Summary
How spatial information is translated into a chemical signal is a fundamental problem in all organisms. The spindle position checkpoint is a prime example of this problem. This checkpoint senses spindle position and, in budding yeast, inhibits the Mitotic Exit Network (MEN), a signaling pathway that promotes exit from mitosis. We find that spindle position is sensed by a system composed of MEN inhibitory and activating zones and a sensor that moves between them. The MEN inhibitory zone is located in the mother cell, the MEN activating zone in the bud and the spindle pole body (SPB), where the components of the MEN reside, functions as the sensor. Only when an SPB escapes the MEN inhibitor Kin4 in the mother cell and moves into the bud where the MEN activator Lte1 resides can exit from mitosis occur. In this manner, spatial information is sensed and translated into a chemical signal.
Keywords: spindle position, spindle position checkpoint, mitotic exit, Kin4, cell polarity, centrosome asymmetry
Introduction
Polarized cell division, the process of dividing the cell along a predetermined axis, is central to the development of complex biological systems. Cell intrinsic and/or extrinsic cues establish these axes and the mitotic spindle must be positioned along them to ensure that each daughter cell receives a full genomic complement. Feedback mechanisms have been identified in cultured rat kidney cells and Drosophila germline stem cells that delay the cell cycle in response to defects in spindle position, but this mechanism is best understood in budding yeast (Cheng et al., 2008; O'Connell and Wang, 2000; Yeh et al., 1995). In yeast, the site of bud formation and therefore cytokinesis is determined during G1. Thus, the axis of division is defined prior to mitosis and the mitotic spindle must be aligned along this mother – bud axis during every cell division. When this process fails, a surveillance mechanism known as the spindle position checkpoint (SPOC) delays mitotic exit to provide the cell with an opportunity to reposition the spindle and thus partition the genetic material equally prior to spindle disassembly and cytokinesis. When this surveillance mechanism fails, cells that mis-position their spindles give rise to mitotic products with too many or too few nuclei (Yeh et al., 1995).
Mitotic exit is controlled by a signaling pathway known as the Mitotic Exit Network (MEN). The central switch of the MEN is the small GTPase, Tem1, which localizes to spindle pole bodies (SPBs, yeast centrosomes) during mitosis (Bardin et al., 2000; Molk et al., 2004; Pereira et al., 2000; Shirayama et al., 1994b). At the SPB, activated Tem1 (presumably Tem1-GTP) transduces a signal through a two kinase cascade ultimately resulting in the activation of the phosphatase, Cdc14. Cdc14 brings about the inactivation of mitotic cyclin-dependent kinases, which results in spindle disassembly, cytokinesis and, ultimately, resetting of the cell to a G1-like state (reviewed in (Stegmeier and Amon, 2004)).
Tem1 is positively regulated by the bud cortex localized protein, Lte1, and negatively by the two-component GTPase activating protein (GAP) complex, Bub2 - Bfa1 (Bardin et al., 2000; Bloecher et al., 2000; Geymonat et al., 2002; Lee et al., 1999; Li, 1999; Pereira et al., 2000; Shirayama et al., 1994a). The GAP complex in turn is inhibited by the Polo-like kinase, Cdc5 (Geymonat et al., 2003; Hu et al., 2001). During metaphase, Tem1 and the GAP complex localize to both SPBs; during anaphase they become concentrated at the SPB that migrates into the bud (Bardin et al., 2000; Fraschini et al., 1999; Lee et al., 1999; Li, 1999; Molk et al., 2004; Pereira et al., 2000).
The SPOC inhibits the MEN at the level of Tem1 activation. The protein kinase Kin4 and the protein phosphatase PP2A-Rts1 are essential for SPOC function (Chan and Amon, 2009; D'Aquino et al., 2005; Pereira and Schiebel, 2005). Kin4 localizes to the mother cell cortex throughout the cell cycle and at the mother spindle pole body in mid-anaphase where it phosphorylates Bfa1 and protects the GAP complex from inhibitory phosphorylation by Cdc5 (Maekawa et al., 2007). In cells with mis-positioned spindles, Kin4 associates with both anaphase SPBs, phosphorylates the GAP complex at both SPBs and thus inhibits the MEN (Maekawa et al., 2007; Pereira and Schiebel, 2005). The protein phosphatase PP2A-Rts1 regulates Kin4’s phosphorylation state and promotes the loading of Kin4 onto SPBs, which is essential for SPOC function (Chan and Amon, 2009).
How the cell monitors spindle position and translates this spatial information to regulate MEN is only partially understood. Emergence of a Tem1 bearing SPB into the bud where Lte1 resides ensures that Tem1 activation by Lte1 only occurs when the spindle elongates along the mother-bud axis (Bardin et al., 2000; Pereira et al., 2000). The importance of spatial restriction of Lte1 to the bud is underscored by experiments that inappropriately place Lte1 in the mother cell thus leading to premature mitotic exit when the spindle is mispositioned (Bardin et al., 2000; Castillon et al., 2003; Geymonat et al., 2009). Fission yeast appears to promote activation of the Tem1 homolog, Spg1, through a strikingly similar mechanism (Garcia-Cortes and McCollum, 2009).
However, restricting Lte1 to the bud cannot be the only mechanism that regulates Tem1 in response to spindle position because Lte1 is only essential for mitotic exit at low temperatures (Adames et al., 2001; Shirayama et al., 1994a). Many other mechanisms have been proposed to act in addition to Lte1 (Adames et al., 2001; D'Aquino et al., 2005; Fraschini et al., 2006). We have proposed the key parallel mechanism to be Kin4 mediated MEN inhibition (D'Aquino et al., 2005) and based on the restriction of Kin4 to the mother cell cortex and Lte1 to the daughter cell cortex, put forward a “zone model” that states that in order for the MEN to signal, a Tem1 bearing SPB must first escape the zone of inhibition or the mother cell where Kin4 resides and then enter the zone of activation or the daughter cell where Lte1 resides. This proposed function of Kin4 has however not been tested.
To shed light on whether and how Kin4 is regulated by spindle position defects we have defined the regions within Kin4 that mediate its MEN inhibitory function and its cortical localization in the mother cell. This analysis identified a C-terminal cortical localization domain and a single amino acid, serine 508, which is necessary for restriction to the mother cell cortex. Mutation of this serine did not increase Kin4 activity or protein levels but led to the association of Kin4 with both the mother and daughter cell cortices, allowing us to test the validity of the zone model of Kin4 function. We find that symmetrically localized Kin4 inhibits the MEN in the absence of spindle position defects but only in the absence of the MEN activator Lte1. These data not only demonstrate that when present in the same compartment, Lte1’s MEN promoting activity dominates over Kin4’s MEN inhibitory activity but also show that Kin4 establishes a MEN inhibitory zone in the mother cell that the MEN bearing SPB must escape in order to promote exit from mitosis. Our data further indicate that Kin4 is not activated by spindle position defects but that Kin4 is active in every cell cycle to restrain MEN signaling in the mother cell. Thus, cells ensure the accurate partitioning of the genetic material between mother and daughter cells by establishing zones of MEN inhibition and activation that allows the translation of spatial cues into a cell cycle signal.
Results
KIN4 acts in every cell cycle
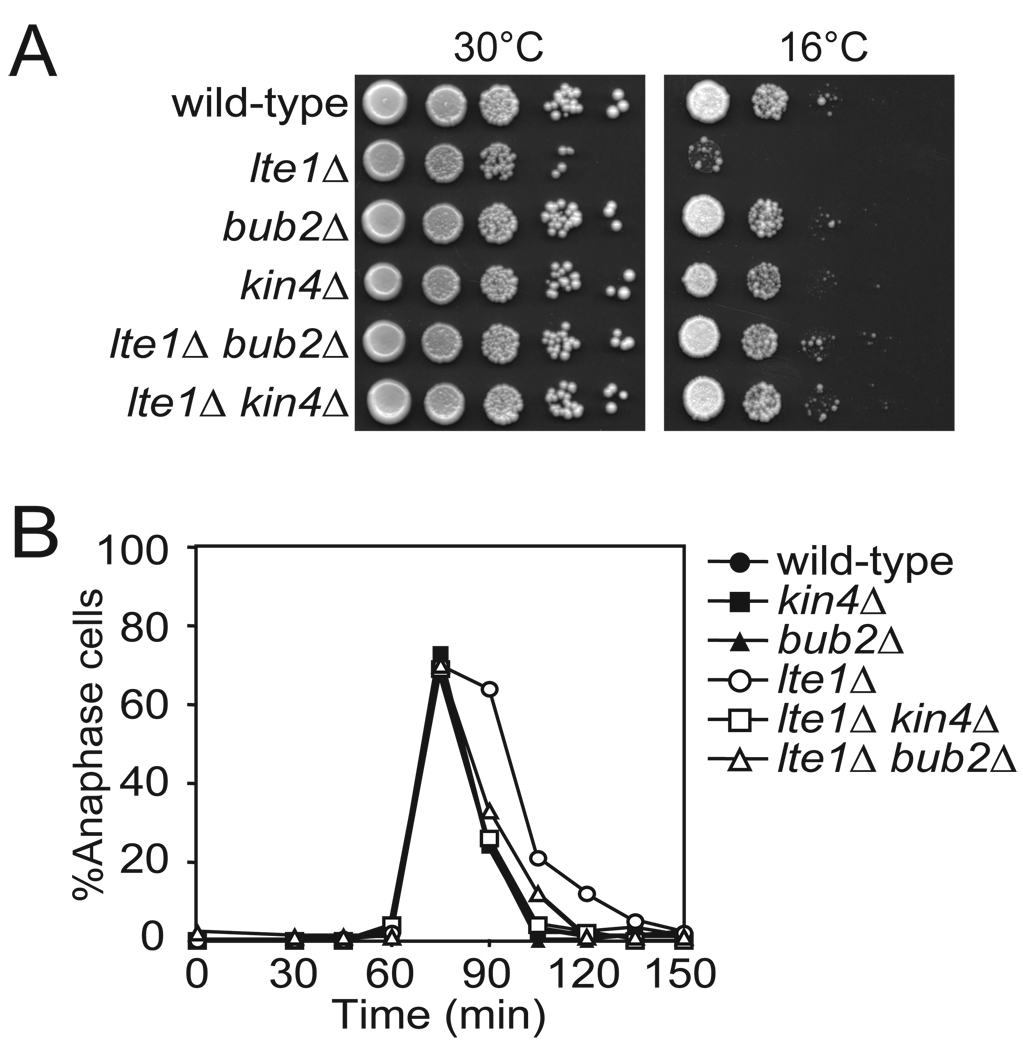
The zone model, where spindle position is sensed and translated by a sensor (Tem1) that moves from an inhibitory (Kin4) to an activating (Lte1) zone makes the following predictions: (1) Kin4 and Lte1 should not be regulated by spindle position but should control the MEN during every cell cycle, (2) targeting Lte1 to the mother cell should promote mitotic exit in cells with mis-positioned spindles (this was indeed found to be the case, (Bardin et al., 2000; Castillon et al., 2003; Geymonat et al., 2009)), and (3) targeting Kin4 to both the mother cell and the bud should inhibit mitotic exit. We tested prediction (1) first. The Cdc14 early anaphase release (FEAR) network is a non-essential pathway that acts in parallel to the MEN to promote timely mitotic exit. Simultaneous deletion of LTE1 and FEAR network components such as SPO12 results in lethality (Stegmeier et al., 2002). The observation that kin4Δ suppresses the lethality of lte1Δ spo12Δ double mutants indicated that KIN4 restrains MEN activity even in an unperturbed cell cycle (D'Aquino et al., 2005). The analysis of lte1Δ kin4Δ double mutants further confirmed this notion. lte1Δ mutants are unable to proliferate at temperatures below 18°C because of an inability to exit from mitosis (Shirayama et al., 1994a) (Figure 1A). Deletion of KIN4 completely suppressed this proliferation defect of the lte1Δ mutant, similar to deletion of BUB2 (Pereira and Schiebel, 2005; Yoshida et al., 2003) (Figure 1A). Furthermore, we found that the 15 minute delay in mitotic exit observed in lte1Δ cells at room temperature was fully suppressed by the deletion of KIN4 (note that this result also demonstrates that LTE1 is required for efficient mitotic exit during every cell cycle) (Adames et al., 2001; Bardin et al., 2000; Jensen et al., 2004) (Figure 1B). The extent of the suppression was similar to that caused by deletion of BUB2 (Figure 1B). Our data indicate that KIN4 antagonizes mitotic exit during every cell cycle.
Figure 1. Kin4 Inhibits the MEN in every Cell Cycle.
(A) Wild-type (A2587), lte1Δ (A24807), bub2Δ (A23045), kin4Δ (A17865), lte1Δ bub2Δ (A24808) and lte1Δ kin4Δ (A24806) cells were spotted on YePAD plates and incubated at 30°C and 16°C. Pictures shown represent growth from 2 days for the 30°C condition and 10 days for 16°C. The first spot represents growth of approximately 3×104 cells and each subsequent spot is a 10 fold serial dilution.
(B) Cells in (A) were arrested in G1 with 5 µg/mL α factor and released into pheromone free media at room temperature. After 70 minutes, 10 µg/mL α factor was added to prevent entry into the subsequent cell cycle. Cells were collected every 15 minutes and stained for tubulin by indirect immuno-fluorescence and for the DNA with DAPI. Anaphase was determined by spindle and nuclear morphology. n ≥ 100 cells.
The N-terminal kinase domain of Kin4 mediates inhibition of mitotic exit
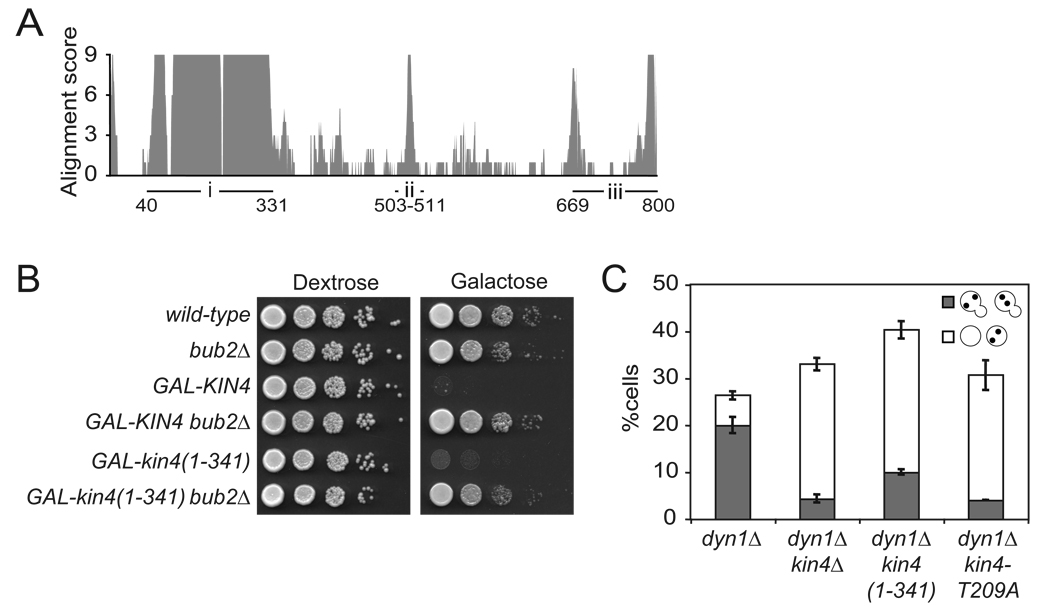
To test the prediction that targeting of Kin4 to the mother and daughter cell cortex inhibits mitotic exit, we first needed to create an allele of KIN4 that localizes to both the mother cell and bud while retaining its MEN inhibitory function. To this end, we characterized the functional domains of Kin4. The kinase activity of Kin4 is absolutely required for its ability to inhibit the MEN (Chan and Amon, 2009). During spindle mis-position, Kin4 must phosphorylate the GAP complex and protect it from inhibitory phosphorylation by Cdc5 to maintain a cell cycle arrest (Maekawa et al., 2007). Previous findings have suggested that the inhibition of the MEN by the SPOC and thus Kin4 is quite potent. Cells that misposition their spindles were observed to delay mitotic exit for over three hours (Yeh et al., 1995). Additionally, overexpression of Kin4 arrests cells in anaphase and is suppressed by bub2Δ indicating that Kin4 is a potent inhibitor of the MEN (D'Aquino et al., 2005) (Figure 2B). We find that overexpression of the kinase domain alone (kin4(1–341)) from the GAL1-10 promoter is lethal (Figure 2A, region i). This lethality is due to an inability to exit mitosis as deletion of BUB2 restored the viability of GAL1-10-kin4(1–341) cells (Figure 2B).
Figure 2. The N-terminal kinase domain of Kin4 mediates MEN inhibition.
(A) The amino acid sequence of Kin4 and its orthologs were aligned using T-Coffee and the alignment score was plotted against the amino acid position. The three major regions of conservation are labeled i, ii and iii with the amino acid positions marked.
(B) Wild-type (A2587), bub2Δ (A1863), pGAL1-10-GFP-KIN4 (A11997), pGAL1-10-GFP-KIN4 bub2Δ (A18792), pGAL1-10-GFP-kin4(1–341) (A23250) and pGAL1-10-GFP- kin4(1–341) bub2Δ (A24113) cells were spotted on plates containing either glucose or galactose and raffinose as in Figure 1A.
(C) dyn1Δ (A17349), dyn1Δ kin4Δ (A17351), dyn1Δ kin4(1–341) (A22262) and dyn1Δ kin4-T209A (A22736) were grown at 14°C for 24 hours. Cells were stained as in Figure 1B. n ≥ 100 cells per sample. Error bars represent SEM.
Despite retaining the ability to inhibit the MEN when overexpressed, we found that kin4(1–341) was unable support checkpoint function when expressed at endogenous levels. Cells lacking dynein (dyn1Δ) frequently mis-position their spindles (particularly at cold temperatures, 14°C) and delay exit from mitosis (Li et al., 1993). Thus dyn1Δ cultures accumulate cells with a mispositioned anaphase spindle bridging the two nuclei entirely in the mother cell body (“arrested” morphology). KIN4 function is required for this delay in exit from mitosis as kin4Δ cells with mis-positioned spindles exit from mitosis prematurely and produce anucleate and multi-nucleated cells (“bypassed” morphology) (D'Aquino et al., 2005; Pereira and Schiebel, 2005). dyn1Δ cultures grown at 14°C for 24 hours accumulated 20% of cells with the arrested morphology and 6.3% of cells with the bypassed morphology (Figure 2C). In dyn1Δ kin4Δ double mutant cultures, 4.3% of cells exhibited the arrested morphology and 28.7% of cells exhibited the bypassed morphology (Figure 2C). As previously reported, the kinase dead allele of KIN4, kin4-T209A, also failed as a checkpoint component (Chan and Amon, 2009) (Figure 2C). The dyn1Δ kin4(1–341) mutant behaved as the dyn1Δ kin4Δ or dyn1Δ kin4-T209A mutant (Figure 2C). This failure of kin4(1–341) to function in the SPOC could not be attributed to defects in kinase activity as levels of associated kinase activity were similar between the full length and truncation constructs as judged by in vitro immuno-precipitation kinase assays (Supplemental Figure 1A). We conclude that the kinase domain alone of Kin4 is a potent inhibitor of the MEN but that critical regions required for regulating its function must reside in Kin4 C-terminal to the kinase domain.
The C-terminus of Kin4 mediates cortical localization
To identify regions of Kin4 outside the kinase domain that might be important for Kin4 function, we aligned the sequence of Kin4 and its orthologs in other members of the Saccharomycetaceae family. This alignment revealed three major regions of high conservation. The highly conserved amino acids 43–313 encode the kinase domain (region i, Figure 2A). Residues 503–511 (region ii) and the C-terminus of the protein (region iii) also contained highly conserved regions (Figure 2A).
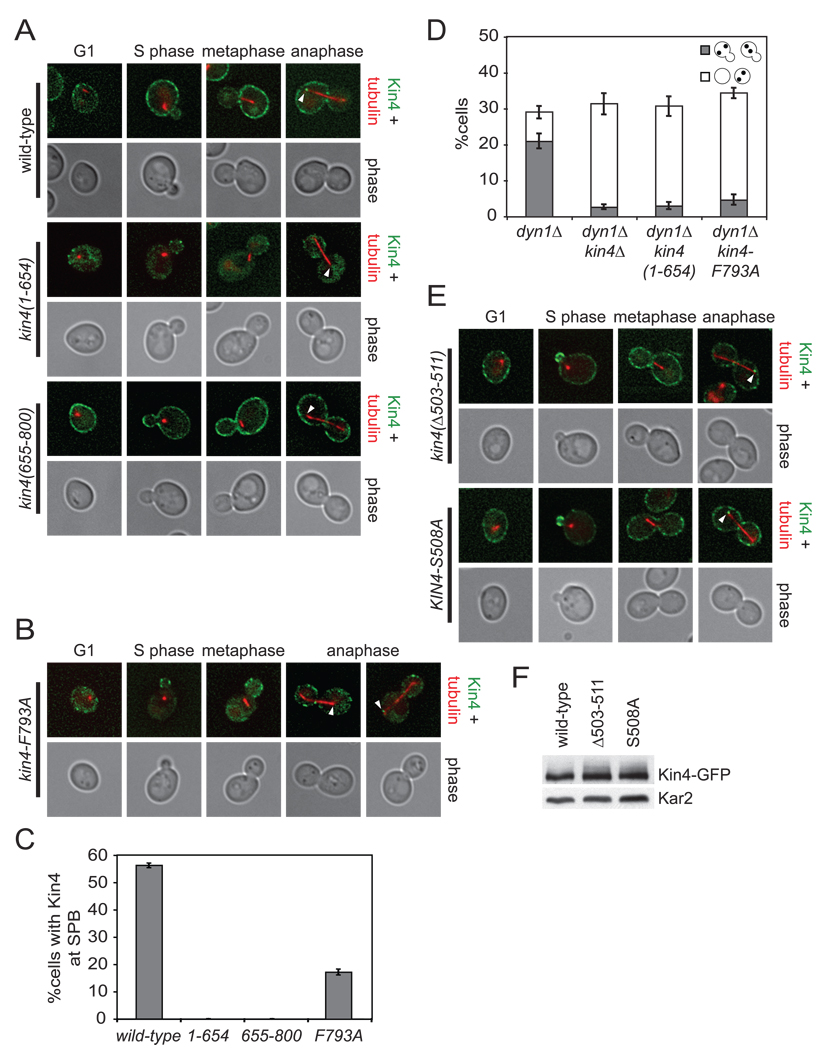
To examine the functions of the C-terminal region, we generated truncations of KIN4 that lack the C-terminal 146 amino acids (kin4(1–654)) and that contain only the C-terminal 146 amino acids (kin4(655–800)) and found that they expressed well (Supplemental Figure 2A). We fused kin4(1–654) to GFP and observed several localization defects. Kin4 normally localizes to the mother cell cortex throughout the cell cycle (D'Aquino et al., 2005; Pereira and Schiebel, 2005) (Figure 3A). Interestingly, kin4(1–654) did not localize to the mother cell cortex but the bud cortex in small budded cells, but this localization did not persist in later stages of the cell cycle (Figure 3A). Full length Kin4 also localizes to the mother cell spindle pole body (mSPB) during anaphase but kin4(1–654) did not (Pereira and Schiebel, 2005) (Figures 3A, C).
Figure 3. Sequences in the C-terminus of Kin4 control asymmetric cortical localization.
(A) Cells expressing an mCherry-Tub1 fusion protein and Kin4-GFP (A19900) or kin4(1–654)-GFP (A21575) or kin4(655–800)-GFP (A21576) were grown to exponential phase and imaged live. The deconvolved GFP signal shown is from 8–10 serial sections. Kin4-GFP is shown in green, mCherry-Tub1 in red. Arrowheads indicate the position of the mother SPB.
(B) Cells expressing kin4-F793A-GFP and mCherry-Tub1 (A21556) were analyzed as in (A).
(C) Cells from (A) and (B) were analyzed over a single cell cycle as described in Figure 1B. Samples were taken at 60, 75, 90 and 105 minutes post release and imaged live. Serial sections spanning the entire cell were collected to ensure imaging of all spindle poles. Loading of Kin4 to the SPBs was judged by co-localization of Kin4-GFP with the ends of the anaphase spindle. n ≥ 100 cells and error bars represent SEM.
(D) dyn1Δ (A17349), dyn1Δ kin4Δ (A17351), dyn1Δ kin4(1–654) (A22263) and dyn1Δ kin4-F793A (A21298) were analyzed as in Figure 2C.
(E) Cells expressing mCherry-Tub1 and kin4(Δ503–511)-GFP (A21555) or Kin4-S508A (A21557) were analyzed as in (A).
(F) Cells expressing mCherry-Tub1 and Kin4-GFP (A19900), kin4(Δ503–511)-GFP (A21555) or Kin4-S508A-GFP (A21557) were lysed and analyzed for expression of the Kin4 fusion protein by western blot analysis. Kar2 was used as a loading control.
kin4(655–800) displayed a different set of localization defects. We observed that kin4(655–800) localized to both the mother and daughter cell cortices in all phases of the cell cycle but failed to load onto either SPB in anaphase (Figures 3A, C). Next we mutated three strictly conserved residues in the highly conserved tail of Kin4 and found that when F793 was mutated to alanine, the C-terminal domain no longer efficiently localized to the cortex (Supplemental Figures 2C, 2D, 2E). Full-length Kin4 carrying this mutation (kin4-F793A) was expressed well but also inefficiently localized to the cortex (Figure 3B, Supplemental Figure 2B). Consistent with cortical localization being required for SPB localization, kin4-F793A loading onto SPBs was also significantly reduced (17% for kin4-F793A vs 56% for wild-type Kin4; Figures 3B, 3C). We next examined the checkpoint function of the kin4(1–654) and kin4-F793A alleles. We found that both alleles behaved as deletion of KIN4 with respect to SPOC function (Figure 3D). We conclude that the C-terminus of Kin4 contains a cortical localization domain but that this domain cannot mediate mother cell restricted cortical localization or SPB localization. Furthermore, this domain is required for Kin4’s checkpoint function.
Amino acids 503–511 are necessary for asymmetric cortical association of Kin4
To determine the function of the conserved nine amino acids at position 503–511 (region ii, Figure 2A), we examined the consequences of deleting this region on Kin4 localization. We found that like wild-type, kin4(Δ503–511) associated with the cortex in G1 cells. However, in small budded cells, the protein was transiently enriched at the bud cortex and was evenly distributed between the mother and bud cortices during the remainder of the cell cycle (Figure 3E). Association of Kin4 with the mSPB was not affected by this internal deletion (Figure 3E). An alanine scan of this region identified a single amino acid substitution that resulted in the same symmetric cortical localization at serine 508 (Figure 3E). This symmetric cortical localization cannot be attributed to increases in protein abundance as kin4(Δ503–511) and Kin4-S508A expressed similarly to wild-type (Figure 3F). We conclude that amino acids 503–511 and specifically S508 are necessary for asymmetric cortical localization of Kin4.
Symmetric Kin4 Delays Mitotic Exit in the absence of LTE1
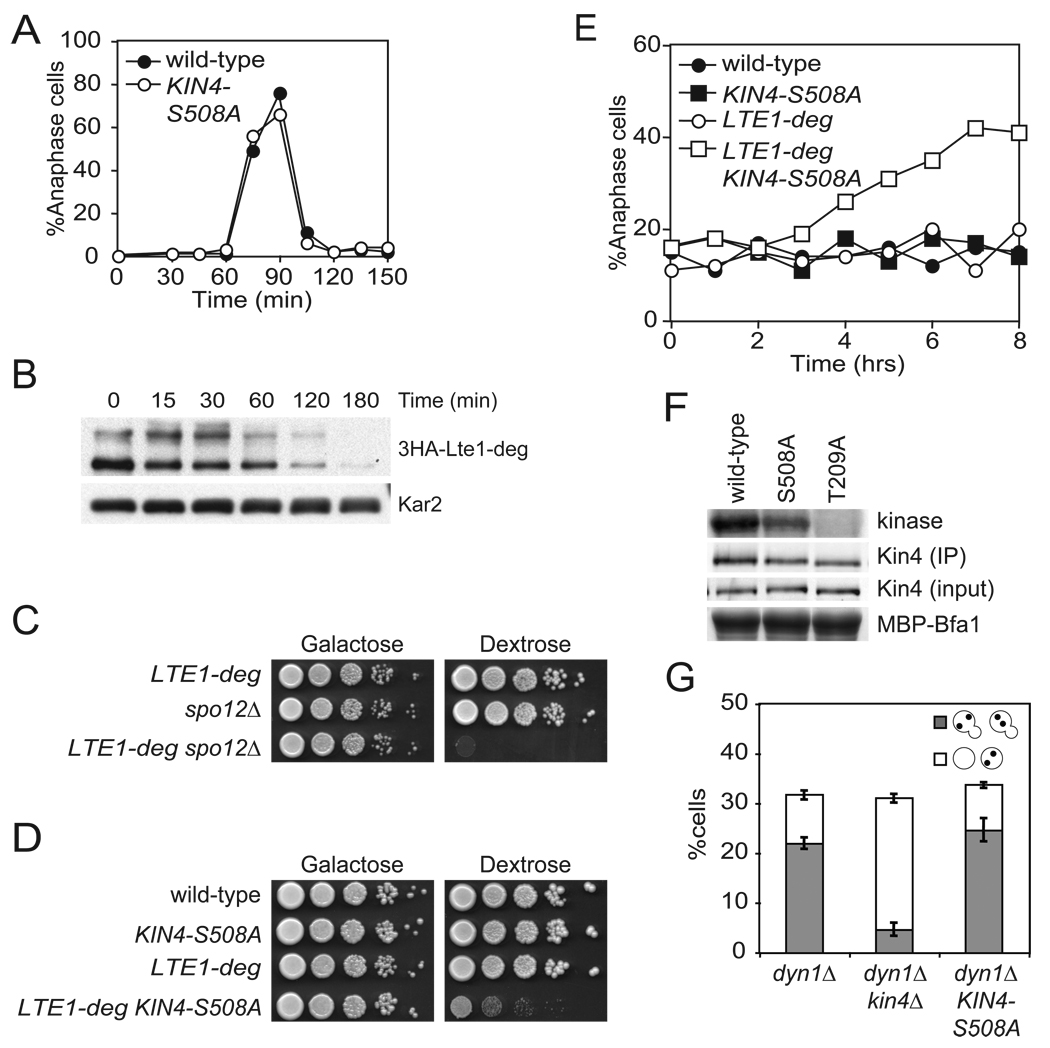
If a Tem1 bearing SPB must escape the zone of Kin4 influence to activate the MEN, then a symmetrically localized Kin4 should delay or prevent exit from mitosis. To test this prediction of the zone model, we released wild-type and KIN4-S508A mutant cells from a pheromone-induced G1 arrest and monitored anaphase entry and exit by spindle morphology. Inconsistent with the prediction, cells expressing KIN4-S508A as the sole source of KIN4 progressed through the cell cycle with wild-type kinetics (Figure 4A).
Figure 4. Kin4 localization in the bud delays mitotic exit cells lacking Lte1.
(A) Wild-type (A2587) and KIN4-S508A (A21299) cells were analyzed as in Figure 1B.
(B) Cells expressing a URL-3HA-Lte1 fusion protein (A23686) from the galactose inducible promoter were grown in YePA + 2% raffinose + 2% galactose (YePA+RG) to exponential phase. 2% glucose was added to the media at time = 0 to repress the GAL promoter and samples were collected for western blot analysis. Kar2 was used as a loading control.
(C) pGAL1-10-URL-3HA-LTE1 (LTE1-deg) (A23686), spo12Δ (A4874) and LTE1-deg spo12Δ (A24543) cells were spotted on plates containing either glucose or galactose and raffinose as in Figure 1A.
(D) Wild-type (A2587), KIN4-S508A (A21299), LTE1-deg (A23686) and KIN4-S508A LTE1-deg (A24084) cells were analyzed as in (C).
(E) Cells in (D) were grown as in (B) and samples were collected and analyzed for spindle and nuclear morphology as in Figure 1B.
(F) Cells expressing either Kin4-3HA (A11779), Kin4-S508A-3HA (A20608) or kin4-T209A-3HA (A22119) were grown to exponential phase and arrested with 15 µg/mL nocodazole for 2 hours. Kin4 associated kinase activity (top, Kin4 kinase), immunoprecipitated Kin4-3HA (second row, Kin4 (IP)), total amount of Kin4-3HA in extracts (third row, Kin4 (input)) and the amount of Bfa1 substrate (as monitored by Coomassie stain) added to the kinase reaction (bottom, MBP-Bfa1) are shown. The band that is shown for Kin4 associated kinase activity and total Bfa1 substrate is the first major degradation product of MBP-Bfa1 as described in Maekawa et al., 2007 and was the dominant signal.
(G) dyn1Δ (A17349), dyn1Δ kin4Δ (A17351), and dyn1Δ KIN4-S508A (A21301) were analyzed as in Figure 2C.
Previous experiments localizing Lte1 to the mother cell showed that this mis-localization is sufficient to bypass the SPOC and prematurely activate the MEN. These experiments not only demonstrated the importance of restricting Lte1 to the bud, but also imply that when Lte1 and Kin4 are in the same cellular compartment, the mitotic exit promoting activity of Lte1 dominates over the mitotic exit inhibitory effects of Kin4. In light of this observation, symmetric Kin4 should not inhibit mitotic exit since Lte1 is still present in the bud. However, if the zone model is correct, symmetrically localized Kin4 should inhibit mitotic exit in cells lacking LTE1. To test this hypothesis, we first constructed a conditional allele of LTE1 by fusing the degradation tag, ubiquitin-arginine-lacZ, to the coding sequence of LTE1 and placed the fusion under the control of the GAL1-10 promoter (LTE1-deg). This fusion protein was efficiently depleted after three hours in glucose at room temperature (Figure 4B). Consistent with efficient depletion, we find that depletion of Lte1 led to loss of viability in cells lacking the FEAR network component SPO12 (Figure 4C).
Under conditions of Lte1 depletion (glucose), KIN4-S508A and LTE1-deg single mutants grew similarly to wild-type cells, but proliferation was severely hampered in the double mutant (Figure 4D). To determine if this proliferation defect was due to a defect in exit from mitosis, we monitored accumulation of anaphase cells in single and double mutants. In glucose, both KIN4-S508A and LTE1-deg cultures behaved as the wild-type and accumulated 10–20% anaphase cells at any given time (Figure 4E). In contrast, LTE1-deg KIN4-S508A double mutant cultures accumulated cells with anaphase spindles over time, with 40% of cells arresting in anaphase after eight hours in glucose (Figure 4E). The mitotic exit inhibitory effect of the KIN4-S508A allele cannot be attributed to increased protein abundance or kinase activity (Figures 3F, 4F). In fact, Kin4 associated kinase activity was slightly reduced in the KIN4-S508A mutant (73.8% ± 4.4% of wild-type activity). Our results show that symmetric Kin4 inhibits mitotic exit in the absence of LTE1. Furthermore, these data indicate that Lte1’s MEN promoting function dominates over Kin4’s MEN inhibitory function. With respect to sensing spindle position, our data show that the daughter bound SPB must escape the zone of Kin4 influence in order for the MEN to signal.
The zone model also predicts that a symmetrically localized Kin4 should have no effect on SPOC function. Since the SPBs of a mispositioned spindle never escape the mother cell, symmetric localization of Kin4 is inconsequential. Indeed, we found that KIN4-S508A retained full checkpoint function (Figure 4G). The findings that symmetric Kin4 inhibits mitotic exit in the absence of Lte1 and retains full checkpoint function support the zone model.
Kar9 and Clb4 prevent Kin4 from loading onto the daughter SPB
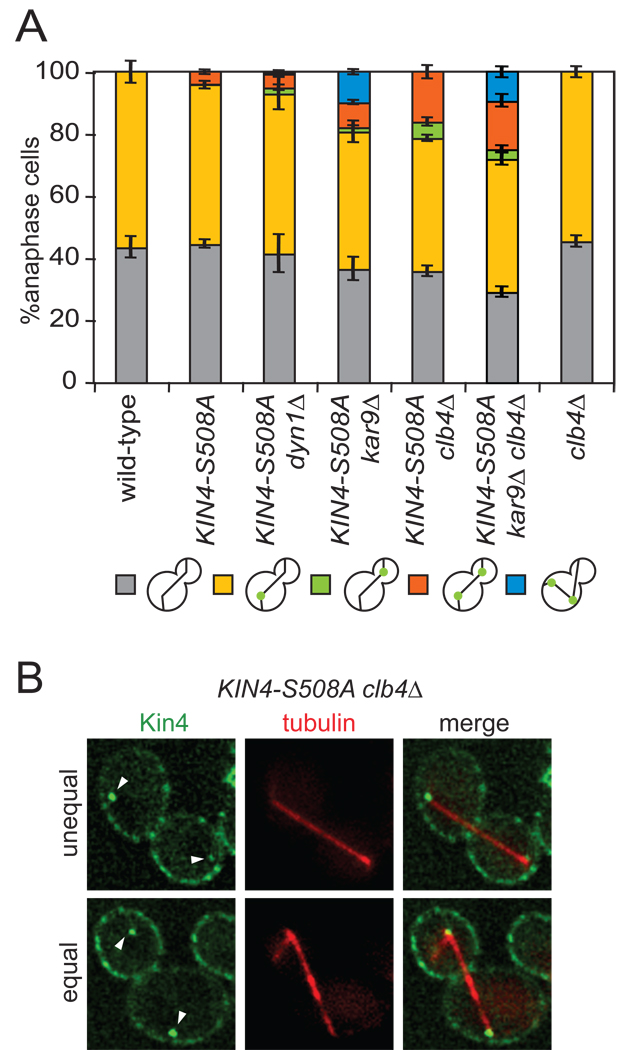
Kin4 association with SPBs is critical for its MEN inhibitory function (Maekawa et al., 2007). Analysis of Kin4-S508A localization revealed that despite the protein associating with the bud cortex, Kin4-S508A loaded onto the daughter cell SPB (dSPB) in only 3.8% of anaphase cells (Figures 3E, 5A). In contrast, Kin4-S508A associated with the mSPB in 51% of anaphase cells (Figure 5A). This finding indicates that additional layers of regulation must exist to exclude Kin4 from the dSPB. The microtubule motor dynein (Dyn1), the cytoskeletal adaptor protein Kar9 and the B-type cyclin Clb4 have all previously been implicated in regulating the asymmetric distribution of proteins between mother and daughter SPBs (Lee et al., 2003; Liakopoulos et al., 2003; Maekawa and Schiebel, 2004; Maekawa et al., 2003). We therefore tested whether deletion of any of these factors affected the association of Kin4-S508A with SPBs. We found no increase in the number of cells with Kin4-S508A at both SPBs in the dyn1Δ mutant (4.5%), a modest increase in the kar9Δ mutant (8%) and the largest increase in the clb4Δ mutant cells (16%; Figure 5A). Analysis of SPB localization in the kar9Δ clb4Δ double mutant showed no additive effects suggesting that KAR9 and CLB4 act in the same pathway to exclude Kin4 from the dSPB (Figure 5A). In cells with Kin4-S508A localizing to both SPBs we often (46 of 70 cells with Kin4-S508A at both SPBs) observed the GFP signal to be stronger on the mSPB suggesting that Kar9 and Clb4 are not the only factors that mediate Kin4 exclusion from SPBs (Figure 5B). Wild-type Kin4 did not load onto dSPBs in clb4Δ mutants suggesting that Kin4 first needs to be targeted to the daughter cell to load onto the dSPB (Figure 5A). We conclude that the dSPB specific factors, Kar9 and Clb4 contribute to the exclusion of Kin4-S508A from the dSPB.
Figure 5. Kar9 and Clb4 prevent symmetric Kin4 from loading to the daughter SPB.
(A) Cells expressing GFP tagged Kin4 and mCherry-Tub1 with the following genetic backgrounds: wild-type (A19900), KIN4-S508A (A21557), KIN4-S508A dyn1Δ (A23052), KIN4-S508A kar9Δ (A23051), KIN4-S508A clb4Δ (A23055), KIN4-S508A clb4Δ kar9Δ (A25794) and clb4Δ (A23249) were analyzed as in Figure 3C. n ≥ 100 cells and error bars represent SEM.
(B) Representative KIN4-S508A clb4Δ cells with symmetric Kin4 localization to the SPBs. The top cell shows an unequal SPB loading pattern whereas the bottom cells shows equal Kin4-GFP intensity at both SPBs. The deconvolved GFP signal shown is from 10 serial sections.
Increased dSPB loading of Kin4 enhances the mitotic exit defect of KIN4-S508A LTE1-deg cells
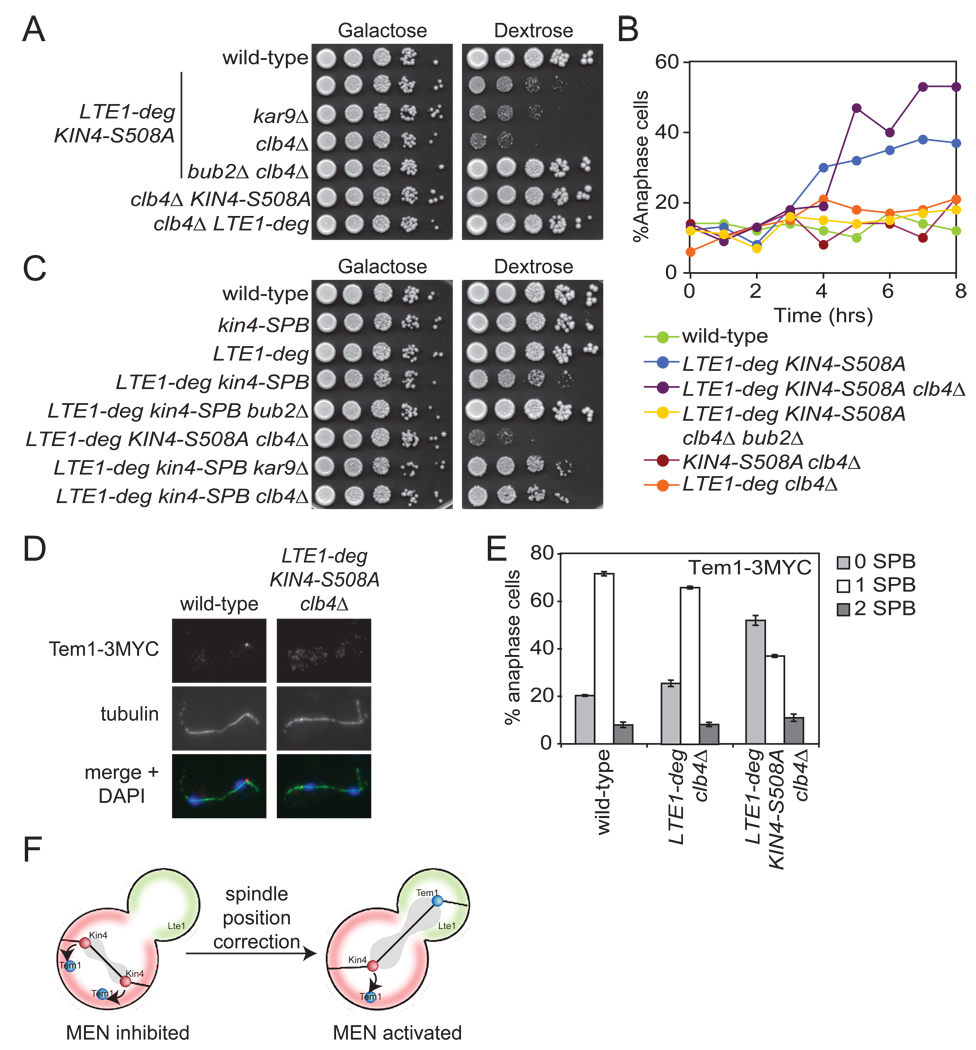
To test whether increased loading of Kin4-S508A onto the dSPB enhances the mitotic exit defect of cells lacking Lte1 and containing symmetric Kin4, we examined the consequences of deleting KAR9 or CLB4 on KIN4-S508A LTE1-deg cells. Deletion of KAR9 mildly enhanced the proliferation defect of KIN4-S508A LTE1-deg mutants (Figure 6A). Deletion of CLB4 enhanced the proliferation defect of KIN4-S508A LTE1-deg cells by 10 fold (Figure 6A, rows 2, 4). Neither KIN4-S508A clb4Δ nor LTE1-deg clb4Δ double mutants displayed any proliferative defects consistent with the notions that Lte1 dominates over Kin4’s inhibition of the MEN and that Kin4 must load onto both SPBs to inhibit mitotic exit (Figure 6A, rows 6, 7). Deletion of BUB2 fully suppressed the proliferative defect of KIN4-S508A LTE1-deg clb4Δ mutants indicating that the proliferation defect is due to MEN inhibition (Figure 6A, row 5). Cytological examination of cells confirmed this. We observed a severe anaphase delay in the KIN4-S508A LTE1-deg clb4Δ mutant that was completely suppressed by deletion of BUB2 (Figure 6B).
Figure 6. Loading of Kin4 onto dSPBs arrests cells in anaphase.
(A) Wild-type (A2587), LTE1-deg KIN4-S508A (A24084), LTE1-deg KIN4-S508A kar9Δ (A24816), LTE1-deg KIN4-S508A clb4Δ (A24086), LTE1-deg KIN4-S508A clb4Δ bub2Δ (A24346), KIN4-S508A clb4Δ (A24083) and LTE1-deg clb4Δ (A24085) cells were analyzed as in Figure 4C.
(B) Wild-type (A2587), LTE1-deg KIN4-S508A (A24084), LTE1-deg KIN4-S508A clb4Δ (A24086), LTE1-deg KIN4-S508A clb4Δ bub2Δ (A24346), KIN4-S508A clb4Δ (A24083) and LTE1-deg clb4Δ (A24085) cells were analyzed as in Figure 4E.
(C) Wild-type (A2587), kin4-SPC72(177–622) (kin4-SPB) (A24586), LTE1-deg (A23686), LTE1-deg kin4-SPB (A24587), LTE1-deg kin4-SPB bub2Δ (A24588), LTE1-deg KIN4-S508A clb4Δ (A24086), LTE1-deg kin4-SPB kar9Δ (A24817) and LTE1-deg kin4-SPB clb4Δ (A24858) cells were analyzed as in Figure 4C.
(D–E) Wild-type (A1828), LTE1-deg clb4Δ (A24805) and LTE1-deg KIN4-S508A clb4Δ (A24761) cells expressing a Tem1-3MYC fusion protein were grown as in Figure 4E. Cells were collected seven hours post glucose addition and stained for tubulin (green) and Tem1-3MYC (red) by indirect immuno-fluorescence and the DNA (blue) was stained with DAPI. Tem1 loading was judged by colocalization of Tem1-3MYC with the ends of the anaphase spindle. n ≥ 100 cells and error bars represent SEM.
(F) The zone model of SPOC function. See text for further details.
A fusion construct between Kin4 and a fragment of the SPB resident protein, Spc72 (kin4-Spc72(177–622), henceforth kin4-SPB) was previously constructed and cells expressing this fusion protein displayed a very subtle delay in mitotic exit but failed to inhibit the MEN in response to spindle position defects (Chan and Amon, 2009; Maekawa et al., 2007). Consistent with the subtle cell cycle delay, we observed no detectable loss in proliferative capacity in the kin4-SPB mutant (Figure 6C, row 2). When combined with the LTE1-deg allele, kin4-SPB led to a subtle proliferation defect that was not nearly as severe as that observed in KIN4-S508A LTE1-deg or KIN4-S508A LTE1-deg clb4Δ cells (Figure 6C, rows 4, 6). This finding suggests that while kin4-SPB is hyper-morphic for SPB loading, it is also hypo-morphic for MEN inhibition once at the SPB. This loss-of-function is likely due to the effects of the protein fusion. Importantly, the kar9Δ and clb4Δ mutations did not enhance the growth defect of LTE1-deg kin4-SPB mutants indicating that the phenotypic enhancement these mutations lend to the KIN4-S508A LTE1-deg mutant is caused by enhanced loading of Kin4-S508A to dSPBs (Figure 6C, rows 7, 8).
To further characterize the effects of loading Kin4 onto both SPBs on MEN function, we examined the localization of Tem1. Tem1 localizes to the dSPB in anaphase. Its loss from this organelle correlates well with Kin4 inhibition of the MEN. In cells with a mis-positioned spindle, Tem1 fails to load onto either SPB. Without KIN4, such cells inappropriately load Tem1 onto both SPBs, presumably triggering premature mitotic exit (D'Aquino et al., 2005). Cells that over-express KIN4 and arrest in anaphase also fail to load Tem1 onto either SPB (D'Aquino et al., 2005). Accordingly, Tem1 association with dSPBs was reduced in KIN4-S508A LTE1-deg clb4Δ mutants during anaphase (Figures 6D, 6E). We conclude that even in the absence of spindle mis-position, Kin4 targeted to both cell cortices and SPBs inhibits mitotic exit in the absence of LTE1.
Discussion
Multiple control mechanisms ensure that Kin4 inhibits the MEN only in the mother cell
The N-terminal kinase domain of Kin4 mediates MEN inhibition. Overexpression of this region of the protein alone is sufficient to inhibit the MEN and hence exit from mitosis. This potent MEN inhibitory activity requires multiple control mechanisms to ensure that inhibition only occurs when the mitotic spindle is mis-positioned and the MEN bearing SPBs are in the mother cell. These regulatory mechanisms are:
Lte1 dominates over Kin4. When both proteins are present in the same cellular compartment, Lte1’s MEN promoting effects trump the MEN inhibitory effects of Kin4. The Lte1-8N allele, which localizes to both the mother cell and bud cortices, promotes exit from mitosis in the mother cell even in the presence of Kin4 (Geymonat et al., 2009). Conversely, the KIN4-S508A mutant only inhibits mitotic exit in the absence of LTE1.
Kin4 is restricted to the mother cell cortex by unknown mechanisms.
Kin4 is excluded from the daughter cell SPB. The dSPB specific factors Clb4 and Kar9 inhibit binding of Kin4-S508A to the dSPB suggesting that SPB asymmetry contributes an additional layer to Kin4 regulation.
While such multiple overlapping mechanisms make their dissection difficult, they highlight the biological importance of restricting Kin4 function.
If Kin4 function is largely governed by localization, how is Kin4 localization controlled? The C-terminal 146 amino acids of Kin4 harbor sequences sufficient for association with the cell cortex. We do not yet know how this region mediates cortex association. It is, however, interesting to note that this cortical localization mechanism is operative in mammalian cells. Kin4 and kin4(655–800) localizes to the cell cortex in 293T cells while the point mutant, kin4(655–800)-F793A failed to do so suggesting that the localization mechanisms are the same in yeast and mammalian cells (Supplemental Figures 2F, G).
Full length Kin4 very transiently localizes to the bud tip during nascent bud formation but subsequently associates with the mother cell cortex (Supplemental Figure 3A). The N-terminal domain of Kin4 appears to contain a bud-cortex targeting domain. kin4(1–341) was found to be associated with the bud cortex throughout the cell cycle (Supplemental Figure 3B). Perhaps kin4(1–341) harbors the signals necessary for initial bud cortex association but lacks the sequences necessary to re-direct Kin4 to the mother cortex and/or clear the protein from the bud cortex. Residues 503–511 appear to contain such sequences. In their absence, Kin4 is present throughout the mother and bud cortices. It is important to note that while many proteins localize exclusively to the bud, only a few proteins show mother cortex selectivity. Such proteins must either defy vectorial transport that directs most newly synthesized proteins to the bud and/or be redirected back to the mother cell by unknown mechanisms (perhaps by selective proteolysis) to yield this unusual localization pattern.
The machinery that restricts Kin4 to the mother cell is not completely efficient. Low levels of Kin4 can be detected in the daughter cell, particularly in anaphase (L. Chan unpublished observations). However, additional mechanisms exist that prevent leaky Kin4 from inappropriately inhibiting the MEN at SPBs. The dSPB-associated proteins Clb4 and to a lesser extent Kar9 exclude Kin4 from the SPB in the bud. Due to Kar9’s critical role in loading Clb4 onto the dSPB and the lack of enhancement in Kin4 loading to both SPBs in the clb4Δ kar9Δ double mutant, we favor a model where Clb4 rather than Kar9 regulates Kin4’s association with dSPBs (Maekawa and Schiebel, 2004). Clb4 could modify SPB components or Kin4 itself to prevent Kin4 from binding to the daughter cell SPB. Alternatively, Clb4’s role could be indirect. The interactions between cytoplasmic microtubules and the bud cortex are increased in clb4Δ mutants (Maekawa and Schiebel, 2004). If symmetric Kin4 loads from the bud cortex to the dSPB via cytoplasmic microtubules, it is possible that increased cortex-microtubule interactions could lead to an increase in Kin4-S508A loading to the dSPB. Much needs to be discovered about the factors responsible for Kin4’s unusual localization pattern, but our domain analysis of Kin4 already demonstrates that their function will be critical for coordinating cell cycle progression and spindle position.
A bud-localized MEN activating zone and a mother cell-localized MEN inhibitory zone drive spindle position sensing
Our results and those of others demonstrate that SPOC function is at least in part based on the spatial restriction of the MEN regulators Kin4 and Lte1. In this “zone model”, for the MEN to become active and to promote exit from mitosis, one of the MEN bearing SPBs must escape the zone of inhibition or the mother cell where Kin4 resides and enter the zone of activation or the daughter cell where Lte1 resides (Figure 6F). Perturbation of either zone leads to mis-regulation of the MEN. When Lte1 is localized in the mother cell, either by overexpression, crippling of the septin ring diffusion barrier or mutations within Lte1, cells with mis-positioned spindles prematurely exit from mitosis (Bardin et al., 2000; Castillon et al., 2003; Geymonat et al., 2009). Conversely, symmetric targeting of Kin4 inhibits exit from mitosis. The observation that Kin4 could only do so in the absence of Lte1 not only indicates that Lte1’s MEN promoting activity dominates over Kin4’s inhibitory effects, it also raises the interesting possibility that in addition to its role in promoting timely mitotic exit, Lte1 restrains any spurious effects of Kin4 that leaks into the bud.
Our analysis of the KIN4-S508A allele has important implications for how we think about the SPOC. The observation that in the absence of LTE1, this allele inhibits exit from mitosis even in cells with correctly positioned anaphase spindles, demonstrates that a spindle mis-position event is not a prerequisite for Kin4’s ability to inhibit the MEN. Thus, Kin4 is not a sensor of spindle position. Rather it is the MEN bearing SPBs and ultimately the GTPase switch, Tem1, that moves and senses its position in space relative to its regulators, Lte1 and Kin4, that allows the MEN to monitor spindle position. The idea that MEN is activated by movement of a SPB out of the Kin4 inhibitory zone and into the bud is supported by several other observations. Tem1 becomes concentrated on the dSPB upon anaphase spindle elongation and is further enhanced by Lte1 (Bardin et al., 2000; Molk et al., 2004; Pereira et al., 2000). Also, the MEN inhibitor, Bub2, is lost from the dSPB after it emerges into the bud (Molk et al., 2004).
Several other models have been proposed for how the MEN is activated and how the SPOC functions to shut down the MEN in response to spindle position defects. A model where disappearance of the Bub2-Bfa1 GAP complex from the mSPB triggers mitotic exit is inconsistent with the demonstration that Kin4 association with SPBs inhibits MEN signaling (Chan and Amon, 2009; Fraschini et al., 2006; Maekawa et al., 2007; Pereira and Schiebel, 2005). An alternate model proposes that the presence of cytoplasmic microtubules in the bud neck contribute to SPOC function (Adames et al., 2001; Moore et al., 2009). Cytoplasmic microtubule-bud neck interactions could contribute to triggering and/or maintenance of the SPOC, but it is important to note that loss of cytoplasmic microtubules as is observed in bim1Δ, cnm67Δ and tub2-401 mutants abolishes checkpoint function in only about 20% of cells that misposition their spindles (Adames et al., 2001). This indicates that the presence of cytoplasmic microtubules in the bud neck is not the major mechanism whereby spindle position defects affect MEN signaling. In contrast, misdirecting Lte1 or Kin4 to the wrong compartment leads to an almost complete bypass of the SPOC and anaphase arrest, respectively.
We do not yet know how Kin4 and Lte1 ultimately control the MEN GTPase Tem1. Kin4 phosphorylates Bfa1 thereby preventing inactivation of the GAP complex by Cdc5 (Maekawa et al., 2007). While this explains how Tem1 activity is controlled it does not explain how Tem1’s association with SPBs is regulated. How Lte1 activates Tem1 is also unclear. Lte1 contains putative guanine nucleotide exchange factor (GEF) domains and it has been proposed that Lte1 may activate MEN signaling by acting as a GEF for Tem1 (Bardin et al., 2000; Pereira et al., 2000). However, a recent study has found no detectable in vitro GEF activity for Lte1 against Tem1 (Geymonat et al., 2009). Elucidating mechanistically how Tem1 receives signals from Kin4 and Lte1 and the role of the GAP complex in this process are important next steps in understanding MEN activation and SPOC function.
Sensing activating and inhibitory zones – a general theme in checkpoint signaling?
The tension sensing aspect of the spindle assembly checkpoint (SAC) shares striking similarities with the SPOC model proposed here (Liu et al., 2009; Tanaka et al., 2002). The SAC senses whether kinetochores are attached to microtubules and whether these attachments are under tension and thus properly bi-oriented. When this is not the case, the SAC arrests cells in metaphase (Lew and Burke, 2003). The conserved protein kinase, Aurora B, severs microtubule-kinetochore attachments that are not under tension by phosphorylating the major microtubule binding activity at kinetochores, the KMN network (Cheeseman et al., 2006; DeLuca et al., 2006). The resulting unattached kinetochores are recognized by the SAC, which shuts down cell cycle progression in metaphase. Aurora B resides at the inner centromere (Liu et al., 2009; Tanaka et al., 2002). When sister kinetochores are not attached by microtubules to opposite poles of the spindle and hence not under tension, the KMN network proteins reside in the Aurora B zone where they are phosphorylated and lose their ability to bind microtubules. When sister kinetochores are properly bi-oriented and thus under tension, they are pulled away from the inner centromere and Aurora B can no longer access its kinetochore substrates (Liu et al., 2009; Tanaka et al., 2002). Thus, kinetochore-microtubule attachments are stabilized and proper bi-orientation is achieved. By analogy to the zone model for spindle position sensing proposed here, Aurora B is not activated by tensionless kinetochores, rather, Aurora B is always active but restricted to a zone where tensionless kinetochores (by virtue of not being under tension) reside. Here too, a cell cycle checkpoint relies on the establishment of a regulatory zone to translate spatial information into a chemical signal.
Experimental Procedures
Yeast Strains and Growth Conditions
All strains are derivatives of W303 (A2587) and are listed in Supplemental Table 1. Specifics of strain construction are detailed in the supplemental experimental procedures. Growth conditions are described in the figure legends.
Plasmid Construction
All plasmids used in this study are listed in Supplemental Table 2. Specifics of plasmid construction are detailed in the supplemental experimental procedures.
Immunoblot Analysis
Immunoblot analysis to determine total amount of Kin4-GFP and Kin4-3HA were as previously described (Chan and Amon, 2009). For immunoblot analysis of Kar2 and 3HA-Lte1, cell lysates were prepared as previously described (Chan and Amon, 2009). Kar2 was detected using a rabbit anti-Kar2 anti-serum (Rose et al., 1989) at 1:200,000 and 3HA-Lte1 was detected using an anti-HA antibody (Covance, HA.11) at 1:1000.
Fluorescence Microscopy
Indirect in situ immunofluorescence methods for Tub1 were as previously described (Kilmartin and Adams, 1984). For immunofluorescence detection of Tem1-3MYC, cells were fixed for 15 minutes in 3.7% formaldehyde, 100mM potassium phosphate pH6.4 buffer and prepared for staining as above. Cells were incubated with a mouse anti-MYC [Covance, 9E10] antibody at 1:1000 and an anti-mouse-Cy3 [Jackson] secondary antibody at 1:1500 each for 2 hours. Live cell imaging techniques are as previously described (Chan and Amon, 2009).
Kin4 Kinase Assays
Kin4 kinase assays were performed as previously described (Chan and Amon, 2009).
Sequence Alignment
Kin4 orthologs were identified by BLAST (Altschul et al., 1997) and sequence alignment analysis was carried out using T-Coffee (Notredame et al., 2000).
Highlights.
Mitotic exit is triggered by the escape of an SPB from the zone of Kin4 influence.
Symmetric targeting of Kin4 inhibits the MEN in the absence of Lte1.
Kin4 localization and function is controlled by modular domains.
Supplementary Material
Acknowledgements
We are grateful to Thomas Schwartz and Brett Williams for advice and Michael Yaffe for reagents. We thank Frank Solomon, Stephen Bell, Fernando Monje-Casas and members of the Amon lab for comments on the manuscript. This work was supported by the National Institutes of Health GM056800 to A. A. and a NSF Predoctoral Fellowship to L. Y. C.. A. A. is also an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adames NR, Oberle JR, Cooper JA. The surveillance mechanism of the spindle position checkpoint in yeast. J Cell Biol. 2001;153:159–168. doi: 10.1083/jcb.153.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Bloecher A, Venturi GM, Tatchell K. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat Cell Biol. 2000;2:556–558. doi: 10.1038/35019601. [DOI] [PubMed] [Google Scholar]
- Castillon GA, Adames NR, Rosello CH, Seidel HS, Longtine MS, Cooper JA, Heil-Chapdelaine RA. Septins have a dual role in controlling mitotic exit in budding yeast. Curr Biol. 2003;13:654–658. doi: 10.1016/s0960-9822(03)00247-1. [DOI] [PubMed] [Google Scholar]
- Chan LY, Amon A. The protein phosphatase 2A functions in the spindle position checkpoint by regulating the checkpoint kinase Kin4. Genes Dev. 2009;23:1639–1649. doi: 10.1101/gad.1804609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aquino KE, Monje-Casas F, Paulson J, Reiser V, Charles GM, Lai L, Shokat KM, Amon A. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell. 2005;19:223–234. doi: 10.1016/j.molcel.2005.06.005. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Fraschini R, D'Ambrosio C, Venturetti M, Lucchini G, Piatti S. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J Cell Biol. 2006;172:335–346. doi: 10.1083/jcb.200507162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Formenti E, Lucchini G, Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cortes JC, McCollum D. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J Cell Biol. 2009;186:739–753. doi: 10.1083/jcb.200902116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, de Bettignies G, Sedgwick SG. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J Cell Biol. 2009;187:497–511. doi: 10.1083/jcb.200905114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Smith SJ, Wheatley E, Rittinger K, Johnston LH, Sedgwick SG. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J Biol Chem. 2002;277:28439–28445. doi: 10.1074/jbc.M202540200. [DOI] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J Biol Chem. 2003;278:14591–14594. doi: 10.1074/jbc.C300059200. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Jensen S, Johnson AL, Johnston LH, Segal M. Temporal coupling of spindle disassembly and cytokinesis is disrupted by deletion of LTE1 in budding yeast. Cell Cycle. 2004;3:817–822. [PubMed] [Google Scholar]
- Kilmartin JV, Adams AE. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Hwang HS, Kim J, Song K. Ibd1p, a possible spindle pole body associated protein, regulates nuclear division and bud separation in Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1449:239–253. doi: 10.1016/s0167-4889(99)00015-4. [DOI] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc Natl Acad Sci U S A. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci U S A. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol. 2007;179:423–436. doi: 10.1083/jcb.200705197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H, Schiebel E. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 2004;18:1709–1724. doi: 10.1101/gad.298704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H, Usui T, Knop M, Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk JN, Schuyler SC, Liu JY, Evans JG, Salmon ED, Pellman D, Bloom K. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol Biol Cell. 2004;15:1519–1532. doi: 10.1091/mbc.E03-09-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Magidson V, Khodjakov A, Cooper JA. The Spindle Position Checkpoint Requires Positional Feedback from Cytoplasmic Microtubules. Curr Biol. 2009 doi: 10.1016/j.cub.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol Cell. 2005;19:209–221. doi: 10.1016/j.molcel.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Tanaka K, Toh-e A. Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast. 1994a;10:451–461. doi: 10.1002/yea.320100404. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Toh EA. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol. 1994b;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ichihashi R, Toh-e A. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J Cell Biol. 2003;161:889–897. doi: 10.1083/jcb.200301128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.