SUMMARY
Pre-mRNA 3′-end formation is an essential step in eukaryotic gene expression. Over half of human genes produce alternatively polyadenylated mRNAs, suggesting that regulated polyadenylation is an important mechanism for post-transcriptional gene control. Although a number of mammalian mRNA 3′ processing factors have been identified, the full protein composition of the 3′ processing machinery has not been determined, and its structure is unknown. Here we report the purification and subsequent proteomic and structural characterization of human mRNA 3′ processing complexes. Remarkably, the purified 3′ processing complex contains ~85 proteins, including known and new core 3′ processing factors and over 50 proteins that may mediate crosstalk with other processes. Electron microscopic analyses show that the core 3′ processing complex has a distinct “kidney” shape and is ~250 Å in length. Together, our data has revealed the complexity and molecular architecture of the pre-mRNA 3′ processing complex.
INTRODUCTION
Polyadenylation is a nearly universal step in eukaryotic gene expression. Poly(A) tails have profound influence on the stability, export, and translation efficiency of mRNAs (Colgan and Manley, 1997; Zhao et al., 1999). In mammals, biochemical studies have shown that pre-mRNA 3′ processing requires four multi-subunit protein complexes, CPSF, CstF, CF I and CF II, in addition to the single subunit poly(A) polymerase (PAP) (Takagaki et al., 1989; reviewed by Colgan and Manley, 1997; Mandel et al., 2008; Zhao et al., 1999). With the aid of RNA polymerase (RNAP) II (McCracken et al., 1997; Hirose and Manley, 1998), these and other factors assemble onto the nascent pre-mRNA to form a macromolecular complex in which the 3′ processing reactions take place. Despite significant divergence in the cis-elements required for 3′ processing between yeast and mammalian mRNAs, most 3′ processing factors are conserved. Interestingly, although 20 polyadenylation factors have been identified in yeast, only 15 have been found in mammals, suggesting either that the yeast machinery is more complex than its mammalian counterpart, or that more mammalian 3′ processing factors remain to be discovered.
Most of our knowledge of 3′ processing has been based on biochemical studies of individual factors. In comparison, the molecular architecture and dynamics of the 3′ processing complex remain poorly understood. First, it is not clear what proteins, in addition to the known 3′ processing factors, constitute the functional 3′ processing complex, and in what stoichiometry. Secondly, indirect evidence suggests that the 3′ processing complex is dynamic and structural and/or compositional rearrangements may occur during the reactions. For example, although all 3′ processing factors are required for cleavage, CPSF and PAP are enough to reconstitute specific polyadenylation on pre-cleaved RNAs (Colgan and Manley, 1997; Zhao et al., 1999). Based on these observations, it is possible that CstF, CF I and CF II dissociate from the 3′ processing complex after cleavage. Finally, structural information is critical for understanding the detailed mechanisms of 3′ processing. Currently, crystal structures are available for several individual 3′ processing factors (Deo et al., 1999; Bard et al., 2000; Martin et al., 2000; Meinhart and Cramer, 2004; Mandel et al., 2006; Perez-Canadillas, 2006; Bai et al., 2007; Legrand et al., 2007; Noble et al., 2007; Qu et al., 2007; Coseno et al., 2008; Grant et al., 2008; Meinke et al., 2008), but the structure of the functional 3′ processing complex is unknown. Studies of 3′ processing complexes have been hampered by the lack of a method for purifying such complexes in their intact and functional form.
Accumulating evidence suggests that all steps of gene expression are highly coordinated (Hirose and Manley, 2000; Maniatis and Reed, 2002; Proudfoot et al., 2002; Bentley, 2005). For example, 3′ processing is necessary both for transcription termination and for the export of mRNAs, although the exact mechanisms are not fully understood. The coupling of different steps in gene expression is often mediated by physical interactions among factors involved in seemingly distinct processes. It has been shown that the 3′ processing factor CPSF is associated with the transcription machinery as early as in the pre-initiation complex through a direct interaction with TFIID (Dantonel et al., 1997), and both CPSF and CstF can remain associated with RNAP II throughout the coding region (Venkataraman et al., 2005; Glover-Cutter et al., 2008;). In addition, interactions between CPSF and a component of the U2 snRNP, SF3b, are important for coupling between splicing and 3′ processing (Kyburz et al., 2006). A comprehensive characterization of the crosstalk between 3′ processing and other cellular processes will be important for better understanding gene regulation on a systems level.
In this study, we purified the human 3′ processing complex and determined its protein composition. Remarkably, the purified complex contains ~85 proteins. In addition to known polyadenylation factors, we identified new essential 3′ processing factors and over 50 proteins that may mediate coupling with other cellular processes. We also visualized the core 3′ processing complex using electron microscopy for the first time and describe its basic features.
RESULTS AND DISCUSSION
Purification of functional human pre-mRNA 3′ processing complex
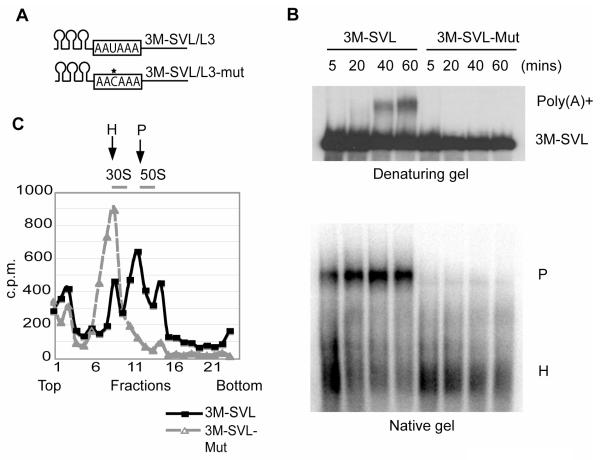
To purify the human pre-mRNA 3′ processing complex, we adopted an RNA-tagging strategy used previously to purify spliceosomal complexes (Jurica et al., 2002; Zhou et al., 2002; Deckert et al., 2006). Briefly, SV40 late (SVL) and adenovirus L3 pre-mRNAs, two commonly used substrates for in vitro 3′ processing analyses (Takagaki et al., 1988), were fused at their 5’ ends to 3 copies of the hairpin that specifically binds to the bacteriophage coat protein MS2 (3M-SVL and 3M-L3, Figure 1A). As controls, we used mutant RNA substrates with single point mutations (U to C) in the highly conserved AAUAAA sequence (3M-SVL-mut and 3M-L3-mut, Figure 1A). In vitro 3′ processing assays showed that as expected this single nucleotide substitution completely abolished cleavage (Figure S1) and polyadenylation (Figure 1B, upper panel). Native gel analyses showed that 3′ processing complexes (P complexes) assembled efficiently on the wild-type RNAs, whereas the mutant RNAs were found almost exclusively in faster-migrating heterogeneous complexes (H complex, Figure 1B, lower panel). Complexes assembled on the wild-type and mutant substrates were further analyzed by glycerol gradient sedimentation (Figure 1C). The RNA distribution profile along the gradient showed that the mutant RNAs were concentrated in a peak of ~30S that corresponds to the H complex. Although a small portion of wild-type RNAs was also present in the same peak, the majority was found in a ~50S peak that corresponds to the 3′ processing complexes (see below).
Figure 1. Characterization of RNA substrates.
(A) A schematic drawing of the RNA substrates. 3MS2 tags (hairpins) and the AAUAAA element in wild-type substrates and AACAAA in mutant substrates (boxes) are shown. The site of the single nucleotide change is marked with an asterisk. (B) Comparison of the wild type and mutant RNA substrates in polyadenylation and complex formation assays. A time course of polyadenylation reactions was analyzed on a denaturing 6% gel (top), or on a 1.5% native agarose gel (bottom), and visualized by phosphorimaging. (C) RNA profiles (measured by radioactivity) on glycerol gradients. The peaks corresponding to the 30S/H and 50S/P complexes, and the peak positions of E. coli 30S and 50S ribosomes from a parallel gradient, are marked.
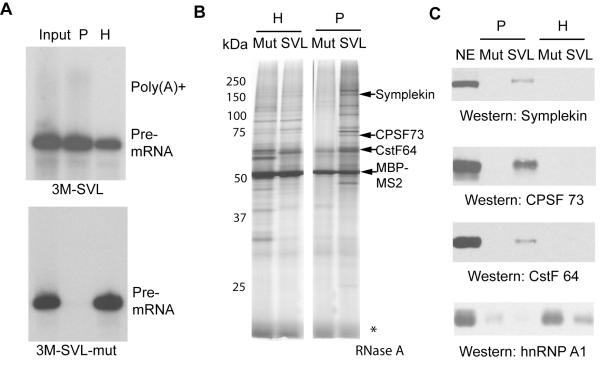
To purify assembled 3′ processing complexes, RNA substrates were first bound to the adaptor protein MBP-MS2 (MBP is maltose-binding protein), and then incubated with HeLa nuclear extract (NE) under polyadenylation conditions (with ATP) to allow assembly of the complexes. Reaction mixtures were then fractionated by glycerol gradient sedimentation as described above. The 30S and 50S fractions were then used for affinity purification with amylose beads. Analysis of the RNAs in the purified complexes showed that the mutant RNA was exclusively found in the 30S/H complexes (Figure 2A, bottom panel). Although some wild-type RNA was also detected in this peak, majority of it was found in the 50S/P complexes (Figure 2A, top panel), consistent with the glycerol gradient profile (Figure 1C). Silver staining of the eluted complexes revealed that a large number of proteins were specifically purified with wild-type substrate in the 50S/P complexes (Figure 2B). The protein profile of the purified 50S/P complex was distinct from that of the purified 30S/H complex (Figure 2B, compare the left and right panel), suggesting that P complexes were effectively separated from the H complexes. Western blotting showed that all 3′ processing factors tested, including CPSF73, CstF64 and symplekin, were specifically detected in the P complexes, but not in the H complexes or with mutant RNA substrates (Figure 2C). In contrast, hnRNP A1, a protein commonly found in H complexes, was highly enriched in the H complex assembled on the mutant substrate. We conclude that we have successfully purified 3′ processing complexes. It is important to point out that under the conditions used there was a significant time window (~20 mins) during which the 3′ processing machinery was fully assembled (Fig 1B, lower panel, from 20 min time point to 40 min) but no significant 3′ processing had occurred (Fig. 1B, top panel). Since we purified the 3′ processing complexes from this time window, the vast majority of the purified complexes were in the pre-cleavage stage.
Figure 2. Purification of 3′ processing complexes.
(A) RNAs from input, and purified 30S/H and 50S/P complexes were purified, resolved and visualized as described above. (B and C) Proteins from the purified 30S/H and 50S/P complexes were resolved by SDS-PAGE and analyzed by silver staining (MBP-MS2 and several western-confirmed bands are labeled) (B) and western blotting (C).
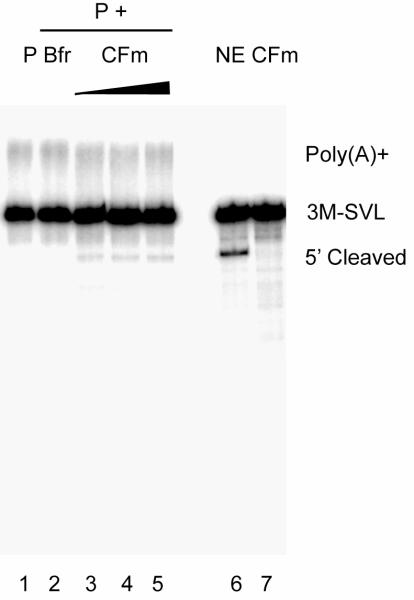
We next wished to determine whether the purified 3′ processing complexes were functional. To this end, we assembled 3′ processing complexes on the 3M-SVL RNA as above. Following purification, the complexes were tested for cleavage activity (in the presence of 3′ dATP, a polyadenylation inhibitor). No significant cleavage was observed with the purified complexes alone (Figure 3, lane 2), possibly indicating that one or more factors were missing or limiting. Indeed, proteomic analyses (see below) indicated that the CF II component Pcf11 was detected at sub-stoichiometric levels and the other CF II subunit, Clp1, was completely missing. We therefore tested whether supplementing the purified complexes with CF components could allow cleavage to take place. To this end, we added partially purified cleavage factor complex (CF) that contains both CF I and II (Takagaki et al., 1989) to the purified 3′ processing complexes, and indeed cleavage products were now detected (Figure 3, lanes 3-5). Therefore, we conclude that the purified 3′ processing complexes were functional in this complementation assay.
Figure 3. Purified 3′ processing complexes are functional in a complementation assay.
Purified 3M-SVL P complexes alone (P, lane 1), supplemented with buffer D (Bfr, lane 2), or increasing amounts of CF fraction (CF, lanes 3-5) were incubated under cleavage conditions. Standard cleavage assays with NE and CF are shown as controls.
It is notable that other highly purified RNA processing complexes, such as the spliceosome, are also not active on their own, but can be activated in complementation experiments analogous to the one described above (Jurica et al., 2002; Zhou et al., 2002; Deckert et al., 2006). Due to the dynamic nature of such complexes, certain factors may be missing at any given time point.
Proteomic analyses of the purified 3′ processing complexes
For proteomic analyses, we purified 3′ processing complexes assembled on the two aforementioned substrates, 3M-SVL and 3M-L3. For comparison, we also purified the H complexes assembled on 3M-SVL-mut RNAs. The protein composition of each complex was determined using the Multidimensional Protein Identification Technology (MudPIT; see Link et al., 1999 and Experimental Procedures). Table 1 lists proteins found in either the SVL and L3 complexes, but only those found in both complexes were considered to be components of the 3′ processing complex.
TABLE 1.
Protein composition of the human pre-mRNA 3′ processing comple
| Protein Name | Accession # | Yeast homolog |
Motifs/notes on functions |
Cal. Mass |
# of peptides |
|
|---|---|---|---|---|---|---|
| L3 | SVL | |||||
| KNOWN POLYADENYLATION FACTORS | ||||||
| CPSF Complex | ||||||
| CPSF160 (CPSF1) | NP_037423 | CFT1 | SFT1 | 160822 | 80 | 72 |
| CPSF100 (CPSF2) | NP_059133.1 | CFT2 | β-CASP | 88487 | 59 | 52 |
| CPSF73 (CPSF3) | NP_057291 | YSH1 | β-CASP | 77486 | 42 | 22 |
| CPSF30 (CPSF4) | NP_006684 | YTH1 | Zinc finger | 30124 | 18 | 13 |
| hFip1 | NP_112179 | FIP1 | 66526 | 23 | 19 | |
| CstF Complex | ||||||
| CstF77 (CSTF3) | NP_001317 | RNA14 | HAT | 82922 | 60 | 54 |
| CstF64 (CSTF2) | NP_001316 | RNA15 | RRM | 60959 | 31 | 22 |
| CstF50 (CSTF1) | NP_001315 | WD repeats | 48358 | 21 | 18 | |
| CF Im Complex | ||||||
| CF Im 68 (CPSF6) | NP_008938 | RRM | 59209 | 15 | 10 | |
| CF Im 59 | NP_079087 | RRM | 52050 | 15 | 12 | |
| Cf Im 25 (CPSF5) | NP_008937 | 26227 | 26 | 21 | ||
| CF IIm Complex | ||||||
| Pcf11 | NP_056969.2 | PCF11 | CID | 173050 | 5 | 3 |
| Other known polyadenylation factors | ||||||
| Symplekin | NP_004810 | PTA1 | 126500 | 51 | 39 | |
| Symplekin variant | BAE06092.1 | 118793 | 48 | 38 | ||
| PAPOLG | NP_075045 | PAP | RRM | 82803 | 2 | 3 |
| PABPC1 | NP_002559.2 | PAB1 | RRM | 70671 | 27 | 33 |
| PABPC4 | NP_003810.1 | RRM | 0783 | 20 | 23 | |
| PABPN1 | NP_004634 | RRM | 31749 | 14 | 12 | |
| (PUTATIVE) HOMOLOGUES OF YEAST POLYADENYLATION FACTORS | ||||||
| CstF64 tau (CSTF2T) | NP_056050.1 | SPAC644.16 (S. pombe) |
RRM | 64436 | 21 | 16 |
| WDR33 | NP_060853 | PFS2 | WD repeats | 145921 | 49 | 45 |
| RBBP6 | NP_008841 | MPE1 | RS, DWNN | 201563 | 4 | 3 |
| PP1 alpha | NP_002699.1 | GLC7 | Phosphatase | 37512 | 2 | 7 |
| PP1 beta | NP_002700 | GLC7 | Phosphatase | 37187 | 2 | 5 |
| DNA DAMAGE RESPONSE FACTORS | ||||||
| DNA-PK | NP_008835 | Kinase | 469093 | 21 | 15 | |
| Ku 70 | NP_001460 | YKU70 | DNA helicase | 69843 | 4 | 7 |
| Ku 86/XRCC5 | NP_066964 | YKU86 | DNA helicase | 82705 | 6 | 13 |
| PARP-1 | NP_001609 | 113135 | 5 | |||
| RNAP II AND ASSOCIATED FACTORS | ||||||
| Pol II large subunit Rpb1 |
NP_000928.1 | RPB1 | Polymerase | 217204 | 2 | 2 |
| Pol II B | NP_000929.1 | RPB2 | 133896 | 2 | ||
| Rpb5 (Pol II E) | NP_002686.2 | RPB5 | 24611 | 2 | ||
| Rpb11 | AF468111_1 | RPB11 | 17208 | 2 | ||
| Transcription Factors | ||||||
| TF II, I | NP_127492.1 | 112416 | 8 | 2 | ||
| TAF15 | AAH46099.1 | RRM, zf-RanBP | 52061 | 6 | 5 | |
| Integrator Complex | ||||||
| INTS2 | NP_065799.1 | 134346 | 2 | 2 | ||
| INTS3 | NP_075391.3 | 118013 | 4 | 9 | ||
| INTS4 | NP_291025.3 | 108171 | 2 | 3 | ||
| INTS6 | NP_036273.1 | 100390 | 2 | 3 | ||
| INTS7 | NP_056249.1 | 106834 | 2 | 2 | ||
| INTS8 | NP_060334.2 | 113088 | 3 | |||
| INTS9 | NP_060720.1 | YSH1 homology | 73815 | 4 | ||
| INTS10 | NP_060612.2 | 82236 | 2 | |||
| PAF Complex | ||||||
| Cdc73 | AAH14351.2 | CDC73 | 46675 | 2 | 2 | |
| FACT Complex | ||||||
| FACT complex large subunit |
NP_009123 | SPT16 | 119914 | 4 | 4 | |
| SPLICING FACTORS | ||||||
| p54nrb | NP_031389.3 | 54232 | 11 | 3 | ||
| PSF | NP_005057.1 | 76150 | 10 | 4 | ||
| PUF60 | NP_510965.1 | RRMs | 59876 | 3 | 9 | |
| UAP56 | NP_004631.1 | SUB2 | RNA Helicase | 48991 | 12 | 9 |
| SF1 | NP_004621.2 | BBP | 68330 | 5 | 5 | |
| SF3A, subunit 1 | NP_005868.1 | PRP21 | Surp | 88886 | 9 | 15 |
| SF3A, subunit 2 | NP_009096.2 | PRP11 | 49256 | 2 | 4 | |
| SF3A60 | CAA57388 | PRP9 | 58777 | 3 | 18 | |
| SF3B, subunit 1 | NP_036565.2 | HSH155 | 145830 | 16 | 34 | |
| SF3B, subunit 2 | NP_006833.2 | CUS1 | 100228 | 2 | 21 | |
| SF3B, subunit 4 | NP_005841.1 | HSH49 | 44386 | 3 | 5 | |
| SF3B, 14 kDa | NP_057131.1 | SNU17 | 14585 | 5 | ||
| Prpf38b | NP_060531.1 | 64441 | 2 | 8 | ||
| SRm300 | NP_057417.2 | RS | 299676 | 13 | 15 | |
| U1 70K | NP_003080.2 | SNP1 | RRM | 51557 | 7 | 7 |
| U2AF65 | NP_009210.1 | MUD2 | RRM | 53501 | 6 | 11 |
| U2AF35 | NP_006749.1 | RRM | 27872 | 2 | 3 | |
| PRP19 | NP_055317.1 | PRP19 | RING, WD | 55181 | 5 | 3 |
| EXOSOME | ||||||
| SKIV2L2 (hMTR4) | NP_056175.2 | MTR4 | DEXD | 117933 | 29 | 28 |
| TRANSLATION FACTORS | ||||||
| eEF1-alpha | NP_001393.1 | TEF1/2 | GTPase | 50141 | 14 | 12 |
| eEF1-gamma | NP_001395.1 | TEF4 | GST | 50119 | 5 | 4 |
| eIF2A | NP_114414.2 | YGR054W | 64990 | 4 | 4 | |
| eIF 4A | NP_001407.1 | TIF1 | Helicase | 46154 | 12 | 6 |
| eIF3 gamma | NP_003747.1 | JAB/MPN | 39930 | 5 | 6 | |
| eIF3S2 | NP_003748.1 | TIF34 | WD | 36502 | 2 | 2 |
| eIF3S5 | NP_003745.1 | PCI | 37564 | 3 | 2 | |
| eIF3S6 | NP_001559.1 | 52221 | 2 | 4 | ||
| eIF3S9 eta | NP_003742.2 | RRM | 92482 | 7 | 7 | |
| eIF3A | NP_003741.1 | RPG1 | 166569 | 16 | 9 | |
| eIF4G1 | NP_886553.2 | TIF4631 | 175460 | 2 | 2 | |
| RACK1/GNB2L1/lung cancer oncogene 7 |
NP_006089.1 | ASC1 | WD | 35077 | 5 | 5 |
| FACTORS WITH KNOWN MOTIFS | ||||||
| RNA binding motif protein 7 |
NP_057174.1 | RRM | 30503 | 4 | 4 | |
| RNA binding motif protein 9/Ataxin- binding protein/Fox-1 |
NP_055124 | RRM | 39515 | 2 | 7 | |
| RNA binding motif protein 25 |
NP_067062.1 | RRM | 100186 | 4 | 16 | |
| RNA binding motif protein 39, isoform a |
NP_909122.1 | RRM | 59380 | 2 | 4 | |
| RNA binding motif protein 39, isoform b |
NP_004893.1 | RRM | 58657 | 2 | 4 | |
| DEAH box polypeptide 3 |
NP_001347.3 | DBP1 | RNA helicase | 7244 | 13 | 9 |
| DEAH box | NP_004387.1 | DBP2 | RNA helicase | 69148 | 4 | 5 |
| polypeptide 5 | ||||||
| DEAH box polypeptide 9 |
NP_001348.2 | RNA helicase | 140958 | 32 | 3 | |
| DEAH box polypeptide 15 |
NP_001349.2 | PRP43 | RNA helicase | 90933 | 3 | 17 |
| DEAH box polypeptide 36 |
NP_065916.1 | RNA helicase | 114776 | 12 | 27 | |
| DEAH box polypeptide 39 |
NP_005795.2 | RNA helicase | 49130 | 8 | 5 | |
| DEAD box polypeptide 6 |
NP_004388.1 | DHH1 | RNA helicase | 82432 | 3 | 2 |
| DEAD box polypeptide 17 (p82) |
NP_006377.2 | RNA helicase | 80273 | 6 | 4 | |
| Gemin3 (DEAD box polypeptide 20) |
NP_009135.3 | RNA helicase | 92213 | 4 | 2 | |
| DEAD box polypeptide 23 |
NP_004809.2 | PRP28 (S. Pombe) |
RNA helicase | 95583 | 4 | 5 |
| ZCCHC8 | NP_060082 | Zinc finger | 79375 | 15 | 16 | |
| EBNA2 coactivator, p100/SND1 |
NP_055205.2 | Staphylococcal nuclease domain, Tudor |
101997 | 12 | 2 | |
| Interferon-induced protein with tetratricopeptide repeats 1 isform 2 |
NP_001539.3 | TPR | 55360 | 9 | 7 | |
| Interferon-induced protein with tetratricopeptide repeats 3 |
NP_001540 | TPR | 55985 | 8 | 5 | |
| OTHER FACTORS | ||||||
| G3BP/Ras-GTPase- activating protein SH3-domain-binding protein |
NP_005745.1 | Endoribonuclease | 52164 | 5 | 2 | |
| JUP protein/ gamma- catenin |
NP_002221.1 | 81745 | 5 | 3 | ||
| PNUTS (PPP1R10) | NP_002705.2 | PP1 regulator | 99058 | 6 | 9 | |
Components of multi-subunit complexes that are present in only one purified complexe are listed and lightly shaded.
Proteins that are abundant in H complexes were designated H complex components and not listed here.
The purified 3′ processing complexes contained ~85 proteins, including nearly all previously known 3′ processing factors. The only exception was Clp1, a component of the CF II complex (de Vries et al., 2000), which plays an unknown role in 3′ processing. Pcf11, another subunit of CF II, also seemed to be present at low levels, as only a small number of unique peptides from this protein were detected. These observations indicate that the association between CF II and the core 3′ processing complex may be weak and/or transient. Interestingly, instead of the canonical PAP (Lingner et al., 1991; Raabe et al., 1991), the related neo-PAP/PAPOLG (Kyriakopoulou et al., 2001; Topalian et al., 2001) was the sole PAP detected. The reason for this is unclear, but is consistent with many studies indicating that PAP is not tightly associated with other processing factors (e.g., (Takagaki et al., 1988)), and may reflect a dynamic association between PAP and the core 3′ processing complex and/or functional redundancy between the two poly(A) polymerases.
The complexes described above were assembled under polyadenylation conditions (with ATP). For comparison, we also purified 3′ processing complexes assembled under cleavage conditions (in the presence of the polyadenylation inhibitor 3′- dATP and without ATP) and analyzed them by mass spectrometry (Figure S2 and Table S1). Again, most RNAs within the purified complexes were unprocessed (Figure S2), indicating the majority of the purified complexes were in the pre-cleavage stage. Comparison between Tables 1 and S1 shows that the protein compositions of the 3′ processing complexes assembled under the two conditions were highly similar. For example, under both conditions, all subunits of CPSF, CstF, and CF I were identified while CF II components were either detected only by a small number of unique peptides or entirely missing. In addition, many other factors, including WDR33, PP1, Rbbp6, and CstF64 tau (see below), were identified in both analyses. These observations suggest that the assembly conditions have very little effect on the general protein composition of the pre-cleavage 3′ processing complexes.
Characterization of new 3′ processing factors
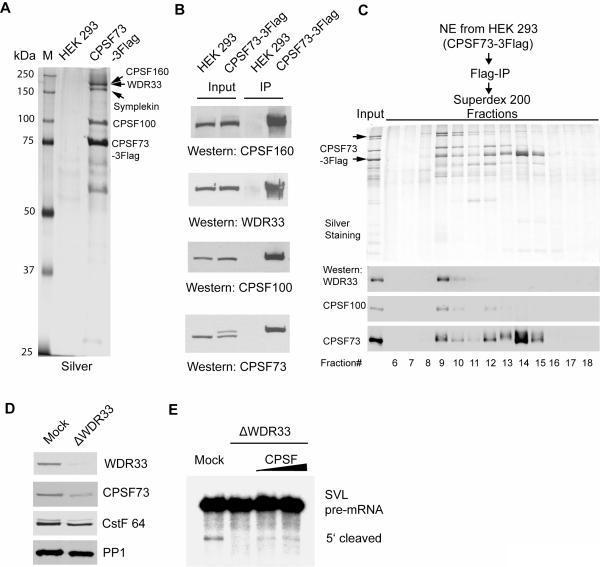
Three proteins identified in our study not previously implicated in mRNA 3′ processing in mammals (WDR33, Rbbp6, and PP1) are known or putative homologues of yeast 3′ processing factors. WDR33/WDC146 is a WD40 repeat-containing protein, and is the putative mammalian homologue of the yeast 3′ processing factor Pfs2 (Ohnacker et al., 2000). To characterize its potential functions in 3′ processing, we first tested whether WDR33 interacts with the CPSF complex since Pfs2 binds strongly to Ysh1, the yeast homologue of CPSF73 (Ohnacker et al., 2000). To this end, we established a stable cell line expressing Flag-tagged CPSF73 and purified CPSF73 and associated proteins by immunoprecipitation (IP) (Figure 4A). We analyzed the CPSF73-containing complexes by mass spectrometry and the identified proteins are listed in Table S2, next to those found in the 3′ processing complexes. All the known subunits of the CPSF complex (CPSF-160, -100, -73 and -30, and Fip1) were identified. Although not detected in CPSF previously purified through multiple chromatographic steps (Bienroth et al., 1991; Murthy and Manley, 1992), symplekin was detected at close to stochiometric levels, consistent with previous studies that it associates with CPSF as well as CstF (Takagaki and Manley, 2000). Strikingly, WDR33 was also present in the CPSF complex, as indicated by the large number of unique peptides detected (Figure S3 and Table S2) and confirmed by western blotting (Figure 4B). The WDR33 band partially overlapped the CPSF160 band on SDS-PAGE (Figure 4A), perhaps explaining in part why WDR33 previously escaped detection. Gel filtration analysis of the purified CPSF complex showed that WDR33 co-eluted with CPSF (Figure 4C).
Figure 4. WDR33 is a bona fide component of the CPSF complex.
(A and B) Chacterization of the immuopurified CPSF73 complex. Proteins from Flag-IP of NE from HEK293 or the CPSF73-3Flag cell line were resolved by SDS-PAGE and stained with silver (A), or analyzed by Western blotting using specific antibodies (B). Protein bands in (A) whose identities were confirmed by western are labeled. (C) Immunopurified CPSF complexes were analyzed by gel filtration using a Superdex-200 column. Proteins from input and each fraction were analyzed by SDS-PAGE and silver staining (top panel) and western (bottom panel). WDR33 is marked by a arrow. (D) Western analyses of the mock-treated (mock) or WDR33-depleted (ΔWDR33) NE. Intensities of individual bands were quantified using the Odyssey Infrared scanner (Li-Cor). (E) WDR33 is essential for 3′ processing. Mock, ΔWDR33 NE alone, or ΔWDR33 supplemented with immunopurified CPSF were used in standard cleavage assays. RNAs were resolved and visualized as described above.
We next immuno-depleted WDR33 from NE to determine whether WDR33 is required for 3′ processing. Quantitative western analyses showed that ~95% of WDR33 was removed (Figure 4D). About two thirds of CPSF73 was also co-depleted, indicating that the majority of CPSF73 is associated with WDR33. CstF64 levels were reduced by about a third while the level of the phosphatase PP1 was not significantly affected. When WDR33-depleted NE was used in 3′ processing assays, both cleavage (Figure 4E) and polyadenylation (Figure S4) were essentially abolished. Add-back of the immunopurified CPSF complex restored cleavage (Figure 4E). Together, these data indicate that WDR33, despite the fact that it had not been previously identified, is a bona fide component of the CPSF complex and suggest that it plays an essential role in mammalian 3′ processing.
Pfs2 was initially suggested to be the yeast equivalent of mammalian CstF50, another WD40 repeat protein with which it shares limited similarity (Ohnacker et al., 2000). This was consistent with the fact that the yeast CstF equivalent, CF IA, lacks a CstF50 homologue (Zhao et al., 1999). However, out data now indicates that the human 3′ processing machinery contains two WD40 proteins, one in CPSF and another in CstF. It is noteworthy that the plant PFS2/WDR33 homologue, FY, has been implicated as playing an important role during floral transition (Simpson et al., 2003).
Rbbp6/PACT is the putative homologue of the yeast 3′ processing factor Mpe1 (Vo et al., 2001). Interestingly, Rbbp6 was originally identified as a p53- and Rb-binding protein, playing important roles in apoptosis, cell cycle, and p53 regulation (Sakai et al., 1995; Simons et al., 1997). Rbbp6 is significantly larger than Mpe1, and contains additional domains, including an arginine/serine-rich (RS) domain that is found in many splicing factors, and a RING-finger-related domain (Pugh et al., 2006). Although we have not directly tested its role in processing, it is likely that Rbbp6, like its yeast counterpart Mpe1, functions in 3′ processing. It is possible that Rbbp6 may link mRNA 3′ end formation to the Rb/p53 pathways and tumorigenesis.
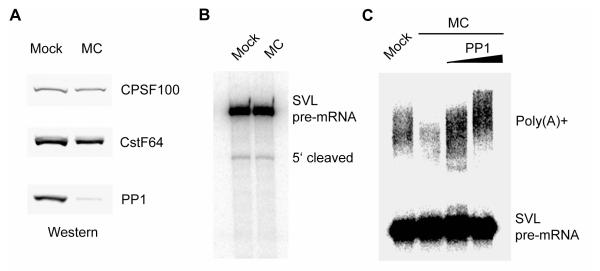
Our results show that the serine/threonine phosphatase PP1 and its regulator PNUTS are components of the human 3′ processing complex (Table 1). The PP1 homolog in yeast, Glc7, is a known 3′ processing factor, and its phosphatase activity is specifically required for polyadenylation, but not for cleavage (He and Moore, 2005). To test if PP1 is involved in mammalian 3′ processing, we depleted PP1 and the related PP2A family phosphatases using microcystin (MC)-conjugated beads (Figure 5A). MC is a specific small-molecule inhibitor of the PP1/2A phosphatases, and we have shown previously that MC-conjugated beads can be used to efficiently deplete these phosphatases from NE (Shi et al., 2006). When mock or MC-treated NE were used in standard in vitro 3′ processing assays, similar levels of cleaved products were observed (Figure 5B). Polyadenylation, however, was significantly reduced in MC-treated NE and add-back of recombinant PP1 restored polyadenylation (Figure 5C), suggesting that PP1 is specifically required for polyadenylation. Therefore, dephosphorylation by PP1 is an evolutionarily conserved step in 3′ processing.
Figure 5. PP1 is required for polyadenylation, but not for cleavage.
(A) Western analyses of mock-treated (Mock) or microcystin-treated (MC) NE. (B) Depletion of PP1/2A phosphatases has no effect on cleavage. Mock and MC NE were used in standard in vitro cleavage assays with SVL as the substrate. Pre-mRNA and the 5’ cleaved products are marked. (C) PP1 is required for polyadenylation. Mock, MC NE alone, or MC NE supplemented with recombinant PP1 were used in standard polyadenylation assays with SVL as the substrate. Pre-mRNA and poly(A)+ RNAs are marked.
We also found CstF64 tau in our purified 3′ processing complex. CstF64 tau is highly homologous to CstF64, and reportedly found only in testis and not expressed in HeLa cells (Wallace et al., 1999). To determine whether CstF64 tau is a component of the CstF complex, we established a HEK293 cell line stably expressing Flag-tagged CstF77 and purified the CstF complex by IP (Figure S5A). Results of mass spectrometry analyses of purified CstF are listed in Table S2, and CstF64 tau was indeed detected (Figure S5B and Table S2). We suspect that CstF64 tau may be a general component of the CstF complex. An intriguing possibility is that CstF complexes may contain either CstF64 or CstF64 tau, and their functions may be partially redundant. This is consistent with earlier observations that a ~90% reduction in CstF64 levels had no significant effect on cell growth (Takagaki and Manley, 1998), and that CstF may function as a dimer in 3′ processing (Bai et al., 2007).
Accumulating evidence suggests that all steps of gene expression are highly coordinated, and coupling of different steps is often mediated by physical interactions among factors involved in seemingly distinct processes (Hirose and Manley, 2000; Maniatis and Reed, 2002; Proudfoot et al., 2002; Bentley, 2005). Consistent with this theme, we identified in our purified 3′ processing complex a large number of proteins that have known or putative functions in transcription and splicing, both of which are known to be connected to 3′ processing. In fact, splicing factors detected in our complexes, such as SF3b, U2AF, and U1-70K, have been shown to associate with specific 3′ processing factors and mediate crosstalk between splicing and polyadenylation (Gunderson et al., 1998; Vagner et al., 2000; Kyburz et al., 2006). We detected RNAP II in the purified 3′ processing complex, consistent with earlier findings that RNAP II is necessary for efficient 3′ cleavage in vitro (Hirose and Manley, 1998). In addition, we identified the PAF complex, a RNAP II-associated transcription elongation factor that was recently shown to function in 3′-end formation of polyadenylated mRNAs in yeast (Penheiter et al., 2005). Most subunits of another RNAP II-associated complex, the Integrator, were also found in the 3′ processing complexes. The Integrator complex was recently shown to function in the 3′ processing of snRNAs and two of its subunits, Ints9 and Ints11, display sequence homology with CPSF 100 and 73 respectively (Baillat et al., 2005). The integrator subunits INTS8, 9 and 10 were missing in the L3 complex. The reason for their absence is unclear, but may be due to slightly lower levels of the Integrator complex in the L3 complexes or the association between these subunits and the rest of the complex might be weak and/or transient. It is currently unclear what, if any, role the Integrator might play in the 3′ processing of pre-mRNAs.
Our study also identified a number of factors that may mediate unexpected connections between 3′ processing and other cellular processes. For example, we found that the DNA-activated protein kinase complex (DNA-PKcs/Ku70/Ku86), well studied for its functions in DNA damage repair, is associated with the 3′ processing complex (Table 1). This is potentially similar to transcription where several DNA repair factors, such as XPB and XPD, are also essential transcription factors as components of the TFII H (Drapkin et al., 1994). These results are also consistent with previous studies showing that 3′ processing is connected to DNA damage response (Kleiman and Manley, 2001; Mirkin et al., 2008) Another intriguing factor associated with the 3′ processing complex was the translation elongation factor and GTPase eEF1 alpha. Interestingly, Tef1, the yeast homologue of eEF1 alpha, co-purifies with the yeast 3′ processing factor CF I (Gross and Moore, 2001). It will be of interest to examine what, if any, roles these and other factors identified in our proteomic analyses play in 3′ processing.
A comparison between our proteomic analyses of the 3′ processing complexes and previously studies of the spliceosome (Jurica et al., 2002; Zhou et al., 2002; Deckert et al., 2006) revealed both similarities and differences. One common theme is that all of these studies strongly support a link between splicing and 3′ processing. An interesting difference is that although the spliceosome includes a number of factors that have been implicated in transcription (e.g. TREX and TAT-SF1) (Zhou et al., 2002), the 3′ processing complex contains RNAP II and RNAP II-associated factors, such as the Integrator and PAF (Table 1). Therefore, splicing and 3′ processing are both connected to transcription, but probably by different factors.
Structural analyses of 3′ processing complexes by EM
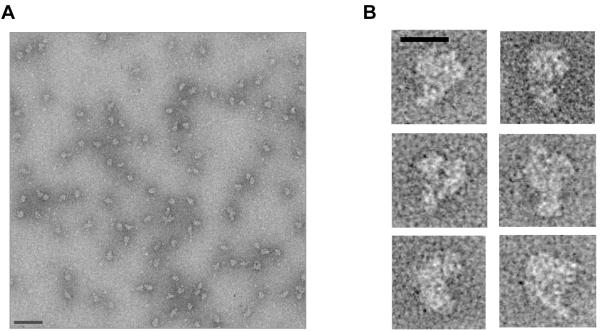
We next wished to begin to analyze the structure of the 3′ processing complex. To this end, the isolated complexes were processed following the GraFix method (Kastner et al., 2008). Briefly, the 3′ processing complexes purifed as described above were subject to a second glycerol gradient sedimentation during which the complexes are centrifuged into increasing concentration of the fixation reagent glutaraldehyde. This step serves to further purify and gently fix the complexes to preserve their integrity, and has been successfully used, for example, to process spliceosomes for electron microscopic (EM) analyses (Deckert et al., 2006; Behzadnia et al., 2007). During this second glycerol gradient sedimentation, the 3′ processing complexes were found again in a ~50S peak (data not shown), indicating that structural integrity of the complex was preserved throughout the purification. The peak fraction was negatively stained with uranyl formate using the carbon sandwich method (Radermacher et al., 1987), and analyzed by EM. A typical raw image of the 3′ processing complex shows monodisperse particles of similar sizes (Figure 6A). We also obtained tilted images of the particles to confirm that they were fully sandwiched between carbon membranes, and that the staining was homogeneous (Fig. S6). Images of representative particles show a distinct “kidney” shape, slightly elongated and bent (Figures 6B). There appears to be a central cavity surrounded by two or more peripheral densities. The maximum dimension of the complex is ~250 Å, which is consistent with its ~50S sedimentation coefficient.
Figure 6. EM analyses of the purified 3′ processing complex.
(A) A typical CCD micrograph of the negatively stained purified 3′ processing complexes. Bar, 100 nm. (B) A gallery of representative particles. Bar, 20 nm.
3,671 molecular images were collected for image processing using both SPIDER (Frank, 1996) and EMAN (Ludtke et al., 1999). After reference-free alignment and classification, 50 (using SPIDER, shown in Fig. S7) and 47 (using EMAN, shown in Fig. S8) two-dimensional class averages were obtained. Although most of the class averages have defined edges and consistent sizes, they seem to lack strong internal features. This could potentially be due to heterogeneity among particles caused by the dynamic nature of this complex and/or the presence of substoichiometric factors. Another possible explanation is that classification of negative stain images focuses on the shape of the boundary, so that small changes in orientation will not affect the class designation yet can result in changes of internal features seen in projection. As a result, internal features of class averages can be blurred. Nonetheless, our results have provided a first view of the polyadenylation complex and revealed its general structural features.
Given the seemingly simple nature of the polyadenylation reaction, it is remarkable that it involves such a large complex. The size of the 3′ processing complex is close to that of the bacterial ribosome large subunit (Radermacher et al., 1987) and the spliceosomal A complex (Behzadnia et al., 2007). The major components of our purified complexes, such as CPSF, symplekin, CstF, and CF I, likely constitute the “core” of the 3′ processing complex seen in the class averages, as their collective molecular weight (~1.1 MDa) is already close to that of the bacterial ribosome large subunit (1.5 MDa) (Radermacher et al., 1987). These core factors, at least one of which, CstF, may be present as a dimer (Bai et al., 2007), likely correspond to some of the observed major densities. For the rest of the factors identified in the proteomics (total molecular weight ~7 MDa), the majority are present at sub-stoichiometric levels and likely contribute to the heterogeneity observed among particles. The structure described here, we believe, corresponds to the core polyadenylation machinery.
In this study, we purified functional human pre-mRNA 3′ processing complexes and determined their protein composition. We detected all but one known 3′ processing factor and identified several new and potentially essential ones. We identified a number of proteins involved in other cellular processes, expanding the view that 3′ processing is integrated with other cellular events. We also visualized the 3′ processing complex for the first time and characterized its basic structural features. Together, our study has provided critical insights into the molecular composition and the structure of the 3′ processing complex, revealing a molecular architecture that is much more complex than previously expected.
EXPERIMENTAL PROCEDURES
Antibodies
Anti-CPSF160, 100, 73, and WDR33 were kindly provided by Orit Rosenblatt and Bethyl Laboratories; anti-CstF64 6A9 was described previously (Takagaki et al., 1990); anti-symplekin was from BD Biosciences; anti-hnRNP A1 was from ImmuQuest.
In vitro 3′ processing assays
Constructs used in this study were derived from pG3SVL-A and pG3L3-A, which contain the SV40 late site and adenovirus 2 L3 poly(A) site, respectively (Takagaki et al., 1988). 3 MS2-binding sequences were as described previously (Zhou et al., 2002), and were inserted between Acc I and Xba I sites before the SVL and L3 sequences. 32P-labeled pre-mRNAs were prepared with SP6 RNA polymerase (Promega) from linearized plasmids. Polyadenylation reactions typically contain: 8pmol radio-labled RNA/ml reaction, 40% NE, 8.8mM HEPES (pH 7.9), 44mM KCl, 0.4 mM DTT, 0.7 mM MgCl2, 1mM ATP, 20 mM creatine phosphate. In cleavage reactions, ATP was omitted, and 0.2 mM 3′ dATP (Sigma), 2.5% PVA, and 40 mM creatine phosphate were added.
Purification of 3′ processing complexes
Radio-labeled RNA substrates were incubated with 50-molar excess of MBP-MS2 adaptor protein for 30 mins on ice. Then the other ingredients of the polyadenylation reaction were added, and the reactions were incubated in 200 μl aliquots at 30°C for 40 mins or otherwise specified time. The reactions were chilled and loaded onto 11ml 10-30% glycerol gradients (20 mM HEPES pH 7.9, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT). The gradients were centrifuged at 22,000rpm for 16 hours in a SW41 rotor, and then 500 μl fractions were manually collected from the top to the bottom. Radioactivity of each fraction was measured using a liquid scintillation counter. Peak fractions were pooled and mixed with amylose beads for 1 hour at 4°C. After washing with wash buffer (20 mM HEPES pH 7.9, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT), the complexes were eluted in wash buffer plus 12 mM maltose. For mass spectrometry analyses, the eluted complexes were treated with RNase A and precipitated before analyses. For purification of complexes assembled on mutant substrates and cleavage complexes, reaction mixtures were loaded onto a Sephacryl S-400 size-exclusion column and RNP-containing fractions were pooled and used for affinity purification with amylose beads (Jurica et al., 2002).
Proteomic analyses of purified 3′ processing complex using multidimensional protein identification technology (MudPIT)
Precipitated protein preparations were dissolved in digestion buffer, digested by trypsin, and analyzed by LC/LC/MS/MS according to published protocols (Link et al., 1999). MS/MS spectra obtained were analyzed by SEQUEST using a non-redundant NCBI protein database. The SEQUEST outputs were then analyzed by DTASelect™ (version 2.0) program. The type of digestion method used was specified (-trypstat for tryptic digests) so as to specifically filter for peptides with trypsin specificity. A user-specified false positive rate was used to dynamically set XCorr and DeltaCN thresholds through quadratic discriminant analysis. This dataset was then further filtered to remove contaminants (i.e. keratin) through the use of Contrast (version 2.0). A minimum of 2 peptides and half tryptic status (−p 2 –y 1) were set in the Contrast.params file. For analyses of the cleavage complexes, mininum peptide number was set to 1.
Immuno-purification of CPSF73- and CstF77-associated proteins
The plasmids CPSF73-3Flag-pCMV14 and Flag-CstF77-pCDNA3.1 were transfected into HEK293 cells, and stable transfectants were selected using G418 (Invitrogen). NE was made from these stable cell lines using standard protocol, and IP was performed using M2 beads (Sigma). For functional analyses, eluted proteins were concentrated using Centricon Y-30 (Millipore) and directly used in in vitro 3′ processing assays.
Immuno-depletion of WDR33
100 μl NE was diluted with equal volume of Buffer D and NP-40 was added to final concentration of 0.1%. The diluted NE was then mixed with either protein G-agarose (mock) or with anti-WDR33-conjugated protein G-agarose (ΔWDR33) for 2 hours at 4°C. The depletion efficiency was measured by quantitative western blotting using the Odyssey infrared scanner (Li-Cor).
Electron Microscopy
A 9 ml polyadenylation reaction was used for purification as described above. Following affinity purification, the complexes were eluted in 300 μl and further treated using the GraFix method (Kastner et al., 2008). Briefly, the eluted complexes were loaded on a 4 ml 10-30% glycerol and 0-0.1% glutaraldehyde gradient, and centrifuged at 51,000 rpm for 2.5 hours in an SW55Ti rotor. Afterwards, 180 μl fractions were taken manually from the top. Negative staining was performed using the carbon-sandwich method (Radermacher et al., 1987). Images were acquired on a JEOL JEM2100F electron microscope operating at 200 kV. Imaging was performed at a set magnification of 30000X under low-dose conditions at an underfocus of 2.3 microns, and images were collected on a Tietz 224HD 2K×2K CCD camera with 24 micron pixel size. The calibrated pixel size was 5.11 A/pixel. Images were processed using the standard SPIDER protocol (Frank, 1996) or the refine2d.py script from EMAN (Ludtke et al., 1999).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. M. Jurica, J. Vilardell, and O. Rosenblatt for providing reagents; KD. Derr and Dr. R. Diaz at the New York Structural Biology Center for technical assistance; Drs. A. Tzagoloff and R. Gonzalez for sharing equipments; B. Reddy for help in the early stage of this study; Dr. V. Vathantham for providing DNA constructs and other members of the Manley lab for helpful discussions. This work was supported by NIH grants GM028983 to J.L.M, P41 RR011823 to J. Y, and R37 GM29169 and GM55440 to J.F. J.F. is a Howard Hughs Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bard J, Zhelkovsky AM, Helmling S, Earnest TN, Moore CL, Bohm A. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science. 2000;289:1346–1349. doi: 10.1126/science.289.5483.1346. [DOI] [PubMed] [Google Scholar]
- Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, Luhrmann R. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem. 1991;266:19768–19776. [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Coseno M, Martin G, Berger C, Gilmartin G, Keller W, Doublie S. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 2008;36:3474–3483. doi: 10.1093/nar/gkn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- Drapkin R, Sancar A, Reinberg D. Where transcription meets repair. Cell. 1994;77:9–12. doi: 10.1016/0092-8674(94)90228-3. [DOI] [PubMed] [Google Scholar]
- Frank J. Three-dimensional electron microscopy of macromolecular assemblies. Academic Press; San Diego, CA: 1996. [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RP, Marshall NJ, Yang JC, Fasken MB, Kelly SM, Harreman MT, Neuhaus D, Corbett AH, Stewart M. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J Mol Biol. 2008;376:1048–1059. doi: 10.1016/j.jmb.2007.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Moore C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc Natl Acad Sci U S A. 2001;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- He X, Moore C. Regulation of yeast mRNA 3′ end processing by phosphorylation. Mol Cell. 2005;19:619–629. doi: 10.1016/j.molcel.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Kyriakopoulou CB, Nordvarg H, Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J Biol Chem. 2001;276:33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- Legrand P, Pinaud N, Minvielle-Sebastia L, Fribourg S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 2007;35:4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W, Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Meinke G, Ezeokonkwo C, Balbo P, Stafford W, Moore C, Bohm A. Structure of yeast poly(A) polymerase in complex with a peptide from Fip1, an intrinsically disordered protein. Biochemistry. 2008;47:6859–6869. doi: 10.1021/bi800204k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin N, Fonseca D, Mohammed S, Cevher MA, Manley JL, Kleiman FE. The 3′ processing factor CstF functions in the DNA repair response. Nucleic Acids Res. 2008;36:1792–1804. doi: 10.1093/nar/gkn005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KG, Manley JL. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- Noble CG, Beuth B, Taylor IA. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 2007;35:87–99. doi: 10.1093/nar/gkl1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnacker M, Barabino SM, Preker PJ, Keller W. The WD-repeat protein pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 2000;19:37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006;25:3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Pugh DJ, Ab E, Faro A, Lutya PT, Hoffmann E, Rees DJ. DWNN, a novel ubiquitin-like domain, implicates RBBP6 in mRNA processing and ubiquitin-like pathways. BMC Struct Biol. 2006;6:1. doi: 10.1186/1472-6807-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Perez-Canadillas JM, Agrawal S, De Baecke J, Cheng H, Varani G, Moore C. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J Biol Chem. 2007;282:2101–2115. doi: 10.1074/jbc.M609981200. [DOI] [PubMed] [Google Scholar]
- Raabe T, Bollum FJ, Manley JL. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional structure of the large ribosomal subunit from Escherichia coli. EMBO J. 1987;6:1107–1114. doi: 10.1002/j.1460-2075.1987.tb04865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Saijo M, Coelho K, Kishino T, Niikawa N, Taya Y. cDNA sequence and chromosomal localization of a novel human protein, RBQ-1 (RBBP6), that binds to the retinoblastoma gene product. Genomics. 1995;30:98–101. doi: 10.1006/geno.1995.0017. [DOI] [PubMed] [Google Scholar]
- Shi Y, Reddy B, Manley JL. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol Cell. 2006;23:819–829. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Simons A, Melamed-Bessudo C, Wolkowicz R, Sperling J, Sperling R, Eisenbach L, Rotter V. PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene. 1997;14:145–155. doi: 10.1038/sj.onc.1200825. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Kaneko S, Gonzales MI, Bond GL, Ward Y, Manley JL. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol Cell Biol. 2001;21:5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo LT, Minet M, Schmitter JM, Lacroute F, Wyers F. Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol Cell Biol. 2001;21:8346–8356. doi: 10.1128/MCB.21.24.8346-8356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci U S A. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.