary oligomers. Concentrations for given experiments are listed in the text and figure legends. Buffers used were 1 M Na+, 10 mM Na · phosphate; or 100 mM Na+, 10 mM Mg2+, 10 mM Na. · PIPES (1,4-piperazinebis(ethanesulfonate)) buffer23 (Sigma). For the self-complementary duplexes we used the buffer conditions of Breslauer.10 The buffer pH with the PIPES system is that of a 2 x stock solution at 25 °C containing the buffer and salts. After the solutions were prepared they were heated to 90 °C and allowed to cool slowly to room temperature prior to the melting experiments.
The melting studies were carried out in Teflon-stoppered 1 cm path length quartz cells under nitrogen atmosphere on a Varian Cary 1 UV– vis spectophotometer equipped with a thermoprogrammer. Absorbance was monitored while temperature was raised from 5 to 80 °C at a rate of 0.5 °C/min; a slower heating rate with this apparatus does not affect the results. The A,T-rich sequences were monitored at 260 nm, and the C,G-rich sequences were followed at 280 nm. In all cases the complexes displayed sharp, apparently two-state transitions, with all-or-none melting from bound complex to free oligomers. Similar results were obtained with heating or cooling cycles. Melting temperatures (Tm) were determined by computer fit of the melting data; the first derivative of the fit is then calculated using 1/T, and the Tm values are reported at 0.971 of the maximum of the first derivative. Uncertainty in Tm is estimated at ±0.5 °C based on repetitions of experiments.
Free energy values were derived by computer-fitting the denaturation data with an algorithm employing linear sloping baselines, using the two-state approximation for melting.9 Fits were excellent, with X2 values of 10−6 or better. Van’t Hoff thermodynamic parameters were previously derived for both core sequences in this study24,10,25 by measuring Tm as a function of concentration (1/Tm vs ln(CT)); in both cases close agreement was seen with the results from curve-fitting, indicating that the two-state approximation is a reasonable one for these specific sequences. Uncertainty in individual free energy measurements is estimated at ±5–10.
Results
Design of Nucleosides
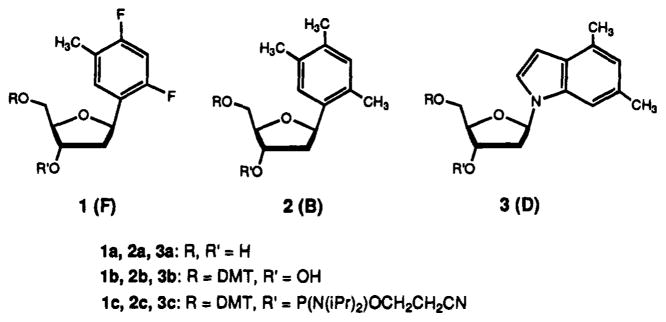
The aromatic rings of nucleosides 1–3 were designed to be the closest possible steric mimics of the natural analogs, without using hydrophilic oxygen or nitrogen-containing groups. Although they are not basic, we refer to these aromatic groups as “bases” in analogy to the natural pyrimidines and purines. Their design, synthesis, and structures are discussed in detail elsewhere.19 Compounds 1 and 2 are thymidine (T) analogs (Figure 1). We considered the best isosteric replacement for C=O functionality of thymine to be the C–F group, since these groups have nearly identical bond lengths.21 The other isosteric replacements in 1 are C–H for N–H, and replacement of N1 with an sp2 carbon. In addition to these groups being steric mimics, they are also isoelectronic with the natural structures. Semiempirical AM1 calculations indicate that 1 and thymine are stericall almost indistinguishable, with all analogous bonds within 0.1 Å of each other in length.19 In fact, compound 1 and T contain the same number of atoms, and differ only by 2 atomic mass units. Since the C–F bonds are undoubtedly polarized, compound 1 is likely to have partial atomic charges similar in sign to those for T, but considerably smaller in magnitude. Compound 2 is an even less polarized version of 1, in which the fluorines have been replaced by methyl groups. Since the methyl groups are larger than the oxygen groups they replace in T, compound 2 is a less exact isostere than compound 1 (see Figure 2).
Figure 1.
The structures of hydrophobic nucleosides 1–3. The nucleoside “bases” are abbreviated F (difluorotoluene), B (trimethylbenzene), and D (dimethylindole).
Figure 2.
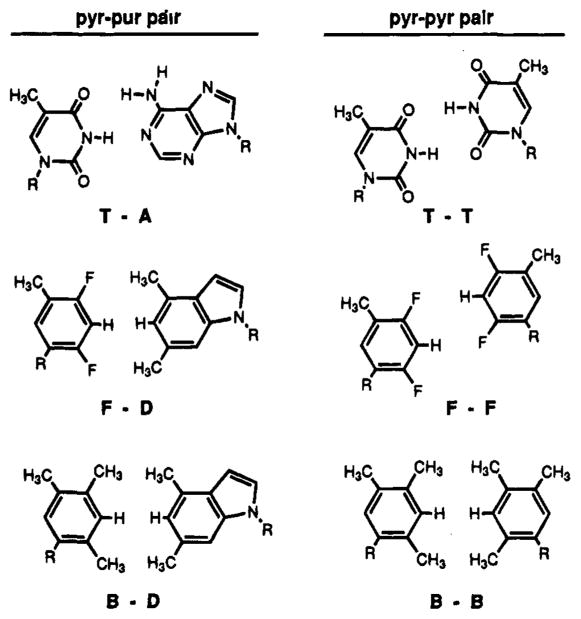
The structures of the T–A and T–T pairs in DNA and proposed structures for the analogous hydrophobic purine-pyrimidine and pyrimidine–pyrimidine pairs.
Indole nucleoside 3 (Figure 1) is a hydrophobic analog of 2-aminodeoxyadenosine, which forms a full 3-hydrogen-bond complementary pair with T. The isosteric replacements made are –CH3 groups for the two exocyclic –NH2 groups; in addition, three C–H groups replace the ring nitrogens N1, N3, and N7. The protons of the C–H groups are expected to be somewhat larger sterically than the nitrogen lone pairs they replace.
Very few reports of similar molecules exist in the literature. Two reports of substituted phenyl-ribosides have been published, but these structures were not incorporated into DNA strand.27,28 There is one report of a thiomethylindole nucleoside, designed for cross-linking purposes, which was incorporated into an oligonucleotide.29 Nitropyrrole- and nitroindole-based nucleosides designed to serve as “universal” pairing compounds were also recently reported.30 In one earlier study by Millican et al.,31 a phenyl-β-deoxy-D-riboside was incorporated into oligonucleotides and paired against the natural bases; in that study, weak pairing was observed; the result was attributed to poor base stacking.31 No previous reports exist, to our knowledge, on the use of hydrophobic nucleoside isosteres in nucleic acids or on the possibility of hydrophobic pairing in DNA.
Hydrogen Bonding Ability
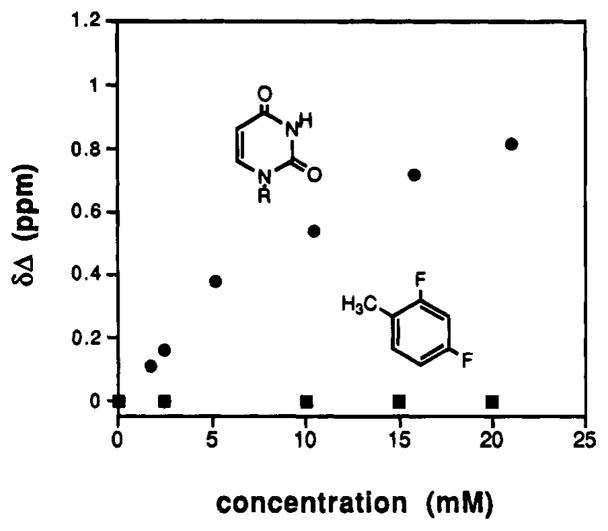
Since there have been reports of C–F groups acting as weak hydrogen bond acceptors32 and aromatic C–H groups as weak hydrogen bond donors,33 it is conceivable that 1 could undergo hydrogen bonding to natural adenine analogs. To explore this possibility, we examined by 1H-NMR the ability of varying concentrations of difluorotoluene (DFT, the “base” of 1) to associate with 9-ethyladenine (EA) in CDCl3 (Figure 3). Results show that there is no significant chemical shift of EA in the presence of DFT at the concentrations studied, whereas a natural T analog, cyclohexyluracil, shows a clear binding curve. Thus, while we cannot rule out very weak interactions, it is likely that 1 will not undergo significant H-bonding with adenine analogs in DNA. This is especially true in the higher-dielectric aqueous medium of the pairing experiments (see below).
Figure 3.
Proton NMR titration of 9-ethyladenine with l-cyclohexy- luracil (●)and with nucleoside l a (■). Experiments were carried out in CDCl3 at 25 °C with 1 mM 9-ethyladenine, following the N4–H resonance on 9-ethyladenine.
Design and Synthesis of Oligodeoxynucleotides
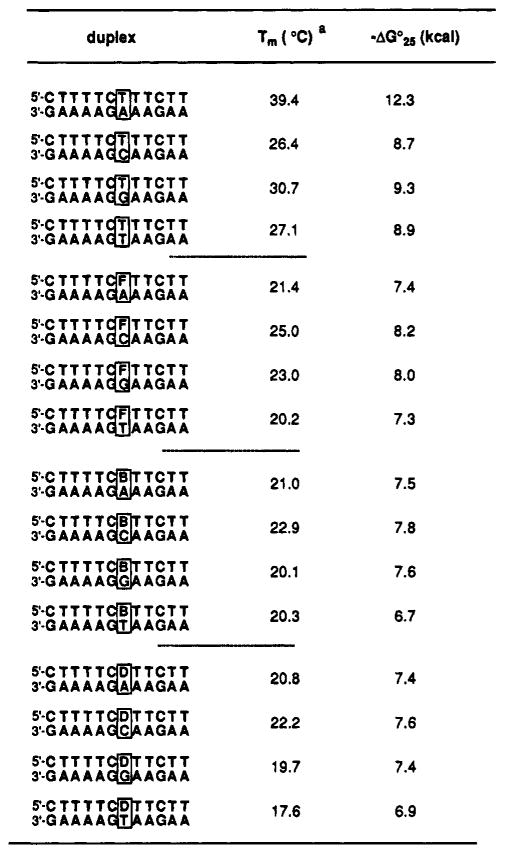
The hydrophobic nucleosides were tested in the context of two different DNA sequences. One is a 12-base pair heteroduplex which is A, T- rich.25 The second is a self-complementary 6-base pair duplex which is C,G-rich.10,26 Both parent duplexes have been studied previously;25,10,26 we used buffer conditions taken from those reports. In the 12-bp duplex series we replaced a central T base or T–A base pair with hydrophobic isosteres 1–3 (Tables 1 and 2). In two experiments we added synthetic nucleosides (at the ends or intemally) while keeping the natural 12-base pairs intact (Table 2, bottom). In the 6-bp self- complementary series we conserved the six C–G base pairs of the core duplex and added one or two additional pairs at the ends or in the center (Table 3) for comparison.
Table 1.
Free Energies and Melting Temperatures for Dodecamer Duplexes Containing a Variable T–X, F–X, B–X, or D–X Base Pair (X = A, T, C, G)
 |
Conditions: 100 mM NaCl, 10 mM MgCl2, 10 mM Na · PIPES, pH 7.0, 1.6 μM each strand.
Table 2.
Stabilities of Hydrophobic Base Pairs. As Measured by Melting Temperatures(Tm (°C)) and Free Energies (−ΔG°25(kcal)) for Duplexes Containing Nucleotides F (1), B(2), or D (3)
 |
Conditions: 100 mM NaCl, 10 mM MgCl2, 10 mM Na. PIPES. pH 7.0,1.5 μM each strand.
Table 3.
Stabilities of Terminal and lnternal Hydrophobic Base Pairs. As Measured hy Free Energies and Melting Temperatures for Duplexes from Self-Complementary Strands Containing Nucleotides F (1). B (2), or D (3)
| duplex | Tm (°C)a | − Δ°25 (kcal) | |
|---|---|---|---|
| core duplex | 5′-CGCGCG | 41.1 | 9.6 |
| 3′-GCGCGC | |||
| external pairs | 5′-TCGCGCGA | 51.1 | 11.8 |
| 3′-AGCGCGCT | |||
| 5′-TCGCGCGT | 52.6 | 12.4 | |
| 3′-TGCGCGCT | |||
| 5′-FCGCGCGF | 62.8 | 13.4 | |
| 3′-FGCGCGCF | |||
| internal pairs | 5′-GCATGCG | 50.0 | 10.9 |
| 3′-GCGTACGC | |||
| 5′-CGCTAGCG | 46.8 | 10.1 | |
| 3′-GCGATCGC | |||
| 5′-CGCTTGCG | 58.9 | --b | |
| 3′-GCGTTCGC | |||
| 5′-CGCTCGCG | 52.4b | -b | |
| 3′-GCGCTCGC | |||
| 5′-CGCFGCG | 33.4 | 8.3 | |
| 3′-GCGFCGC | |||
| 5′-CGCFFGCG | 33.6 | 8.5 | |
| 3′-GCGFFCGC | |||
| 5′-CGCDBGCG | 33.6 | 8.4 | |
| 3′-GCGBDCGC | |||
| 5′-CGCBDGCG | 31.7 | 8.2 | |
| 3′-OCGDBCGC | |||
| 3′-CGCDFGCG | 30.6 | 8.1 | |
| 3′-GCGFDCGC |
Conditions: 1 M NaCl, 10 mM Na phosphate, pH 7.0,6.0 μM strand concentration.
Predominant structure is a concentration-independent unimolecular hairpin.
The synthesis of phosphoramidites of 1–3 from the nucleosides and their incorporation into oligodeoxynucleotides were straightforward. Tritylation of the 5′-OH and subsequent phosphitylation at the 3′-position (Figure 1) proceeded in nonnal fashion. All three are stable compounds and do not have protecting groups to complicate the oligonucleotide synthesis or deprotection, and so standard deprotection and purification procedures were sufficient. All were incorporated in high yield (>95% stepwise yield), and we confirmed their intact incorporation by proton NMR studies of short model oligomers containing 1, 2, or 3. HPLC analysis of nucleosides from enzymatically digested oligomers was not possible; our studies showed that snake venom phosphodiesterase cleaves oligomers only as far as the nonnatural residues, indicating that 1, 2, and 3 in DNA all inhibit this nuclease. The phenyl base analogs are expected to be highly stable compounds; however, since indoles are somewhat more reactive, we also confirmed the intact incorporation of 3 in a short oligonucleotide by FAB mass spectral analysis, which showed a single parent peak of the expected mass.
Pairing of 1–3 with the Natural Bases
We first examined the properties of compounds 1–3 when paired with the four natural bases in the center of a 12-base pair duplex (Table I and Figure 4). The duplex association affinities were measured by thermal denaturation experiments; monitoring the mixtures at 260 nm allowed determination of melting temperature (Tm) values (conditions: 100 mM NaCl, 10 mM MgCl2, 10 mM Na PIPES, pH 7.0). Free energies for the complexes at 25 °C were estimated by a nonlinear least-squares curve-fitting algorithm with linear sloping baselines.9 All 16 complexes showed well-behaved, apparently two-state melting. It has been previously shown that the two-state model fits the behavior of this parent duplex reasonably well.25
Figure 4.
The pairing of hydrophobic nucleosides 1–3 with the natural bases and with themselves in the center of a 12-base pair duplex, as measured by thermal melting temperature (see Table 1 for conditions).
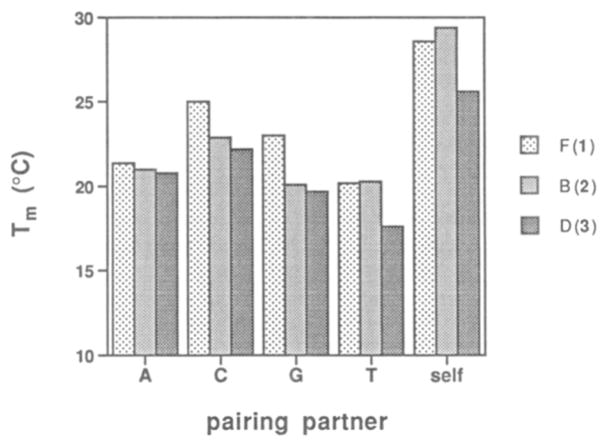
Table 1 shows the data for all possible pairs of 1–3 (F. B, D, respectively) with the four natural bases. For comparison, we also tested the pairing properties of the natural base T under identical conditions. Results show that T, as expected34,25 (first four entries in Table 1), pairs with A with a selectivity of 9–13°C in Tm and 3.0–3.6 kcal/lmol in free energy. The correctly paired (T–A) duplex has a free energy (25 °C) of −12.3 kcal/mol under these conditions. Interestingly. the close isostere F has very different behavior (second set of entries in Table 1). All four duplexes containing this base are considerably weaker than the fully paired T–A duplex, with Tm values 14–19 °C lower and free energies 4–5 kcal/mol less favorable. This is even true for the isosteric F–A pair, which. although presumably able to adopt a standard T–A base pair geometry, is actually one of the weaker pairs of the four. Comparison among these four shows little overall difference between them, with a Tm range of only 4.8 °C and in free energy, 0.7 kcal/mol.
Examination of the last two series in Table 1, involving B–X and D–X pairs (where X = A. T, C, G). reveals very similar behavior to that seen for the F nucleoside. The overall duplex binding affinities are again both low and relatively nonselective among the four natural pairing partners.
Overall comparison of the three hydrophobic compounds paired with natural bases (Figure 4) shows remarkably parallel behavior. On average, the duplexes involving pairs of nucleoside F are stronger, with Tm values ~2 °C higher than for the other two. The duplexes containing the B and D nucleosides have affinities similar to each other. with the B cases perhaps having a slight advantage. The three nucleosides show about the same order of base pairing preferences for the natural bases. although this preference is small. This order is C > T > A > G. The natural T base has a considerably different order of preference. namely. A > G > T > C. and the natural A base is of course expected to have a pairing preference for T. The F and B nucleosides are T analogs. while the D nucleoside is an analog of A; nonetheless. results show that all three of these analogs have similar pairing properties with the natural bases.
Hydrophobic Pairing of 1–3 in Heteroduplexes
After examining the pairing affinities of compounds 1–3 with the natural bases. we then tested their behavior when paired with each other. We tested all 9 possible combinations of F. B, and D in the 12-base pair duplex context. Table 2 compiles the Tm and free energy data for these cases, and Figure 4 shows the self-paired cases compared to the cases paired with A. C. T. and G, described above.
Comparison of the self-paired duplexes (having pairs F–F. B–B. D–D) with the previous data (involving pairs with A, T, G, C) shows a striking preference of the hydrophobic bases for themselves over the natural bases (see Figure 4). This selectivity in Tm values is 4–8 °C for the F nucleoside. 6–9 °C for the B nucleoside. and 3–8 °C for the D nucleoside. In free energy terms. the selectivities for self-pairing are 1–2kcal/mol (Table 2), corresponding to a difference in association constant of up to 20-fold at 25 °C.
Interestingly. while the F, B, and D nucleosides are selective for hydrophobic over hydrophilic partners, they are relatively nonselective among each other (see Table 2). The overall range of Tm values seen for the nine cases is only 4.5 °C. and the free energy range is 1.1 kcal/mol. The top three pairs in stability are the F–B, B–B, and F–F cases; the least stable cases are found to be the five cases involving the D nucleoside.
We also examined two additional duplexes. in which four F–F pairs were placed internally or at the ends of the core 12- base pair duplex (see Table 2, bottom). This parent duplex has a Tm of 39.4 °C and a free energy of −12.3 kcal/mol. Interestingly. it is found that the specific placement of these four pairs makes a large difference in overall stability. Internal placement in the center of the sequence slightly lowers its affinity relative to the unmodified duplex. with a drop in Tm of 1.7 °C and a loss in free energy of 0.8 kcal/mol. When the four F–F pairs are placed at the ends of the duplex, however. a large stabilization is seen. with an increase in Tm of 11 °C, and a 2.6 kcal/mol more favorable association energy.
Hydrophohic Pairing in Self-complementary Duplexes
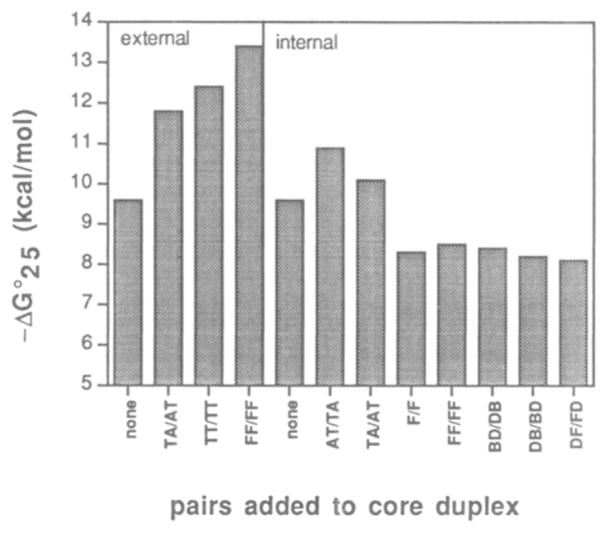
We then examined several hydrophobic pairs in the context of a second sequence. which contains the core duplex arising from the self-complementary d(CG), sequence.10 Single or double pairs were added either in the center or at the ends of the duplex. and we tested the natural T–A or T–T pairs in the same context for comparison of affinities. The conditions used were those reported by Breslauer.10 with a buffer containing 1 M NaCl and 10 mM phosphate at pH 7.0.
The results are given in Table 3. and are compared graphically in Figure 5. In general. the data are in accord with those seen in the 12-base pair heteroduplex context. The hydrophobic pairs examined are the F–F, D–B. and D–F pairs. As seen for the previous cases. the hydrophobic pairs are considerably more stabilizing when placed at the ends of the core duplex than when in the center. Addition of the F–F pair in the center of the 6-bp core decreases stability by 8 °C in Tm or 1.3 kcal/mol of free energy. Significantly. addition of a second F–F pair results in essentially no change. This is consistent with the small effect seen for insertion of four consecutive F–F pairs in the center of the heteroduplex (Table 2).
Figure 5.
Pairing of hydrophobic nucleosides 1–3 with themselves and each other in self-complementary duplex, as compared to natural T–A and T–T pairs. All duplexes contain the same core sequence d(CG)3, and are substituted externally or internally with the pairs shown See Table 3 for conditions and complete sequences.
For comparison. we tested the effects of inserting natural double mismatches. TT/TT and TC/CT. at the center of the d(CG)3 core sequence. Results show that these two cases give melting temperatures which are independent of concentration over the range 5–20 mM; this implies a unimolecular hairpin structure for both (presumably with loops of T–T or T–C. respectively). The hydrophobic double insertions did not show this behavior. As a result, we cannot compare these mismatches to the hydrophobic pairs. Also for comparison. we tested two canonical base pairs. TA/AT and AT/TA. in the same position (see Table 3); we find that the double pairs stabilize the core duplex by 5.7–8.9 °C in Tm, or 0.5–1.3 kcal/mol. Thus, the two central F–F pairs are not as stabilizing in this position as are two T–A pairs.
The other three central insertions. involving DB/BD. BD/DB, and DF/FD pairs. all show similar affinities (Table 3 and Figure 5). The overall Tm and free energy values are similar to the FF/FF case. with the DB/BD (PuPy/PyPu) case showing a small advantage over the BD/DB (PyPu/PuPy) case. A similar effect is seen for the duplexes involving central T–A pairs. in which the AT/TA case is more stable than the TA/AT case. The best hydrophobic double insertion. the DB/BD case. destabilizes the core duplex by 7.5 °C in Tm and 1.2 kcal/mol in free energy when placed in the center of the duplex.
Results are quite different when the nonnatural nucleosides are placed at the end positions of the duplex. The F nucleoside was tested in this context because of its close similarity in shape to the natural thymidine. The addition of one F–F pair at each end of the duplex stabilizes it by 21.7 °C in Tm and −3.8 kcal/mol in free energy (Table 3). This degree of stabilization is much larger than the stabilization due to the (presumably) isosteric T–T pairs (an increase of 11.5 °C in Tm or −2.8 kcal/mol). In fact, the two F–F pairs give substantially greater stabilization than two canonical T–A pairs, which give the smaller stabilization of 10.0 °C in Tm and −2.2 kcal/mol in free energy. Interestingly, in this context the T–T pairs appear to be at least as stabilizing as the correctly matched T–A pairs.
Summary of Results
The results of these experiments can be summarized as follows: First, for the structures in this study, placement of single hydrophobic pairs within a standard DNA duplex is destabilizing, with a level of destabilization roughly equivalent to that of a DNA mismatched pair. Insertion of additional such pairs, however, is not incrementally destabilizing. Second, all the hydrophobic nucleosides prefer to pair with similarly hydrophobic compounds rather than the natural bases, with a selectivity of up to 4–5 kcal/mol. Third, hydrophobic pairs at the ends of duplexes are considerably more stabilizing than the natural base pairs studied. Finally, the isosteres of pyrimidine–pyrimidine pairs (i.e., F–F, B–B, F–B) are more stable than pairs involving the purine isostere D, although the pairing selectivity among these bases is not as high as selectivity among the natural bases.
Discussion
Internal Hydrophobic Pairs
Figure 2 illustrates the structures of the natural T–A and T–T pairs,35 and possible analogous structures for the hydrophobic isostere pairs. No direct evidence for pairing structure involving these nonnatural analogs is yet available, but indirect thermodynamic evidence may be useful in implicating certain features. First, since the thermodynamic stability of these pairs at isolated internal positions is somewhat better than standard DNA mismatches, it seems likely that the bases are stacked in the helix (as are DNA mismatches) rather than looped out in solution; such looping would otherwise result in a considerable energetic cost due to loss of important stacking interaction14,10 One exception may be the hydrophobic–hydrophilic pairs such as F–A; the energy for such pairs is 4–5 kcal less favorable than that for standard pairs such as T–A. It is possible that either the F or the A might be unstacked (looped out) in such a case, although we consider other explanations more likely. Possible alternative reasons for this energetic disparity are discussed below.
Several features of the hydrophobic pairs parallel behavior seen in natural DNA or RNA containing A, G, C, and T(U) bases. As seen for DNA mismatched pairs, single mismatches at internal positions are often destabilizing, while addition of additional mismatches is often less so.36 In addition, mismatches at the ends of helices are usually stabilizing (due to base stacking),10 rather than destabilizing. In addition, as with the natural bases, the hydrophobic bases are selective in pairing, although less so than the natural cases.
Not all properties of the hydrophobic pairs are analogous to properties of the natural pairs, however. Among these differences are the aforementioned low selectivity among the various hydrophobic pairs, and the weaker pairing affinity in the center of a DNA duplex. The origin of destabilization of the hydrophobic pairs as single central mismatches in DNA may be explained by two possibilities: First, since there is no hydrogen bonding to attract the bases to each other, the pairs must rely on base stacking interactions with the nearest neighbors for stabilization. One possible explanation is therefore that these compounds do not base stack well; however, this explanation is unlikely (at least for some of the bases), since addition of the F base at the ends of the helix is even more stabilizing than addition of natural bases, and additional stacking data also indicate that 2 and 3 also stack with high efficiency.26
A second explanation, which we consider more likely, is that in addition to lacking hydrogen bonds (at a cost of ~1–2 kcal relative to natural base pairs), none of the new base pairs can adopt an ideal pyrimidine–purine base pair geometry. This would lead to the observed single-placement destabilization in the center of a DNA duplex; the lack of destabilization at the end positions is then explained by greater freedom to adopt non- B-like conformations.
To best adopt a B-form duplex shape, it is likely that a standard pyrimidine–purine base pair geometry (or something closely approximating it) is probably required. Of the three nucleotides reported here, only the difluorotoluene (F) pyrimidine analog is a very close approximation in shape to a natural pyrimidine. It is interesting to note in this regard that a single F–F pair has almost the same stability as the analogous T–T pair at an internal position (Table 1). The trimethylbenzene (B) pyrimidine, with two methyl groups on the pairing face, is somewhat larger sterically. In addition, the dimethylindole (D) purine analog is larger than the natural adenine base because of the use of C–H groups to replace ring nitrogens. Thus, when in pairs in a helix, a given base pair containing the B or D bases would be wider than the corresponding natural base pairs. The pairing energies measured here are consistent with this idea; all of the pyrimidine–purine pair analogs (F–D, B–D) are among the more destabilizing cases. In fact, the most stable hydrophobic pairs seen here are the pyrimidine–pyrimidine analogs (F–F, B–F, B–B). The B–B pair in particular is a lengthened pyrimidine–pyrimidine analog, since the face of each B residue is extended outward by one bond relative to T. It is interesting to note that this appears to be the most stable pair of the group; presumably, outward extension of the pair brings it closer in geometry to the pyrimidine–purine optimum distance.
We note that as additional F–F pairs are inserted into the center of the core duplexes, the incremental destabilization disappears. The first F–F pair insertion in the hexamer core sequence is destabilizing by 8 °C in Tm and 1.3 kcal/mol in free energy. Addition of the second F–F pair is neither stabilizing nor destabilizing. Addition of four pairs (data from the dodecamer core) almost completely recovers the original duplex strength, indicating that beyond the first pair or two, even F–F pairs are incrementally stabilizing. This is consistent with the idea that such “pyr–pyr”-like pairs are destabilizing in the context of pyrimidine–purine DNA but not in the context of pyrimidine–pyrimidine-type duplexes. The result raises the intriguing possibility that one may be able to construct duplexes which consist mostly or even entirely of such pairs.
External Hydrophobic Pairs
It seems likely that efficient base stacking may be responsible for the high degree of stabilization by the hydrophobic F–F pairs at the ends of duplexes.26,10 It is intriguing that the difluorotoluene “base” may display better stacking characteristics than the natural thymine base, when its size and shape are nearly identical. It is possible that this is due to increased hydrophobicity, favoring desolvation by base stacking; studies of dinucleotide stacking have shown solvophobic characteristics for this interaction, although not of the classical entropy-driven type.11 It should be noted that recent model studies of π–π stacking also argue against a classical hydrophobic interaction,13c and indicate that electrostatic factors may contribute as well.12a Studies are currently underway in our laboratory26 to characterize in detail the base stacking properties of these and additional base analogs, in an effort to better understand the importance of factors such as hydrophobicity, polarizability, and electrostatics in contributing to this noncovalent interaction.
Our findings involving the aromatic bases 1 and 2 are not in accord with the conclusion given by Millican et al.31 in studying an unsubstituted phenyl nucleoside in DNA. The authors in that study attribute poor pairing with natural bases to weak base stacking propensity of the phenyl group, although they did not present experimental evidence specifically addressing its stacking behavior. Although it is certainly possible that the ring substituents in our structures may have a significant effect, we believe that a better explanation for the observed weak pairing with natural bases in that study and the present one is the cost of desolvation of the natural pairing partners (see below). Interestingly, a nitropyrrole nucleoside was recently reported30 to be relatively nonselective in pairing with natural bases, a finding consistent with the present results.
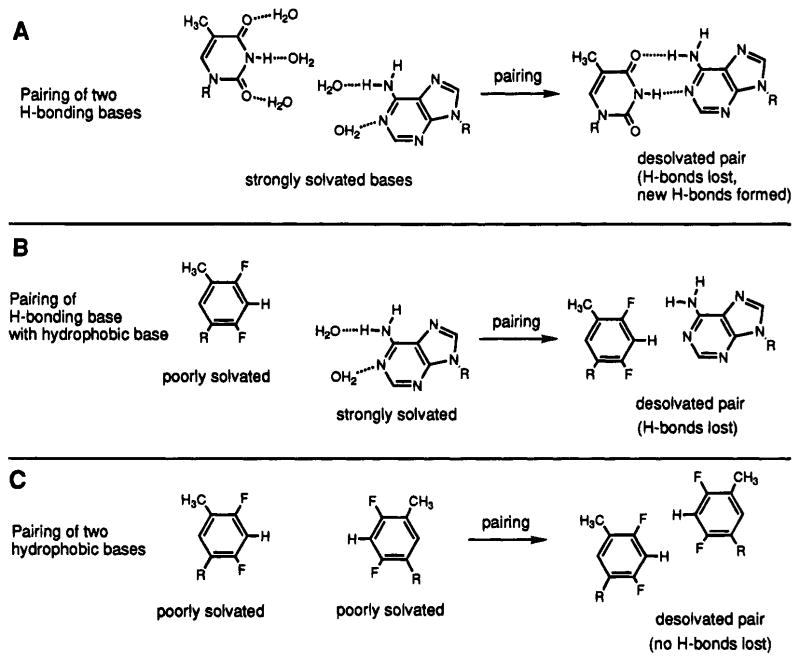
Solvation/Desolvation Effects
The general selectivity of all three hydrophobic isosteres in the present study for each other rather than for the natural bases is perhaps best explained by solvation and desolvation effects in base pairing (Figure 6). A good experimental example of this effect is seen in the comparison of the T–A, F–A, and F–F pairs (see Tables 1 and 2, and Figure 4). Mutating a single T–A pair out of 12 to the potentially isosteric F–A pair results in a 5 kcal/mol destabilization, a large decrease in free energy which corresponds to 40% of the entire duplex affinity. This value is clearly much larger than can be explained by the estimated 1–2 kcal/mol value5,9 given for hydrogen bonding in a U–A pair. One possible explanation is that the base stacking propensity of the F base is also poor; however, the finding that F residues at the ends of helices are stabilizing26 clearly rules this out.
Figure 6.
Illustrations of three different pairing situations seen in this study (A–C). It is proposed that the first (A, natural H-bonded pairing) and third (C, hydrophobic pairing) are more favorable because there are no uncompensated desolvations in base pair formation. The second (B, hydrophobic–hydrophilic pairing) is disfavored by ~5 kcal/mol relative to the first because of energetically costly desolvation of the natural base.
Our hypothesis for this ~5 kcal of destabilization is that it arises from the cost of desolvation of the hydrogen bond donors and acceptors of adenine during formation of an F–A pair (Figure 6). Prior to the formation of a normal T–A pair, the hydrogen bonding groups are each solvated by water molecules.37 Pair formation entails the loss of these solvating waters (breaking two H-bonds) concomitant with the formation of the two expected new hydrogen bonds of the base pair.38 The overall hydrogen bonding free energy is therefore low because H-bonds are both broken and formed. By contrast, when a hydrophobic–hydrophilic F–A pair is being formed the F “base” desolvation cost is presumably neutral (or even favorable), while the adenine desolvation cost remains high. If the pair is formed isosterically with the natural pair, the adenine must be desolvated without formation of new compensating bonds. This cost might be estimated independently by experimental free energy values for hydrogen bond formation in low- polarity media.39 The value of −2.7 kcal/mol has been reported for the cyclohexyl-U/9-ethyladenine pair in chloroform.40 This value is somewhat smaller than ours; however, weak H-bonding interactions with the chloroform solvent are likely to lower that value relative to true non-H-bonding environments. Another possible explanation is that in the context of other base pairs a single pairing interaction is entropically less disfavored than in the case of small-molecule dimerizations. In any case, the value is closer to our experimental value than is the aqueous base pair H-bond strength of 0.8–1.3 kcal/mol.5,6
An alternative, but related, explanation for our observed ~5 kcal of destabilization in hydrophobic–hydrophilic pairs is that when a given pair faces the prospect of desolvation, this costly occurrence is avoided instead by looping one of the two bases out of the helix so that the hydrophilic base can remain solvated. The expected energetic cost of this type of conformational change is unclear at present, but it would clearly be large because of loss of two nearest-neighbor interactions. Although we currently favor the first explanation, structural studies should clarify this issue in the future.
Double Mutation Rescues Stability
This “avoidance of desolvation” hypothesis leads to the prediction that a hydrophobic base would prefer to pair with another poorly solvated base, and that is what we observe experimentally. Mutating the unstable F–A pair to an F–F pair results in an increase in Tm of 7.2°C and in free energy, −1.5 kcal. This is despite the more unfavorable pyrimidine–pyrimidine geometry in the F–F case. This hydrophobic–hydrophobic pairing preference is thus best described not as a special affinity between hydrophobic groups, but instead as an aversion to the cost of desolvation of hydrogen-bonding groups. This kind of phenomenon has been observed in mutagenesis studies of proteins,41 in which an internal hydrogen bond is present between two groups in the protein interior. For example, the protein T4 lysozyme contains an internal threonine residue (Thr 157) which is hydrogen bonded to the backbone amide of Asp 159.41 Replacement of the threonine side chain with a group such as the isoleucine side chain destabilizes the protein folded structure by +2.9 kcal/mol, presumably for similar reasons as those given here.41 This value is similar in magnitude to our measured destabilization; to our knowledge this effect has not been previously observed in nucleic acids.
The results of the present study underscore the high thermodynamic cost of desolvating a hydrogen-bonding group in water. From this standpoint it is not surprising that even mismatched pairs in DNA are commonly hydrogen bonded to each other.35 Our finding that the isosteric F–F and T–T pairs are approximately equally stable suggests that hydrogen bonding is not intrinsically necessary for stable pairing, and thus may be more important in influencing the selectivity of pairing rather than the affinity. This is consistent with the finding that our nucleosides are significantly less selective in pairing when choosing between various hydrophobic structures, even though they vary quite considerably in shape. Thus, it is not so much shape selectivity that determines pairing selectivity in DNA as it is complementarity in hydrophilic groups.
Prospects for Hydrophobic Bases and Base Pairs
The current hydrophobic base pairs are quite stabilizing in some contexts, but less so in others. More studies are needed to understand better the origin of the various noncovalent interactions involved in pair formation with these molecules. In this vein, we are currently studying in detail the base stacking propensities of these and related compounds.26 The structure of these pairs in DNA is also a primary topic of study. In addition, there may be a need for design and synthesis of new isosteres which might form even more stable pairs; especially useful for that purpose would be a better isostere for purines.
There has been recent interest in the development of “universal” DNA bases which can be useful in primers for DNA amplification.30 Such bases are designed to pair about equally well with all four natural bases. Our results with 1–3 show that this pairing nonselectivity may generally hold true for such hydrophobic compounds. Whether 1–3 specifically can serve as such universal bases in PCR amplification remains to be seen.
With the current data in hand, however, we can envisage several potential uses for these DNA base and base pair analogs. One use, now underway, is as test molecules in the study of hydrogen bonding and base stacking in nucleic acids. Another potential use is as probes for noncovalent interactions between DNA-binding small molecules and their DNA target. Protein–DNA interactions might be examined in a similar fashion. In addition, we have recently found that some of these nucleotides can be incorporated into oligonucleotide strands by DNA polymerases.42 If such incorporation were to occur efficiently, addition of a hydrophobic pair to the standard A, T, G, C repertoire might allow expansion of the genetic code.43
Conclusions
We have incorporated nonpolar isosteres of the natural bases adenine and thymine in DNA and examined their base pairing properties. We find that in intemal contexts a hydrophobic pair can exhibit pairing stability as good as, or slightly better than, the T–G mismatch pair. At the ends of helices they can be more stabilizing than a canonical A–T base pair. In addition, the hydrophobic base analogs are significantly selective for pairing with hydrophobic partners (by ~20-fold) rather than the natural bases. The energetic penalty for hydrophobic–hydrophilic mismatched pairing in one case is found to be −5 kcal/mol; we attribute this to the cost of desolvation of the hydrophilic partner. Such nonpolar analogs may be of general use in probing noncovalent interactions in biological systems involving DNA.
Acknowledgments
We thank Prof. Samuel Gellman for helpful discussions, and we thank the Office of Naval Research for support. E.T.K. gratefully acknowledges awards from the Young Investigator Programs of the Arnold and Mabel Beckman Foundation, the Office of Naval Research, and the Army Research Office, a Teacher-Scholar Award from the Dreyfus Foundation, and an Alfred P. Sloan Foundation Fellowship.
References
- 1.Watson JD, Crick FHC. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Schultz GE, Schirmer RH. Principles of Protein Structure. Springer-Verlag; New York: 1979. pp. 149–165. [Google Scholar]
- 3.(a) Pabo CO, Sauer RT. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]; (b) von Hippel PH. Science. 1994;263:769–770. doi: 10.1126/science.8303292. [DOI] [PubMed] [Google Scholar]
- 4.(a) Klotz IM, Frantzen JS. J Am Chem Soc. 1962;84:3461–3466. [Google Scholar]; (b) Cantor CR, Schimmel PR. Biophysical Chemistry Part I: The Conformation of Biological Macromolecules. W. H. Freeman; San Francisco: 1980. pp. 277–279. [Google Scholar]
- 5.Turner DH, Sugimoto N, Kierzek R, Dreiker SD. J Am Chem Soc. 1987;109:3783–3785. [Google Scholar]
- 6.SantaLucia J, Kierzek R, Turner DH. Science. 1992;256:217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- 7.Breslauer KJ, Frank R, Blocker H, Marky LA. Proc Natl Acad Sci USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantor CR, Schimmel PR. Biophysical Chemistiy Part III: The Behavior of Biological Macromolecules. W. H. Freeman; San Francisco: 1980. pp. 1117–1133. [Google Scholar]
- 9.Petersheim M, Turner DH. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 10.Senior M, Jones RA, Breslauer KJ. Biochemistry. 1988;27:3879–3885. doi: 10.1021/bi00410a053. [DOI] [PubMed] [Google Scholar]
- 11.Lowe MJ, Schellman MJ. J Mol Biol. 1972;65:91–109. doi: 10.1016/0022-2836(72)90494-9. [DOI] [PubMed] [Google Scholar]
- 12.Cozzi F, Cinquini M, Annuziata R, Siegel JS. J Am Chem Soc. 1993;115:5330–5331. [Google Scholar]; (b) Hunter CA. Angew Chem, Int Ed End. 1993;32:1584–1586. [Google Scholar]
- 13.(a) Leonard NJ. Acc Chem Res. 1979;12:423–429. [Google Scholar]; (b) Rotello VM, Viani EA, Deslongchamps G, Murray BA, Rebek J. J Am Chem Soc. 1993;115:797–798. [Google Scholar]; (c) Newcomb LF, Gellman SH. J Am Chem Soc. 1994;116:4993–4994. [Google Scholar]
- 14.Colocci N, Distefano MD, Dervan PB. J Am Chem Soc. 1993;115:4468–4469. [Google Scholar]
- 15.(a) Jernigan RL, Sarai A, Ting KL, Nussinov R. J Biomol Struct Dyn. 1986;4:41–48. doi: 10.1080/07391102.1986.10507645. [DOI] [PubMed] [Google Scholar]; (b) Hunter CA. J Mol Biol. 1993;230:1025–1054. doi: 10.1006/jmbi.1993.1217. [DOI] [PubMed] [Google Scholar]
- 16.(a) Maroun RC, Olson WK. Biopolymers. 1988;27:561–584. doi: 10.1002/bip.360270403. [DOI] [PubMed] [Google Scholar]; (b) Ornstein RL, Rein R, Breen DL, Macelroy RD. Biopolymers. 1978;17:2341–2360. doi: 10.1002/bip.1978.360171005. [DOI] [PubMed] [Google Scholar]; (c) Friedman RA, Honig B. Biopolymers. 1992;32:145–159. doi: 10.1002/bip.360320205. [DOI] [PubMed] [Google Scholar]
- 17.(a) Maroun RC, Olson WK. Biopolymers. 1988;27:585–603. doi: 10.1002/bip.360270404. [DOI] [PubMed] [Google Scholar]; (b) Sarai A, Mazur J, Nussinov R, Jernigan RL. Biochemistry. 1988;27:8498–8502. doi: 10.1021/bi00422a030. [DOI] [PubMed] [Google Scholar]
- 18.Uhlmann E, Peyman A. Chem Rev. 1990;90:543–584. [Google Scholar]
- 19.Schweitzer BA, Kool ET. J Org Chem. 1994;59:7238–7242. doi: 10.1021/jo00103a013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gait MJ, editor. Oligonucleotide Synthesis. IRL Press; Oxford: 1984. pp. 45–49. [Google Scholar]
- 21.(a) Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1984. p. 123. [Google Scholar]; (b) Weast RC, editor. CRC Handbook of Chemistry and Physics. 6. CRC Press; Boca Raton: 1979. p. F216. [Google Scholar]
- 22.Beaucage SL, Caruthers MH. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 23.Borer PN. In: Handbook of Biochemistry and Molecular Biology. 3. Fasman GD, editor. I. CRC Press; Cleveland: 1985. p. 589. [Google Scholar]
- 24.The PIPES buffer has the lowest pKa vs temperature dependence of the Good buffers: Good NE, Winget GD, Winter W, Connolly TN, Izawa S, Singh RMM. Biocieemistry. 1966;5:467–477. doi: 10.1021/bi00866a011.
- 25.Kool E. J Am Chem Soc. 1991;113:6265–6266. [Google Scholar]
- 26.Schweitzer BA, Chaudhuri NC, Shiels CJ, Kool ET. Manuscript in preparation. [Google Scholar]
- 27.Klein RS, Kotick MP, Watanabe KA, Fox JJ. J Org Chem. 1971;36:4113–4116. doi: 10.1021/jo00825a022. [DOI] [PubMed] [Google Scholar]
- 28.Sharma RA, Bobek M, Bloch A. J Med Chem. 1975;18:473–476. doi: 10.1021/jm00239a006. [DOI] [PubMed] [Google Scholar]
- 29.Coleman RS, Dong Y, Arthur JC. Tetrahedron Lett. 1993;34:6867–6870. [Google Scholar]
- 30.(a) Nichols R, Andrews PC, Zhang P, Bergstrom DE. Nature. 1994;369:492–493. doi: 10.1038/369492a0. [DOI] [PubMed] [Google Scholar]; (b) Loakes D, Brown DM. Nucleic Acids Res. 1994;22:4039–4043. doi: 10.1093/nar/22.20.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millican TA, Mock GA, Chauncey MA, Patel TP, Eaton MAW, Gunning J, Cutbush SD, Neidle S, Mann J. Nucleic Acids Res. 1984;12:7435–7453. doi: 10.1093/nar/12.19.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Vinogradov SN, Linnell RH. Hydrogen Bonding. Van Nostrand Reinhold; New York: 1971. pp. 124–135. [Google Scholar]; (b) Jones DAK, Watkinson JG. J Chem Soc. 1964:2366–2370. [Google Scholar]
- 33.Jorgensen WL, Severance DL. J Am Chem Soc. 1990;112:4768–4774. [Google Scholar]
- 34.Ahoul-ela F, Koh D, Tinoco I. Nucleic Acids Res. 1985;13:4811–4825. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kouchakdjian M, Li BFL, Swan PF, Patel DJ. J Mol Biol. 1988;202:139–155. doi: 10.1016/0022-2836(88)90526-8. [DOI] [PubMed] [Google Scholar]
- 36.(a) SantaLucia J, Kierzek R, Turner DH. Biochemistry. 1991;30:8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]; (b) Nikonowicz EP, Gorenstein DG. Biochemistry. 1990;29:8845–8858. doi: 10.1021/bi00489a048. [DOI] [PubMed] [Google Scholar]
- 37.Schneider B, Cohen D, Berman HM. Biopolymers. 1992;32:725–750. doi: 10.1002/bip.360320703. [DOI] [PubMed] [Google Scholar]
- 38.Alden CJ, Kim SH. J Mol Biol. 1979;132:411–434. doi: 10.1016/0022-2836(79)90268-7. [DOI] [PubMed] [Google Scholar]
- 39.Pimentel GC, McClellan AL. The Hydrogen Bond. W. H. Freeman; San Francisco: 1960. pp. 206–225. [Google Scholar]
- 40.Kyogoku Y, Lord RC, Rich A. J Am Chem Soc. 1967;89:496–504. doi: 10.1021/ja00979a005. [DOI] [PubMed] [Google Scholar]
- 41.Alber T, Dao-pin S, Wilson K, Wozniak JA, Cook SP, Matthews BW. Nature. 1987;330:41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- 42.Schweitzer BA, Kool ET. 1995 Manuscript submitted. [Google Scholar]
- 43.Bain JD, Switzer C, Chamberlin AR, Benner SA. Nature. 1992;356:537–539. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]