Abstract
Salmonella enterica serotype Typhimurium (S. Typhimurium) causes acute gut inflammation by using its virulence factors to invade the intestinal epithelium and survive in mucosal macrophages. The inflammatory response enhances the transmission success of S. Typhimurium by promoting its outgrowth in the gut lumen through unknown mechanisms. Here we show that reactive oxygen species generated during inflammation reacted with endogenous, luminal sulphur compounds (thiosulfate) to form a new respiratory electron acceptor, tetrathionate. The genes conferring the ability to utilize tetrathionate as an electron acceptor produced a growth advantage for S. Typhimurium over the competing microbiota in the lumen of the inflamed gut. We conclude that S. Typhimurium virulence factors induce host-driven production of a new electron acceptor that allows the pathogen to use respiration to compete with fermenting gut microbes. Thus, the ability to trigger intestinal inflammation is crucial for the biology of this diarrhoeal pathogen.
Introduction
S. Typhimurium is an invasive enteric pathogen associated with diarrhoea, acute intestinal inflammation and the presence of neutrophils in stool samples 1. The pathogen triggers intestinal inflammation by employing two type III secretion systems (T3SS-1 and T3SS-2) that enable S. Typhimurium to invade the intestinal epithelium and survive in mucosal macrophages 2. Recent studies suggest that acute intestinal inflammation enhances growth of S. Typhimurium in the intestinal lumen 3-5. The resulting increase in numbers establishes the pathogen as a prominent species in the gut, thereby enhancing its transmission success 6. However, the mechanisms by which S. Typhimurium can overgrow other microbes in the hostile environment of the inflamed gut remain uncharacterized.
The ability of S. Typhimurium to overgrow other microbes under certain in vitro growth conditions has been exploited for enrichment methods that facilitate its isolation from biological samples containing competing microbes. A commonly used approach, known as tetrathionate enrichment, was developed in 1923, and is based on the ability of S. Typhimurium to use tetrathionate as a terminal electron acceptor 7. It is widely believed that tetrathionate respiration is not important during infection, because there are neither known sources of tetrathionate in the mammalian host, nor does a S. Typhimurium mutant deficient for tetrathionate respiration exhibit reduced virulence in a mouse model of typhoid fever 8(Figure S1). These observations suggested that tetrathionate respiration encoded by the ttrSR ttrBCA gene cluster (Figure S1A) might be most important when free-living bacteria grow in tetrathionate-containing environments such as soil or decomposing carcasses 9.
S4O6 2- availability in the gut
A fresh look at sulphur metabolism in the inflamed intestine suggested an alternative to this conventional wisdom (Figure 1). Colonic bacteria produce large quantities of hydrogen sulfide (H2S), a highly toxic compound. The cecal mucosa protects itself from the injurious effects of H2S by converting it to thiosulfate (S2O3 2-) 10,11 (Figure 1A). While thiosulfate is therefore likely to be present in the intestinal lumen, this compound cannot be used as an electron acceptor by the ttrSR ttrBCA gene cluster 12. However, tetrathionate broth used for enrichment of Salmonella serotypes contains thiosulfate, not tetrathionate (S4O6 2-). Prior to use of the medium, thiosulfate is oxidized to tetrathionate by addition of the strong oxidant iodine (Figure 1A). We reasoned that oxidation of thiosulfate might occur during intestinal inflammation, a condition accompanied by neutrophil transmigration into the gut lumen (Figure 1B) and production of nitric oxide radicals (NO) and reactive oxygen species 13.
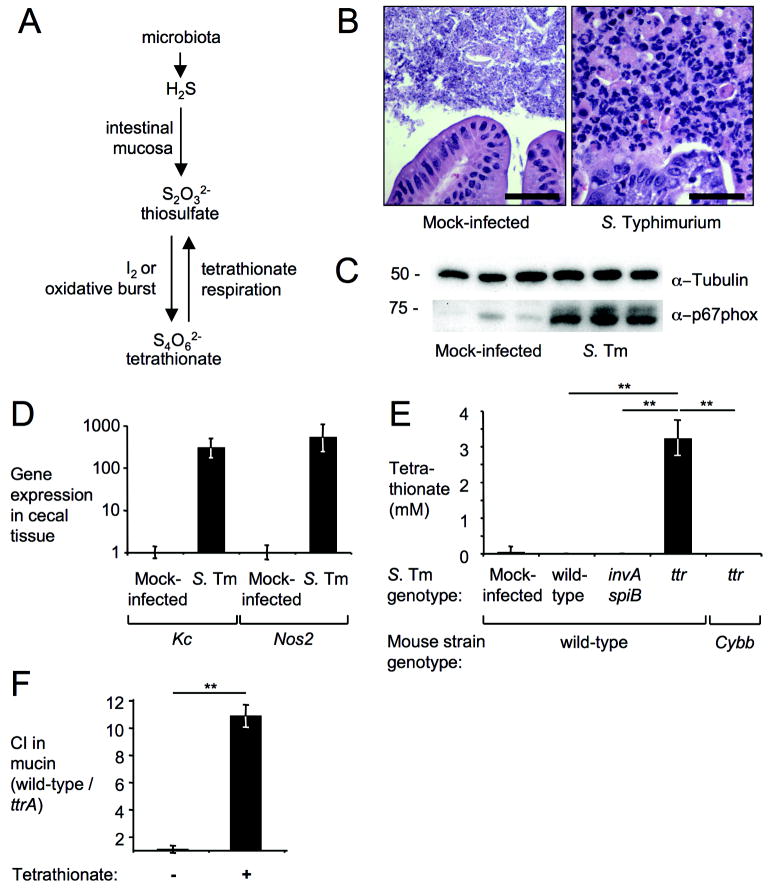
Figure 1. Tetrathionate becomes available during inflammation.
(A) Schematic of intestinal sulfur metabolism. (B-E) Samples from a mouse colitis model four days after infection with S. Typhimurium (S. Tm) or mock-infection. (B) H&E stained cecal sections. Scale bar, 100 μm. (C) Detection of NADPH oxidase (α–p67phox) or tubulin (α–tubulin) in cecal extracts (n=3). (D) Expression of Kc and Nos2 in cecal RNA samples (n≥3) using qRT-PCR (fold-increases over mock-infection). (E) Tetrathionate detected in cecal contents using LC-MS (n≥3). (F) Competitive indices (CI) for anaerobic growth in mucin broth with (+) or without (-) tetrathionate (n=3). (D-F) Bars represent geometric means ± standard error. **, P < 0.01.
To test this idea, we measured the formation of tetrathionate in vivo using a mouse colitis model 14. Compared to mock-infected animals, infection of mice (C57BL/6) with S. Typhimurium resulted in acute cecal inflammation (Figure 1C, 1D, and Figure S2). Infection with a mutant deficient for tetrathionate respiration (ttr mutant) was accompanied by increased tetrathionate levels, which were detected in cecal contents by reverse phase high performance liquid chromatography (HPLC) coupled with mass spectrometry (MS) (Figure 1E). S. Typhimurium causes intestinal inflammation by employing two type III secretion systems, T3SS-1 and T3SS-2, which mediate epithelial invasion and macrophage survival, respectively 15. Inactivation of T3SS-1 (through a mutation in invA) and T3SS-2 (through a mutation in spiB) renders S. Typhimurium unable to trigger intestinal inflammation in the mouse colitis model 16 (Figure 2). Tetrathionate was not detected in mice infected with an invA spiB mutant (P < 0.01), suggesting that inflammation is required for generating tetrathionate in the intestine. Furthermore, tetrathionate did not accumulate during infection with the S. Typhimurium wild-type strain (P < 0.01), which raised the possibility that the ttr genes might promote consumption of this electron acceptor during infection.
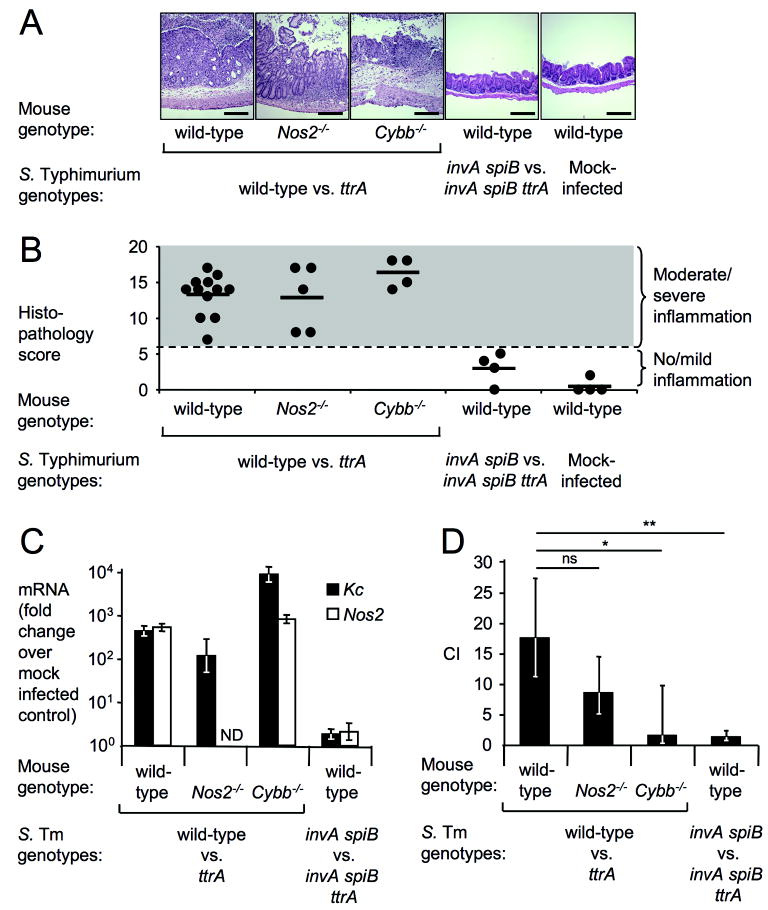
Figure 2. Tetrathionate respiration confers growth advantage.
(A-D) Samples from a mouse colitis model (n indicated in B) four days after infection with S. Typhimurium (S. Tm) or mock-infection. (A) H&E stained cecal sections. Scale bar, 400 μm. (B) Blinded histopathology scoring showing averages (bars) and individual scores (circles). (C) Kc (closed bars) and Nos2 (open bars) expression in cecal RNA samples using qRT-PCR (fold-increases over mock-infection). (D) Competitive indices (CI) of indicated S. Typhimurium strains determined by recovering bacteria from colon contents. (C-D) Bars represent geometric means ± standard error. *, P < 0.05; **, P < 0.01; ns, not significant, ND, not determined.
S4O6 2- promotes growth in the gut
To investigate the growth benefit conferred by tetrathionate respiration in vitro, the S. Typhimurium wild-type strain and a ttrA mutant (Figure S1A and B) were co-cultured in tetrathionate broth in the presence or absence of oxygen (Figure S1C). When thiosulfate was not oxidized to tetrathionate by the addition of iodine, the wild-type strain and the ttrA mutant grew equally well. However, in the presence of iodine, tetrathionate respiration promoted outgrowth of the S. Typhimurium wild-type strain under anaerobic and microaerobic, but not under aerobic growth conditions. A tetrathionate concentration of 2.5 mM was sufficient to promote outgrowth of the wild-type strain (Figure S1D) (P < 0.01). Co-culture of the S. Typhimurium wild-type strain and the ttrA mutant in mucin broth resulted in enrichment for the wild type only in the presence of tetrathionate (Figure 1F) (P < 0.01). Collectively, these data suggested that tetrathionate respiration might provide a benefit during the anaerobic growth conditions encountered in vivo, e.g. in the intestinal mucus layer.
To test this idea, mice were infected with an equal mixture of the S. Typhimurium wild-type strain and a ttrA mutant (Figure 2). S. Typhimurium infection resulted in prominent intestinal inflammation (Figures 2A and B) and increased mRNA levels of Kc, encoding a neutrophil chemoattractant, and Nos2, encoding inducible nitric oxide synthase (iNOS) (Figure 2C). A marked enrichment for the S. Typhimurium wild-type strain was observed 4 days after infection in the colon contents (Figure 2D), suggesting that tetrathionate respiration provided an advantage during growth in the lumen of the inflamed gut. In contrast, both strains were recovered in similar numbers from the spleen in a mouse model of typhoid fever (Fig S1F), suggesting that tetrathionate was not available for growth at systemic sites. We next validated our results using a bovine ligated small intestinal (ileal) loop model in which S. Typhimurium causes acute mucosal inflammation (Figure 3)17. Upon infection with an equal mixture of the S. Typhimurium wild-type and a ttrA mutant, higher numbers of the wild-type strain were associated with the mucus fraction and with the intestinal mucosa, while equal numbers of both strains were recovered from the luminal fluid 8 hours after infection. These data suggested that the selective advantage conferred by tetrathionate respiration was greatest in close proximity to the inflamed mucosal surface.
Figure 3. Tetrathionate respiration promotes growth of S. Typhimurium in close proximity to the mucosal surface.
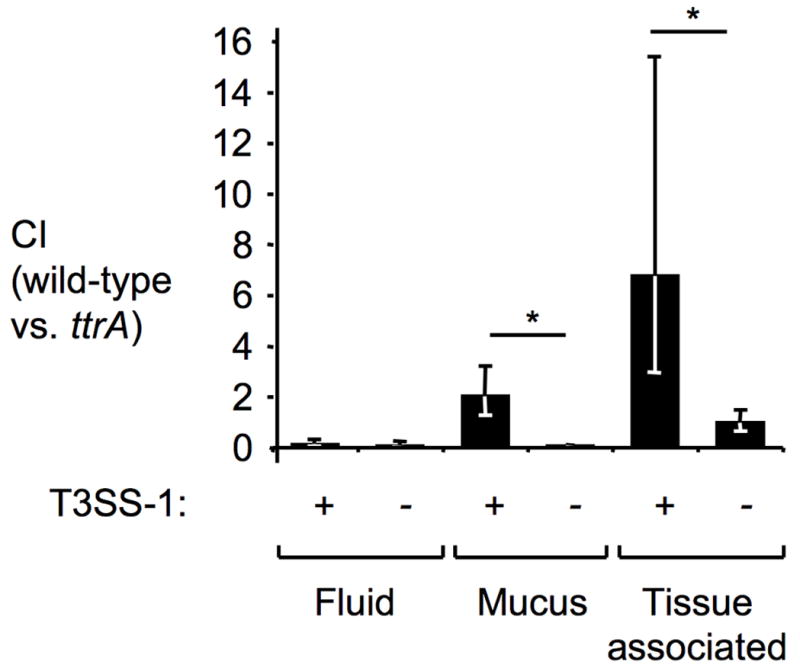
Bovine ligated ileal loops (n=3 animals) were infected with a mixture of S. Typhimurium T3SS-1 proficient (+) strains (wild-type [AJB715] versus ttrA mutant[SW661]) or T3SS-1-deficient (-) strains (invA mutant [SW737] versus invA ttrA mutant [SW736]) and samples collected 8 hours after infection from the luminal fluid, mucus scrapings and tissue punches (tissue-associated bacteria). Bars represent geometric means ± standard error. *, P ≤ 0.05.
To determine whether tetrathionate respiration provides a colonization advantage in the absence of inflammation, mice were infected with an equal mixture of an invA spiB mutant and an invA spiB ttrA mutant. Mice infected with this mixture neither developed intestinal pathology nor exhibited elevated levels of Nos2 or Kc mRNA (Figures 2A-C). Equal numbers of both strains were recovered from colon contents (Figure 2D). During the early stages of infection modelled in bovine ligated ileal loops, intestinal inflammation is largely dependent on T3SS-1 17. Infection with an equal mixture of an invA mutant and an invA ttrA mutant resulted in equal recovery of both strains from bovine ligated ileal loops (Figure 3). Collectively, these data suggested that tetrathionate respiration provided no growth benefit in the absence of intestinal inflammation.
Oxygen radicals generate S4O6 2-in vivo
Induction of a respiratory burst in blood leukocytes resulted in oxidation of thiosulfate to tetrathionate (Figure S1G). To determine whether iNOS or NADPH oxidase are required for tetrathionate respiration in vivo, Nos2-deficient mice and Cybb (gp91phox)-deficient mice were infected with an equal mixture of the S. Typhimurium wild-type strain and the ttrA mutant. S. Typhimurium infection resulted in marked intestinal inflammation (Figure 2A and B) and increased mRNA levels of Kc (Figure 2C). While enrichment for wild-type bacteria was detectable in Nos2-deficient mice, no enrichment for the S. Typhimurium wild-type strain was observed in Cybb-deficient mice (Figure 2D) (P < 0.05). Thus, oxygen radicals produced by NADPH oxidase may be more important than nitric oxide radicals in promoting tetrathionate respiration in vivo. Infection of Cybb-deficient mice with a ttr mutant was not accompanied by production of tetrathionate (Figure 1E). Collectively, these data suggested that the respiratory burst of phagocytes recruited during inflammation stimulates growth of S. Typhimurium in the gut by providing a terminal electron acceptor.
Outgrowth by S4O6 2- respiration
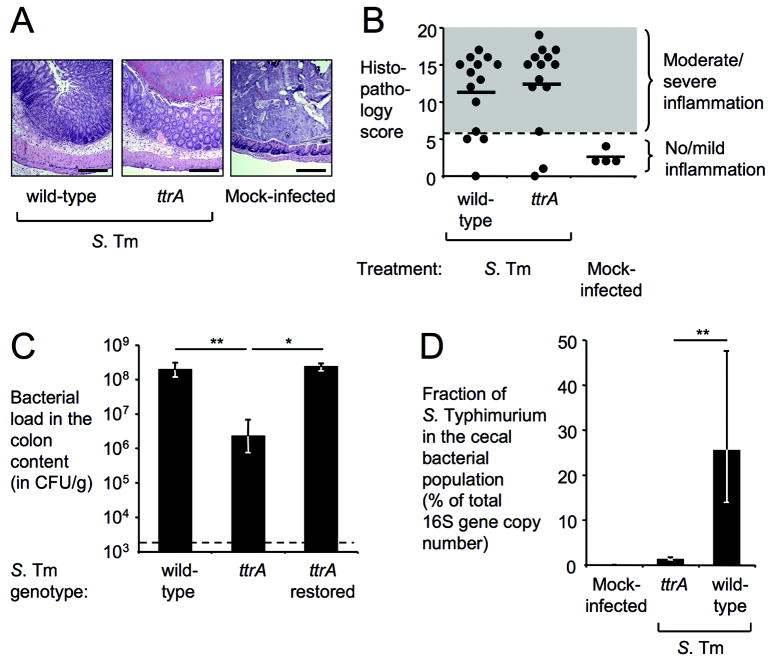
Under anaerobic conditions, microbes compete for high-energy resources that are available for fermentation, but fermentation end products cannot be further utilized. By reducing tetrathionate, S. Typhimurium is able to use fermentation end products that can only be respired, providing a substantial selective advantage. To test the magnitude of this growth advantage, we measured the effect of tetrathionate respiration on the abundance of S. Typhimurium in intestinal contents (Figure 4). Mice were inoculated with the S. Typhimurium wild-type strain or a ttrA mutant and bacteria were recovered four days after infection. The S. Typhimurium wild-type strain was recovered in approximately 80-fold higher numbers (P < 0.01) than the ttrA mutant (no tetrathionate respiration) (Figure 4A-C). Restoration of tetrathionate respiration in the ttrA mutant by homologous recombination re-established growth at the level of the wild-type strain. Analysis of the microbiota composition indicated that the Typhimurium wild-type strain, but not the ttrA mutant, was able to outcompete other bacteria inhabiting the cecum (Figure 4D and Figure S3). These results suggested that the ability of S. Typhimurium to outgrow the microbiota during inflammation depends on tetrathionate respiration.
Figure 4. Tetrathionate respiration increases the abundance of S. Typhimurium in the intestinal lumen.
(A-C) Samples from a mouse colitis model (n indicated in B) four days after infection with S. Typhimurium (S. Tm) or mock-infection. (A) H&E stained cecal sections. Scale bar, 400 μm. (B) Blinded histopathology scoring showing averages (bars) and individual scores (circles). (C) Recovery of S. Typhimurium from colon contents. (D) Fraction of S. Typhimurium as percentage of the cecal bacterial population using 16S rRNA gene qRT-PCR (wild-type n=6, ttrA mutant n=6, mock-infected n=4). (C-D) Bars represent geometric means ± standard error. *, P < 0.05; **, P < 0.01.
Discussion
An important recent conceptual advance in bacterial pathogenesis is the demonstration that enteric pathogens can utilize host responses to outgrow the intestinal microbiota, but the mechanisms were not clear 3,4,18. Here we show that S. Typhimurium gains a growth advantage in the competitive environment of the gut by utilizing a virulence factor-induced electron acceptor generated by the host respiratory burst. These data suggest that tetrathionate respiration provides a significant selective advantage, because enrichment for S. Typhimurium during growth in the inflamed gut leads to increased transmission by the fecal-oral route 6. The selective advantage conferred by tetrathionate respiration is likely an important reason why S. Typhimurium causes gastrointestinal disease, since this property places virulence factors (i.e. T3SS-1 and T3SS-2) that are required for inducing the inflammatory host response needed for the formation of tetrathionate in vivo, under selection. This may also explain why the ability to reduce tetrathionate is among a constellation of functions found in most Salmonella isolates and is historically used as a criterion for identification of Salmonellae. It is noteworthy that the ttr gene cluster is also present in the enteric pathogen Yersinia enterocolitica, but is absent from a close relative, Y. pestis, which does not colonize the intestine 19.
Methods Summary
Bacterial strains and plasmids used are listed in Supplementary Table 1. S. Typhimurium was routinely cultured in LB broth or on LB agar plates. Construction of tetrathionate respiration deficient mutants is described in the Supplementary Methods. Tetrathionate broth (BD Biosciences) or mucin broth (0.05 % hog mucin [Sigma-Aldrich] in minimal media supplemented with 40 mM sodium tetrathionate as indicated) was inoculated with 100 colony forming units(CFU) /ml of each strain and incubated at 37°C for 16 h either with aeration, statically or anaerobically as indicated. All animal experiments were approved by the Institutional Animal Care and Use Committees at the University of California, Davis (mouse experiments) or the Texas A&M University (calf experiments). Ligated ileal loop surgery was performed as described previously 17. A S. Typhimurium mouse colitis model has been described 14. Groups of 10-12 week old, female mice (C57BL/6, B6.129S-Cybbtm1Din/J, B6.129P2-Nos2tm1Lau/J; The Jackson Laboratory) were orally infected with S. Typhimurium and tissue samples collected 4 days later. Bacterial numbers were determined by spreading serial 10-fold dilutions of tissue homogenates on selective media. The competitive index was calculated by dividing the number of wild-type cells by the number of mutant cells and normalized by the input ratio. Formalin fixed, Hematoxylin and Eosin (H&E) stained cecal sections were examined for signs of inflammation (Supplementary Figure 2). Tetrathionate concentration of cecal extracts was measured by RP-LC-MS. To measure relative expression levels of Kc and Nos2 mRNA, total RNA was isolated from the cecum using TRI reagent (Molecular Research Center), reverse transcribed (TaqMan reverse transcription reagents; Applied Biosystems) and SYBR-Green (Applied Biosystems) based real-time PCR performed using the primers listed in Supplementary Table 2. Fold changes in mRNA levels measured by real-time PCR, tetrathionate concentrations, and bacterial numbers underwent logarithmic transformation before ANOVA analysis followed by Student’s t-test.
Supplementary Material
Acknowledgments
We would like to thank Hiutung Chu for providing real-time PCR primers, Vladimir Tolstikov for performing the LC-MS analysis, Mariana Xavier for assistance with histopathology and Valerie Gerriets for technical assistance. Work in AJB’s laboratory is supported by Public Health Service Grants AI040124, AI044170, AI073120, AI076246, and AI088122. P. T. was supported by a stipend from the Department of Microbiology, Chiang Mai University, Thailand.
Footnotes
Contributions S. E. W. contributed to the experimental design, constructed bacterial strains and contributed to Figures 1C, 1D, 1E, 2C, 2D, 3D, 3E, S1A, S1B, S1E, S1F and S3. P. T. contributed to Figures 1F, S1C, S1D and assisted with mouse experiments. M. G. W. assisted with mouse experiments and performed cloning experiments. B. P. B. contributed to Figures 1B, 2A, 2B, 3B, 3C, and S2. D. L. H. constructed bacterial strains. R.W.C. and J.M.R. contributed to Figure 3A. L.G.A. performed the ligated loop surgery. C.L.B., L.G.A., R. M. T., J. R. R. and A. J. B. provided financial support for the study and contributed to the experimental design. S. E. W. and A. J. B. were responsible for the overall study design and for writing the manuscript.
Competing financial Interests The authors declare no competing financial interests
Supplementary Information accompanies the paper on www.nature.com/nature.
Contributor Information
Sebastian E. Winter, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Parameth Thiennimitr, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA; Department of Microbiology, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand.
Maria G. Winter, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Brian P. Butler, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Douglas L. Huseby, Department of Microbiology, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Robert W. Crawford, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Joseph M. Russell, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Charles L. Bevins, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
L. Garry Adams, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX 77843, USA.
Renée M. Tsolis, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
John R. Roth, Department of Microbiology, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Andreas J. Bäumler, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Literature
- 1.Harris JC, Dupont HL, Hornick RB. Fecal leukocytes in diarrheal illness. Ann Intern Med. 1972;76:697–703. doi: 10.7326/0003-4819-76-5-697. [DOI] [PubMed] [Google Scholar]
- 2.Santos RL, et al. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stecher B, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barman M, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infection and immunity. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekirov I, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infection and immunity. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawley TD, et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infection and immunity. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller L. Un nouveau milieu d’enrichissement pour la recherche du Bacille Typhique at Paratyphique. Comptes Rendus des Seances de la Societe de Biologie et de ses Filiales. 1923;89:434–437. [Google Scholar]
- 8.Hensel M, Nikolaus T, Egelseer C. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol Microbiol. 1999;31:489–498. doi: 10.1046/j.1365-2958.1999.01190.x. [DOI] [PubMed] [Google Scholar]
- 9.Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol Microbiol. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 10.Furne J, Springfield J, Koenig T, DeMaster E, Levitt MD. Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem Pharmacol. 2001;62:255–259. doi: 10.1016/s0006-2952(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 11.Levitt MD, Furne J, Springfield J, Suarez F, DeMaster E. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J Clin Invest. 1999;104:1107–1114. doi: 10.1172/JCI7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinsley AP, Berks BC. Specificity of respiratory pathways involved in the reduction of sulfur compounds by Salmonella enterica. Microbiology. 2002;148:3631–3638. doi: 10.1099/00221287-148-11-3631. [DOI] [PubMed] [Google Scholar]
- 13.Reinders CA, et al. Rectal nitric oxide and fecal calprotectin in inflammatory bowel disease. Scand J Gastroenterol. 2007;42:1151–1157. doi: 10.1080/00365520701320505. [DOI] [PubMed] [Google Scholar]
- 14.Barthel M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsolis RM, Adams LG, Ficht TA, Baumler AJ. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infection and immunity. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raffatellu M, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell host & microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, et al. SipA, SopA, SopB, SopD and SopE2 act in concert to induce diarrhea in calves infected with Salmonella enterica serotype Typhimurium. Infect Immun. 2002;70:3843–3855. doi: 10.1128/IAI.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Thomson NR, et al. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS genetics. 2006;2:e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- 21.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 22.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis RW, B D, R RJ. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press; New York: 1980. [Google Scholar]
- 24.Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter SE, et al. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype typhimurium infection. Infection and immunity. 2009;77:1904–1916. doi: 10.1128/IAI.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter SE, Raffatellu M, Wilson RP, Russmann H, Baumler AJ. The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell Microbiol. 2008;10:247–261. doi: 10.1111/j.1462-5822.2007.01037.x. [DOI] [PubMed] [Google Scholar]
- 27.Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingsley RA, et al. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: identification of intestinal colonization and persistence determinants. Infection and immunity. 2003;71:629–640. doi: 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.