Abstract
Insulin-like growth factor (IGF)-binding protein -5 (IGFBP5), an important member of the IGF axis involved in regulating cell growth and differentiation, acts by modulating IGF signaling and also by IGF-independent mechanisms. We identified IGFBP5 by microarray analysis as a gene differentially regulated during N-(4-Hydroxyphenyl)-retinamide (4HPR)-induced neuronal differentiation of human retinal pigment epithelial (RPE) cells. IGFBP5 is expressed in human RPE cells, and its expression, mRNA as well as protein, is greatly decreased during the 4HPR-induced neuronal differentiation. Exogenous IGFBP5 does not block the neuronal differentiation indicating that IGFBP5 down-regulation may not be a prerequisite for the neuronal differentiation. IGFBP5 down-regulation, similar to neuronal differentiation, is mediated by the MAPK pathway since U0126, an inhibitor of MEK1/2, effectively blocked it. The overexpression of transcription factor CCAAT/enhancer binding protein-β (C/EBPβ) inhibited the 4HPR-induced down-regulation of IGFBP5 expression and the neuronal differentiation of RPE cells. The deletion of C/EBP response element from IGFBP5 promoter markedly decreased the basal promoter activity and abolished its responsiveness to 4HPR treatment in reporter assays, suggesting that the expression of IGFBP5 is regulated by C/EBP. Thus, our results clearly demonstrate that the IGFBP5 expression is down-regulated during 4HPR-induced neuronal differentiation of human RPE cells through a MAPK signal transduction pathway involving C/EBPβ.
Keywords: RPE, IGFBP5, MAPK, C/EBP, Retinoic acid
Introduction
Retinal degeneration is involved in several common eye diseases, such as retinitis pigmentosa, glaucoma and age-related macular degeneration (AMD). The health of the retinal photoreceptors is critically dependent upon the retinal pigment epithelium (RPE), a monolayer of non-neuronal cells with no apparent heterogeneity, located between the neural retina and the choroid (Bok, 1993; de Jong, 2006). The regeneration of retinal photoreceptors is beyond the current capabilities because, like other neuronal cells, the terminally differentiated retinal neuronal cells will not reenter the cell cycle. However, number of studies using vertebrate model system has demonstrated that the RPE is capable of transdifferentiation into cell types other than RPE under certain conditions (Sakaguchi et al., 1997; Yan et al., 2001; Zhao et al., 1997). One of the most striking phenomenon of RPE transdifferentiation is the development of neuronal retinal cells in vitro by basic fibroblast growth factor (Pittack et al., 1991). We have also shown recently that N-(4-hydoxyphenyl)retinamide (4HPR or fenretinide) induces neuronal type differentiation in cultured human RPE cells (Chen et al., 2003; Samuel et al., 2008).
A number of factors that regulate neuronal differentiation have been identified. In particular, the insulin-like growth factors (IGFs) are recognized stimulators of growth and differentiation in many cells types, including neuronal cells (Stewart and Rotwein, 1996). These biological activities of IGFs are mediated through IGF receptors type I and II (IGF-IR and IGF-IIR, (Beattie et al., 2006)). In addition, the activity of IGFs is controlled, in part, by a family of six high-affinity insulin-like growth factor binding proteins (IGFBP1 to 6). These are ubiquitously expressed, soluble extracellular proteins and their biological roles extend beyond merely influencing IGFs half-life in circulation or transporting IGFs to target tissues. The ability of IGFBP5 to bind IGFs with high affinity limits the bio-availability of free IGF to bind to IGF receptor, and thereby augment or inhibit IGF actions in different conditions. Although IGFBPs are structurally related, each IGFBP has an individual expression pattern and exerts different functions depending on the physiological or pathological conditions (Schneider et al., 2000). The relative concentrations of IGFBPs in a tissue, its posttranslational modification, and its cleavage by specific proteases are also the major determinants of the actions of IGFBPs (Beattie et al., 2006).
IGFBP5 is the most conserved IGFBP family member between different species. The human IGFBP5 gene consists of four exons spanning 33 kb of DNA and encodes a 34 kD protein (Allander et al., 1994). IGFBP5 is expressed at different levels in many human cell types, and one of its striking features is its cell- and tissue-type-dependent function (Beattie et al., 2006). IGFBP5 can bind to membrane components of many cells to mediate the biological activities independent of IGFs (Miyakoshi et al., 2001). In addition, IGFBP5 binds to its putative receptor and activates downstream signaling via MAPK (mitogen-activated protein kinase) and ERK1/2 (extracellular-signal-regulated kinase 1/2) signaling pathways (Kuemmerle and Zhou, 2002). The function of IGFBP5 also seems to be dependent on the cellular context since it can either stimulate or inhibit cell growth and differentiation in several cell types (Cobb et al., 2004; Salih et al., 2004). Furthermore, the expression of IGFBP5 is regulated by a network of stimuli acting through different transduction pathways (Yeh et al., 1998). Noticeably, CCAAT/enhancer binding proteins (C/EBP), a family of basic leucine zipper transcription factors known to be involved in the differentiation of several cell types, are implicated in the transcriptional control of IGFBP5 (Cesi et al., 2005).
RPE cells are able to produce variety of cytokines and growth factors that may play a role not only in the development, differentiation, and survival of retinal cells but also in several intraocular pathological conditions (Hayashi et al., 1996; Hicks, 1991). The RPE possesses receptors for IGFs and secretes IGF1 and IGF2 as well as IGFBPs (Yang and Chaum, 2003). The importance of cell specific expression of IGFBPs within the eye is to modulate the biological activity of IGFs. IGFBP5 secreted by RPE cells into the interphotoreceptor matrix can modulate IGF levels, which may affect neovascularization of the retina and iris (Punglia et al., 1997). The local expression of IGFBP5 in the ganglion and bipolar layer of neuronal retina regulate IGF1-mediated retinal neurogenesis in fish (Otteson et al., 2002). By microarray analysis we indentified IGFBP5 as a gene that is differentially expressed during 4HPR-induced neuronal differentiation of RPE cells. Here we present evidence that IGFBP5 is expressed in human RPE cells, and that its expression, mRNA and protein, are greatly decreased during the neuronal differentiation of RPE cells induced by 4HPR. We show that the regulation appears to be at the level of transcription and that it is mediated through C/EBPβ.
Materials and methods
Materials
4HPR (N-(4-hydroxyphenyl)retinamide or fenretinide) was obtained from Biomol (Plymouth Meeting, PA). Monoclonal anti-IGFBP5 and anti-β-actin were from US Biologicals (Swampscott, MA). Recombinant Human IGFBP5, a 245 amino acid residue recombinant mature human IGFBP5, was from R&D Systems (Minneapolis, MN). The enhanced chemiluminescence (ECL) detection system and peroxidase-conjugated anti-rabbit and anti-mouse antibodies were from GE Healthcare Life Sciences (Piscataway, NJ). Dual-Luciferase® Reporter Assay System was purchased from Promega (Madison, WI). U0126 was purchased from Sigma (St. Louis, MO). C/EBPβ expression vector was kindly provided by Dr. P. Johnson of NCI, Frederick, USA. The wt IGFBP5 and C/EBP truncated IGFBP5 promoter reporter constructs were a kind gift of Dr. G. Raschella of ENEA Research Center Casaccia, Rome, Italy.
Cells and Culture Conditions
Human retinal pigment epithelial cells (ARPE-19 cells) obtained from ATCC (Manassas, VA) were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing nutrient mixture F12 (Cellgro, VA) supplemented with 5% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 µg/ml) as described previously (Samuel et al., 2008). Cells were seeded at a density of 2 × 105 cells/ml in complete medium and allowed to grow overnight. The culture medium was replaced next day with fresh serum-free medium containing penicillin (100 U/ml) and streptomycin (100 µg/ml) before adding 1 µM of 4HPR. U0126, a MEK1/2 inhibitor, or recombinant IGFBP5 were added 1 h prior to the addition of 4HPR. Treatments were performed under subdued light and other conditions as reported previously (Samuel et al., 2001). All compounds were dissolved at a concentration of 10 mM in DMSO before adding to the cell culture medium. The controls received the same amount of DMSO. The cells were maintained at 37°C in a humidified environment of 5% CO2 in air.
Analysis of neurite outgrowth
Cells were examined using an inverted microscope (model IX 70; Olympus, Tokyo, Japan) every day using criteria similar to our earlier report (Chen et al., 2003; Samuel et al., 2008). Briefly, the cells were judged to be differentiated when the length of their processes was longer than the diameter of the soma or at least two neurites extending from the soma. Cells bearing bidirectional or multidirectional neurite-like processes were counted in minimum 10 randomly selected fields. The percentage of differentiation was calculated from the number of cells that showed neurite outgrowth divided by the total number of cells in each field. Three dishes were used in each experiment, which was repeated three times.
Microarray analysis
Total RNA, 100 ng, was amplified according to Affymetrix’s small sample protocol, and 20 µg of cRNA was then hybridized on each human genome U133 plus 2.0 GeneChip. After hybridization, GeneChip array was washed, stained with streptavidin-PE (Molecular Probes), amplified with biotinylated anti-streptavidin antibody and scanned with an argon ion Confocal Laser at 570 nm (Affymetrix). Affymetrix GeneChip Operating software was used for absolute expression and to normalize the gene expression levels between any two samples. Data were then imported into GeneSpring software 7.2 (Silicon Graphics) for chip normalization, filtering and cluster analysis.
Western Immunoblot Analysis
Equal amounts of total protein (50 µg) from each sample were subjected to SDS-polyacrylamide gel electrophoresis using 4–12% NUPAGE Bis-Tris gels and then transferred to a nitrocellulose membrane (Invitrogen). After blocking in 5% non-fat milk in Tris-buffered saline (TBS) containing 0.05% Tween 20 for 1 h, the membranes were incubated overnight at 4°C with monoclonal anti-IGFBP5 at 1:1000 dilutions (US Biological). Peroxidase-conjugated anti-mouse IgG antibody (1:5000) was used as secondary antibody. Immunocomplexes were visualized by a chemiluminescence method using the ECL Plus Western blotting Detection Kit (Amersham Biosciences).
Quantitative Real-Time RT-PCR
For quantitative real-time RT-PCR, 2 µg of total RNA extracted from ARPE-19 cells with RNeasy Protect Mini Kit (Qiagen) was reverse transcribed using High Capacity cDNA Archive Kit (Applied Biosystems). After reverse transcription, 5 µl of cDNA preparations were used as templates for quantitative real-time PCR performed on an Applied Biosystems 7500 Real-Time PCR System using TaqMan Universal PCR Master Mix and other reagents from Applied Biosystems following manufacturer’s protocols. Each PCR reaction was set up in 50 µl using validated TaqMan probe and primers specific for calretinin (assay ID Hs00418693_m1) and IGFBP5 (assay ID Hs01052296_ml). Human GAPDH gene (catalog number 4326317E) was used as endogenous control. The gene specific probe was labeled with reporter dye FAM, and the endogenous control GAPDH was labeled with a different reporter dye VIC at the 5’ end. Gene amplification data were analyzed with an Applied Biosystems 7500 System Sequence Detection Software version 1.2.3. The results were expressed as n-fold induction or inhibition in gene expression relative to endogenous control calculated using the ΔΔCT method.
Transient transfection and luciferase Assay
The transient transfections were performed on 1 × 106 cells (ARPE-19) by electroporation using the Cell Line Nucleofector® V Kit and the Nucleofector™ II instrument (Amaxa) according to the manufacture’s protocol. For C/EBPβ overexpression, C/EBPβ gene inserted as EcoRI-HindIII fragment into the pcDNA3.1 vector was used. As control vector pcDNA3.1/V5-His-TOPO lacZ vector (Invitrogen) was used. Cells were plated on 6-well plates, and culture medium was changed 18 h after transfections to serum-free medium, and treated with 1µM 4HPR for 72 h. For luciferase assay, 2 µg each of human wild type IGFBP5 promoter (wt IGFBP5-prom) reporter construct, or C/EBP binding site truncated IGFBP5 promoter (mut IGFBP5-prom) reporter construct were co-transfected with 15 ng of Renilla luciferase construct (pRLCMV). The pGL3-basic vector was used as a control. After 24 h post-transfection, the cells were treated with 4HPR (1 µM) for a further 48 h before harvest. The cells were lysed in 250 µl of 1 × Passive Lysis Buffer and stored at −20°C until assayed. Luciferase assays were performed using Dual-Luciferase Reporter Assay System (Promega) according to manufacture’s instructions, and was expressed as relative luciferase activity by normalizing firefly luciferase activity against Renilla luciferase activity.
Statistical analysis
All values have been expressed as mean ±SD, n = 4. For statistical significance, paired Student’s t-test in Excel was used. P < 0.05 denotes statistically significant differences. The results shown are representative of 3 or more independent experiments.
Results
Identification of IGFBPs as a differentially expressed gene in 4HPR-induced neuronal differentiation of RPE cells
Microarray analyses to identify candidate genes affected by 4HPR-induced neuronal differentiation of RPE cells were performed using GeneChip Human Genome U133 plus 2.0 arrays. Total RNA was isolated from control cultures and from cells after 72 h of stimulation with 1 µM of 4HPR. Although 4HPR treatment would likely regulate the expression of genes earlier than 72 h, this time point was selected for study since 4HPR-induced neuronal differentiation was prominent at this time point in this cells (Samuel et al., 2008). The data are summarized in Table 1. Among these genes, IGFBP5 was down regulated by more than 16-fold. Thus, in this study, we particularly focused on the characterization of 4HPR-induced expression of IGFBP5.
Table 1.
List of selected genes differentially expressed during microarray analysis in 4HPR-induced neuronal differentiation of human RPE cellsa
| Symbol | Gene Name | Gene Function | Fold Change |
|---|---|---|---|
| Down-regulated genes | |||
| MME | Membrane-metallo-endopep | cell-cell signaling, proteolysis | −18 |
| IGFBP5 | Insulin-like growth factor BP | cell growth, signal transduction | −16 |
| LEPR | Leptin receptor | energy metabolism, development | −4 |
| GAS6 | Growth-arrest-specific 6 | cell proliferation, signal transduction | −2 |
| COL11A1 | Collagen, type X1, α1 | cell-cell adhesion, organization | −2 |
| INSIG1 | Insulin-induced gene 1 | cell proliferation, metabolism | −2 |
| Up-regulated genes | |||
| IL-8 | Interleukin 8 | cell cycle arrest, proliferation | 27 |
| HSPA1A | Heat Shock 70kDa | heat shock response | 13 |
| GPX3 | Glutathione peroxidase | response to lipid hydroperoxide | 8 |
| ABCA1 | ATP-binding cassette-1 | phagocytosis, cholesterol metabolism | 8 |
| DHRS3 | Dehydrogenase/reductase | fatty acid metabolism, vision | 7 |
| GLRX | Glutaredoxin | electron transport | 6 |
| HMOX1 | Heme oxygenase 1 | heme oxidation | 5 |
| IGFBP6 | Insulin-like growth factor BP | cell growth, proliferation | 4 |
| EDF1 | Endothelial differentiation F1 | cell growth/ or maintenance | 3 |
| FABP5 | Fatty acid binding protein 5 | epidermal differentiation, transport | 2 |
| TIMP1 | Tissue inhibitor of MP 1 | cell proliferation, proteolysis | 2 |
ARPE-19 cells in culture were treated with 1 µM of 4HPR for 72 h. Total RNA was isolated from control and treated cells, and gene expression was analyzed using the GeneChip Human Genome U133 Plus 2.0 array (Affymetrix).
Down-regulation of IGFBP5 expression in 4HPR-induced neuronal differentiation
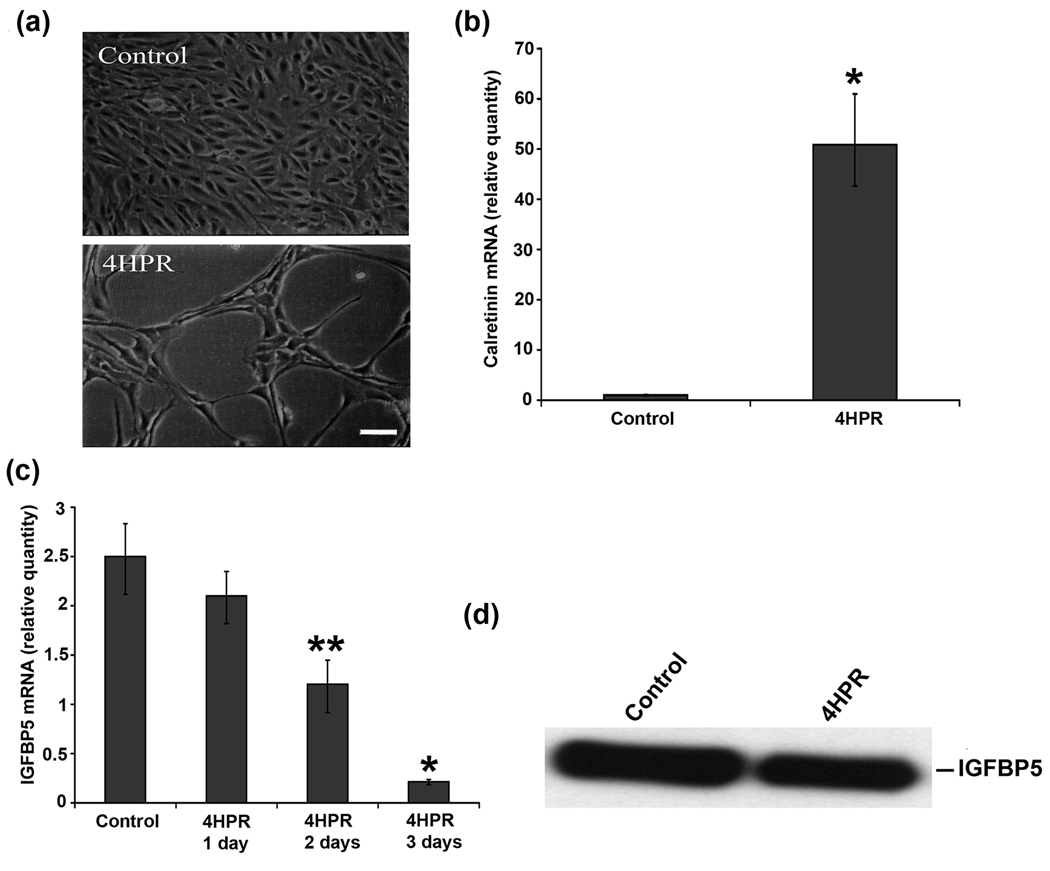
After treatment with 1 µM 4HPR for 72 h, the morphology of ARPE-19 cells was examined by phase-contrast microscopy. As shown in Fig. 1(a), the treatment resulted in visible changes in cell morphology such as shrinkage of the cell body and appearance of processes longer than the cell body. This morphological change produced long processes that are characteristic of neurites, and is similar to our earlier report (Samuel et al., 2008). To further corroborate the observed morphological changes with neuronal differentiation, we analyzed the expression of calretinin, a Ca2+-binding protein normally expressed in retinal ganglion cells and other retinal neurons (Nag and Wadhwa, 1999; Pochet et al., 1989), by RT-PCR as reported earlier (Samuel et al., 2008). Calretinin mRNA expression was markedly increased with 4HPR treatment as expected (Fig. 1b), supporting the light microscopy observations.
Fig.1. IGFBP5 is expressed differentially in 4HPR-induced neuronal differentiation of human RPE cells.
Cultured cells were treated with 1 µM 4HPR for 3 days, and the total RNA preparations were analyzed by real-time quantitative PCR as described under Materials and Methods. The values are mean ± SD, n = 4. *P < 0.001 compared with control. **P < 0.01 compared with control. Panel a, Phase-contrast microscopy analysis of ARPE-19 cells treated with 4HPR. Scale bar = 100 µm. Panel b, calretinin mRNA expression is induced by 4HPR. Panel c, the time-dependent decrease in IGFBP5 mRNA expression by 4HPR. Panel d, 4HPR-induced decrease in expression (protein) of IGFBP5. Equal amount of serum-free culture medium from control and 4HPR treated ARPE-19 cells collected after 72 h was concentrated 50-fold, and then analyzed by Western blot using antibody specific for IGFBP5.
To confirm the results of the microarray analysis, we examined the expression of IGFBP5 in the 4HPR-induced neuronal differentiation of ARPE-19 cells. The expression of IGFBP5 mRNA was decreased with 4HPR treatment in a time-dependent manner (Fig. 1c). Significant decrease in IGFBP5 expression was observed after 48 h of treatment, and more than 90% decrease over the control after 72 h. To see whether the differential expression of IGFBP5 mRNA by 4HPR results in altered protein expression, we performed Western blot analysis on conditioned media from 4HPR treated cells. We find that 4HPR treatment decreased the expression of IGFBP5 protein (Fig. 1e).
MAPK pathway regulates IGFBP5 expression during 4HPR-induced neuronal differentiation of RPE cells
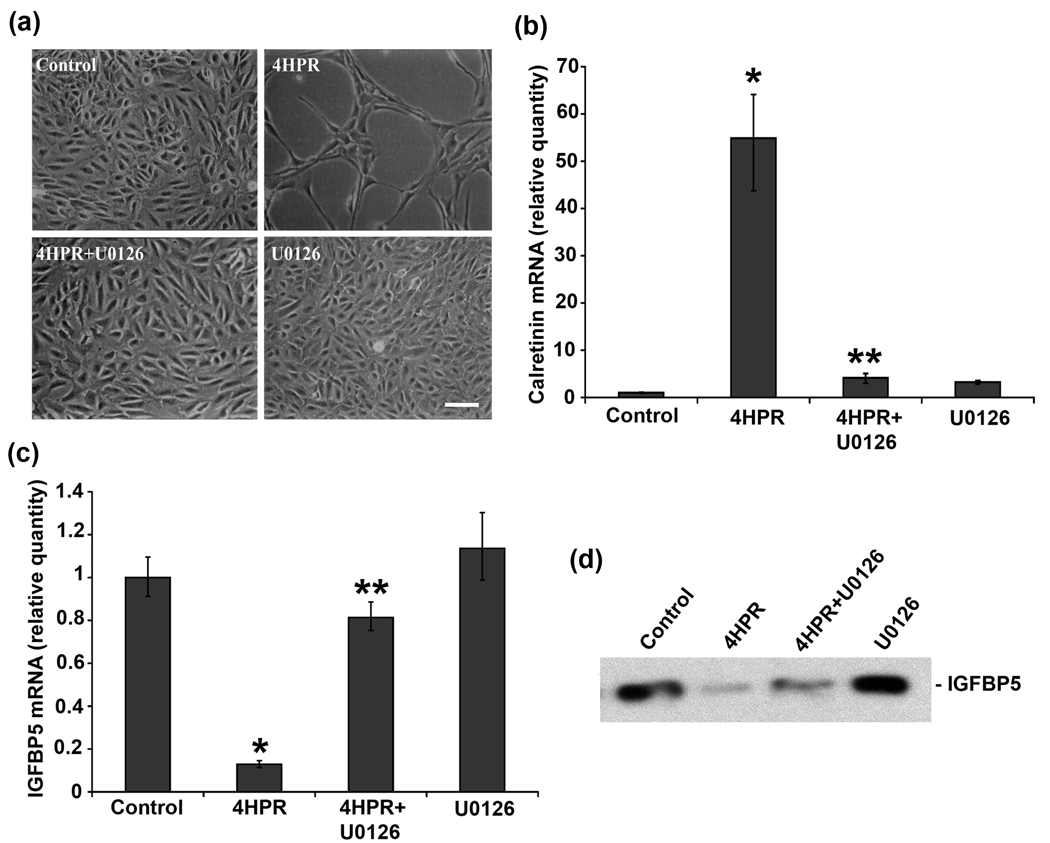
It is known that MAPK signaling cascades play a crucial role in regulating mammalian cell growth and differentiation (Pearson et al., 2001). To address whether the MAPK pathway is involved in 4HPR-induced IGFBP5 expression, we used U0126, which specifically inhibits both the inactive and active forms of MEK1/2 (Favata et al., 1998). In correlation with our earlier observation, 4HPR treatment induced neurite outgrowth in ARPE-19 cells (Samuel et al., 2008), and the cells pretreated with 1 µM of U0126 effectively blocked the neuronal differentiation induced by 4HPR (Fig. 2a). As expected, the neuronal differentiation induced by 4HPR was associated with an increase in calretinin expression which was completely blocked when cells were pretreated with U0126 (Fig.2b).
Fig.2. MEK1/2 inhibitor U0126 blocked the decrease of IGFBP5 mRNA expression induced by 4HPR.
ARPE-19 cells in culture were pretreated with 1 µM of U0126, for 1 h followed by incubation with 4HPR for additional 72 h. Total RNA extracted from treated cells was analyzed by real-time quantitative PCR as described under Materials and Methods. The values are mean ± SD, n = 4. *P < 0.001 compared with control; **P < 0.001 compared with 4HPR treatment. Panel a, Phase-contrast microscopic analysis of the inhibition of 4HPR-induced neuronal differentiation of ARPE-19 cells by U0126. Scale bar = 100 µm. Panel b, the inhibition of 4HPR-induced calretinin mRNA expression by U0126. Panel c, the inhibition of 4HPR-induced decrease of IGFBP5 mRNA expression by U0126. Panel d, the inhibition of 4HPR-induced decrease of IGFBP5 expression (protein) by U0126. Equal amount of serum-free culture medium from control and 4HPR treated ARPE-19 cells collected after 72 h was concentrated 50-fold, and then analyzed by Western blot using antibody specific for IGFBP5.
As shown in Fig. 2c, the expression of IGFBP5 was reduced by 90% in differentiating RPE cells treated with 4HPR for 72 h. This 4HPR-induced down regulation of IGFBP5 expression was blocked in cells pretreated with U0126. Next, we performed Western blot analysis in the conditioned media from cells treated with U0126 (1 µM) 1 h prior to 4HPR treatment. As expected, the down regulation of IGFBP5 mRNA expression translated into a corresponding decrease in IGFBP5 protein expression, and the decrease was inhibited by U0126 pretreatment (Fig. 2d). U0126 by itself did not affect the IGFBP5 expression in ARPE-19 cells.
4HPR-induced neuronal differentiation is independent of IGFBP5 expression
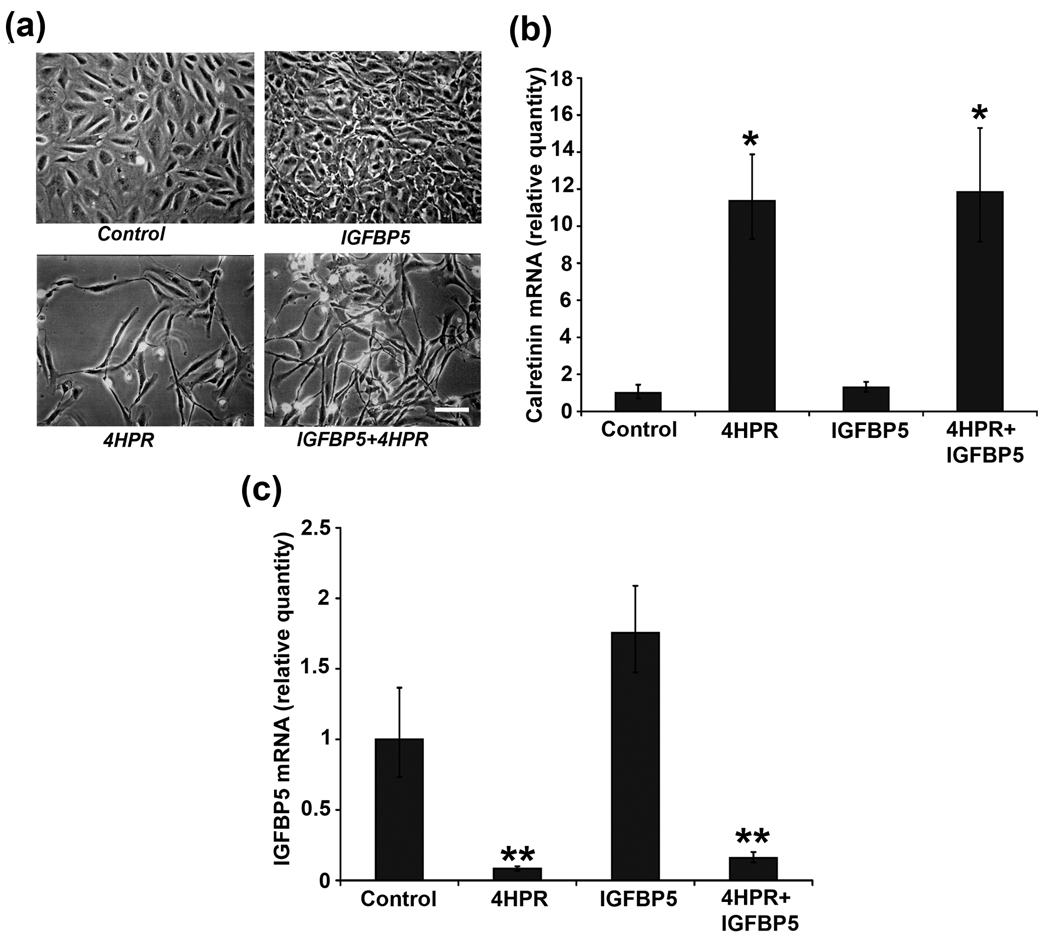
To further elucidate whether the 4HPR-induced neuronal differentiation is dependent on IGFBP5 expression, exogenous recombinant IGFBP5 was added to the cells and morphological changes and the expression of calretinin were analyzed by phase-contrast microscopy and by RT-PCR, respectively. Addition of 100 ng/ml of recombinant IGFBP5 did not inhibit 4HPR-induced neuronal differentiation but rather augmented the differentiation of ARPE-19 cells (Fig. 3a). Also, the administration of recombinant IGFBP5 increased the proliferation of RPE cells when compared to control.
Fig.3. Recombinant IGFBP5 is unable to block 4HPR-induced neuronal differentiation.
ARPE-19 cells in culture were treated with recombinant IGFBP5 (100 ng/ml) for 5 h followed by incubation with 4HPR for additional 72 h. Total RNA extracted from cells treated in the presence or absence of recombinant IGFBP5 was analyzed by real-time quantitative PCR as described under Materials and Methods. The values are mean ± SD, n = 4. *P < 0.001 compared with control. **P < 0.01 compared with control. Panel a, Phase-contrast microscopic analysis of 4HPR-induced neuronal differentiation of ARPE-19 cells after the addition of recombinant IGFBP5. Scale bar = 100 µm. Panel b, 4HPR-induced calretinin mRNA expression was not blocked by the addition of IGFBP5 recombinant protein. Panel c, IGFBP5 recombinant protein addition did not block 4HPR-induced decrease of IGFBP5 mRNA expression
In addition, as expected, a significant induction in calretinin expression was observed with 4HPR treatment (Fig. 3b). This increase in calretinin expression was not blocked by the exogenous recombinant IGFBP5 protein. Further, the cells treated for 72 h with IGFBP5 alone did not induce the expression of calretinin.
The IGFBP5-dependent and –independent effect on neuronal differentiation was further demonstrated by analyzing the expression of IGFBP5 mRNA in 4HPR treated cells in the presence or absence of recombinant IGFBP5 (100 ng/ml) for 72 h. As shown in Fig. 3c, 4HPR treatment greatly decreased the expression of IGFBP5 during the neuronal differentiation. This decrease in IGFBP5 mRNA expression was not blocked by the addition of recombinant IGFBP5.
C/EBPβ mediates the neuronal differentiation of RPE cells induced by 4HPR
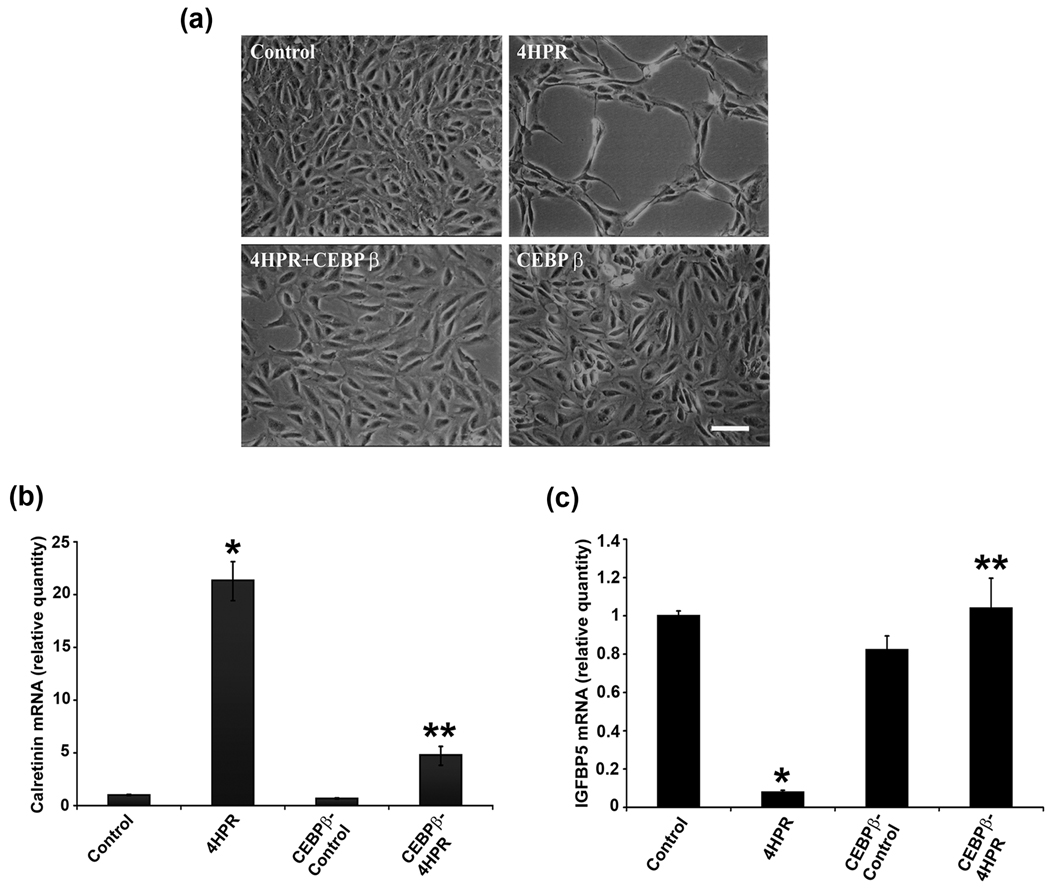
As CCAAT/enhancer binding proteins (C/EBP) play a pivotal role in numerous cellular responses (Ramji and Foka, 2002), and in particular, C/EBPβ is induced during the early phase of differentiation (Williams et al., 1995), we investigated the expression of C/EBPβ during 4HPR-induced neuronal differentiation of RPE cells by real time RT-PCR analysis (Fig. 4a). Following a more than 3-fold increase in C/EBPβ expression during first 1–2 days differentiation there was a time-dependent decrease in expression. After 3 days of treatment the expression level decreased below basal level, and decreased further by 6 days. To address whether C/EBPβ is involved in 4HPR-induced neuronal differention, cells were transfected with C/EBPβ expression vector and after each transfected cell line was treated with 4HPR for 72 h, the morphology was examined by phase contrast microscopy. As shown in Fig. 4b, cells overexpressing C/EBPβ did not undergo neuronal differentiation induced by 4HPR. The inhibition was specific since cells treated with transfection reagent alone underwent the neuronal differentiation induced by 4HPR. Cell transfected with C/EBPβ alone for 72 h retained their normal epithelial morphology and appeared viable.
Fig.4. 4HPR-induced neuronal differentiation is blocked by C/EBPβ overexpression.
ARPE-19 cells were treated with 1µM 4HPR for indicated time, and the total RNA preparations were analyzed by real-time quantitative PCR as described under Materials and Methods. The values are mean ± SD, n = 4. *P < 0.001 compared with control. **P < 0.001 compared with 4HPR treatment. Panel a, the time-dependent change in C/EBPβ mRNA expression during 4HPR-induced neuronal differentiation of RPE cells. Panel b, C/EBPβ overexpression blocked 4HPR-induced neuronal differentiation. Cells in culture were transfected with C/EBPβ expression vector, and after 24 h post-transfection the cells were treated with 1 µM 4HPR for 72 h, and then analyzed by phase contrast microscopy. Scale bar = 100 µm. Panel c, C/EBPβ transfection blocked calretinin mRNA expression induced by 4HPR. Cells were transfected with C/EBPβ expression vector and treated as described in panel b. Panel d, C/EBPβ expression inhibits 4HPR-induced decrease of IGFBP5 mRNA expression. Cells were transfected with C/EBPβ expression vector and treated as described in panel b.
To confirm that overexpression of C/EBPβ blocks neuronal differentiation, we analyzed the expression of calretinin. Treatment of ARPE-19 cells with 4HPR induced the expression of calretinin as expected (Fig. 4c), but was reduced in cells overexpressing C/EBPβ compared to untransfected 4HPR treated cells. These data support the light microscopy findings.
Apart from taking active part in the differentiation of several cell types, C/EBPβ also regulates the expression of IGFBP5 during RA treatment (Cesi et al., 2005). To study whether C/EBPβ play a role in IGFBP5 expression during 4HPR-induced neuronal differentiation, cells transfected with C/EBPβ expression vector were treated with 1 µM of 4HPR, and were analyzed by RT-PCR after 72 h (Fig. 4d). Interestingly, when C/EBPβ was overexpressed, 4HPR was not able to decrease IGFBP5 expression as observed with 4HPR treatment in differentiating cells. In addition, the expression level of IGFBP5 was similar to that of control in C/EBPβ overexpressed cells after 4HPR treatment. The inhibition was specific, since the cells transfected with C/EBPβ alone did not show inhibition of IGFBP5 expression.
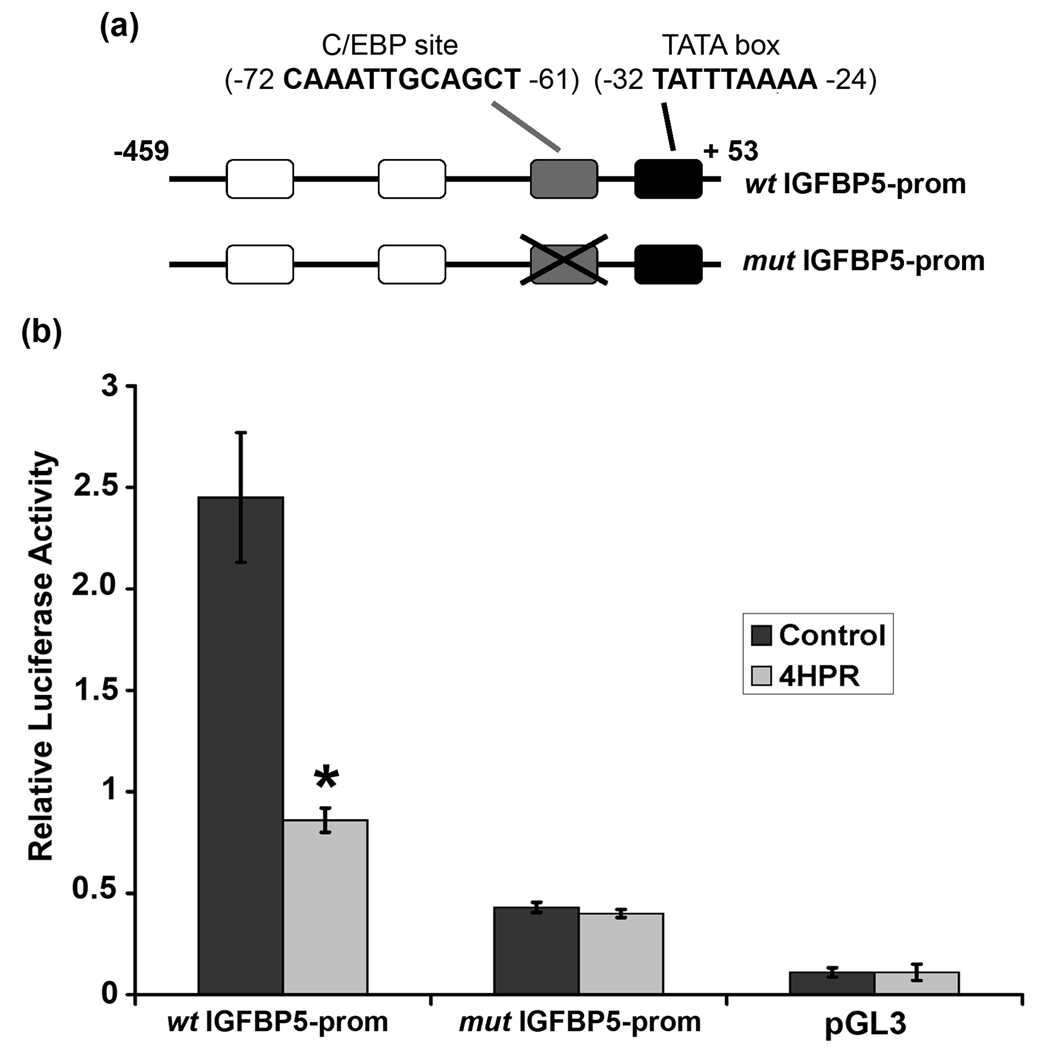
C/EBP transactivate the proximal region of the human IGFBP5 promoter
Analysis of the proximal promoter region of the human IGFBP5 reveals a known putative C/EBP binding site at position −72 to −61 (Fig. 5a). To assess whether C/EBP could be, at least in part, involved in transcriptional regulation of IGFBP5, we carried out luciferase assays in ARPE-19 cells with a reporter vector containing −459 to +59 of the human IGFBP5 promoter. ARPE-19 cells were transfected with 2 µg of the human wild type IGFBP5 promoter reporter vector (wt IGFBP5-prom) in the presence or absence of 4HPR. We observed that the activity of the human IGFBP5 promoter was decreased approximately 2.5 fold in 4HPR treated cells compared to untreated cells after 72 h posttransfection (Fig. 5b). The promoter activity of the pGL3-Basic vector in transfected ARPE-19 cells was very low and was not affected by 4HPR treatment. In order to test whether the C/EBP response element located between nucleotide −72 to −61 plays a role in the 4HPR-induced differentiation, we transfected a deletion construct in which the C/EBP response element in the human IGFBP5 promoter was deleted (mut IGFBP5-prom). Reporter activity of this construct was greatly reduced compared to wt IGFBP5 promoter and was not affected by 4HPR treatment (Fig. 5b). However, mut IGFBP5 promoter construct had 4-fold higher activity than the pGL3-Basic vector.
Fig.5. C/EBP response element regulates IGFBP5 transcription induced by 4HPR.
Panel a, scheme of the proximal IGFBP5 promoter. White, grey and black boxes represent the binding sites of Myb and C/EBP and the TATA box, respectively. C/EBP binding site is truncated in mut IGFBP5-prom construct, and not in wt IGFBP5-prom construct. Panel b, C/EBP mediates 4HPR-induced transactivation of IGFBP5. The ARPE-19 cells growing in monolayer were transiently transfected with wt IGFBP5-prom or mut-IGFBP5-prom reporter constructs. After 24 h post transfection, the cells were treated with 1 µM 4HPR for additional 72 h before measuring luciferase activity. Relative luciferase activity is expressed as the ratio of firefly/Renilla luciferase activities. The values are mean ± SD, n = 4. *P < 0.001 compared with control.
Discussion
In this study, we observed that the expression of IGFBP5 was decreased drastically during 4HPR-induced neuronal type differentiation of human RPE cells. An increase in IGFBP5 expression has been associated with the onset of differentiation in osteoblasts (Thrailkill et al. 1995), and as a differentiation-promoting factor in skeletal muscle cells (Ren et al., 2008; Thrailkill et al., 1995). In contrast, overexpression of IGFBP5 inhibited the myogenic differentiation of mouse C2 myoblasts (Mukherjee et al., 2008). This suggests that the IGFBP5 effects can be either growth inhibitory or stimulatory, depending on the cellular context. We observed that during the neuronal differentiation, the expression of IGFBP5 was decreased in a time-dependent manner. This result substantiates an observation that IGFBP5 expression increases during the early stage and diminishes in terminal differention (Lin et al., 2002). Further, the observed decrease in IGFBP5 mRNA expression correlates with the levels of IGFBP5 protein secretion into culture medium during the neuronal differentiation.
The expression of IGFBP5 decreases during 4HPR-induced neuronal differentiation of human RPE cells, suggesting a potential role for this protein in this process. However, we observed that exogenous recombinant IGFBP5 did not impair the neuronal differentiation of RPE cells or the associated increase in calretinin expression after 4HPR treatment. On the other hand, it augmented the 4HPR-induced neuronal differentiation of RPE cells. The increase in calretinin expression was also not blocked by exogenous IGFBP5. It has been shown that, exogenous recombinant IGFBP5 reversed the impairment of neuronal differentiation in neuroblastoma cells (Tanno et al., 2005). Their study also suggests that an optimal level of IGFBP5 is necessary and a prerequisite for the neuronal differentiation of neuroblastoma cells. Further, IGFBP5 functions as an IGF-independent cytokine and administration of IGF1 or IGF2 recombinant protein neither inhibited the neuronal differentiation nor decreases the calretinin expression (data not shown). Together these data indicate that the decrease in IGFBP5 expression that we observed appears to be due to neuronal differentiation and not a prerequisite for the neuronal differentiation of human RPE cells.
It is known that MAPK signaling cascades play a crucial role in regulating mammalian cell growth and differentiation. We have reported earlier that the 4HPR-induced neuronal differentiation of human RPE cells was mediated through a signal transduction pathway involving MAPKs (Samuel et al., 2008). In the present study, we observed that U0126, a potent inhibitor of the dual-specific protein kinase MEK1 and MEK2, not only blocked the 4HPR-induced neuronal differentiation of human RPE cells and the expression of neuronal marker, calretinin, but also blocked both decrease in IGFBP5 mRNA expression and protein in the conditioned media. This correlates with an earlier observation in rat intestinal smooth muscle cells where IGF-1 modulates IGFBP5 expression exclusively via the MAPK pathway (Xin et al., 2004). The downstream regulators of the MAPK intracellular signaling cascades include members of the CCAAT/enhancer binding protein (C/EBP), ATF/CREB and the AP-1 family (Kuemmerle and Zhou, 2002; Umayahara et al., 1999). In particular, C/EBPs are a family of basic leucine zipper transcription factors which take part in differentiation of several cell types (Ramji and Foka, 2002). In the present study, we observed a time-dependent change in the expression of C/EBPβ during 4HPR-induced neuronal differentiation. Interestingly, the expression of C/EBPβ increased during the early and mid stages of differentiation but diminished during the terminal differentiation, as assessed by the neuronal outgrowth and expression of the neuronal marker calretinin. A similar result was also observed in the case of C/EBPα (data not shown). The induction during the early stage and the decrease during the later phase of differentiation are concordant with our observation of high and low level expression of calretinin and IGFBP5, respectively. This transcriptional activation and/or repression of genes may be necessary for the maintenance of the differentiated state of RPE cells. Such a situation is seen in non-neural cells like 3T3-L1, in which C/EBPβ expressed early in the differentiation program and is thought to be responsible for transcriptional activation and for the maintenance of the differentiated states of adipose and hepatic tissues (Darlington et al., 1998; Umek et al., 1991). In addition, we have demonstrated earlier that the phosphorylation of ERK1/2 is necessary for the neuronal differentiation of RPE cells (Samuel et al., 2008). Once ERK1/2 is phosphorylated, it activates the phosphorylation of C/EBPβ downstream of the signaling cascade to mediate activation and/or repression of gene expression in response to growth factors via different mechanisms (Chen et al., 1996).
The family members of C/EBPs act in cell-type specific manner to promote and/or repress transcription of genes essential for terminal differentiation. Here we show that overexpression of C/EBPβ in RPE cells blocks the neuronal type differentiation and calretinin expression induced by 4HPR. Forced expression of C/EBPβ has been shown to induce proliferation, and regulate the expression of differentiation-specific genes in a wide range of cell types (Johnson, 2005). Thus, it is possible that C/EBPβ overexpression might block the neuronal differentiation through a similar mechanism. In the present study, we have also observed that overexpression of C/EBPβ not only blocks the neuronal differentiation but also 4HPR-induced down regulation of IGFBP5. In neuroblastoma cells, the transcriptional regulation of IGFBP5 during RA-induced differentiation depends on the binding of either C/EBPα and/or C/EBPβ to their regulatory site (Cesi et al., 2005). The same study also reported that both C/EBPα and β binds TATA box in basal growth conditions in vitro and in vivo. Thus, it is interesting to speculate that overexpression of C/EBPβ relieves the repression of IGFBP5 expression by binding to their response elements, and/or through the activation of yet other unknown transcription factors.
The effect of C/EBPs on cell proliferation and differentiation is mediated by its ability to bind upstream CCAAT elements to regulate gene expression (Tenen et al., 1997). However, studies have provided evidence that these factors also regulate cell fate by protein-protein interaction independent of their transcriptional activity (McKnight, 2001). In our experiments with human RPE cells, we observed that the 459 base pair human IGFBP5 promoter has approximately 2.5 fold increase in basal transcriptional activity compared to the basic vector. This basal transcription activity of the IGFBP5 promoter was markedly diminished after 4HPR treatment. This is consistent with a study in neuroblastoma cell line that the C/EBP-responsive element, located between −72 to −61 relative to the transcription start site, had a repressive effect on IGFBP5 transcription after RA treatment (Cesi et al., 2005). The same study has also pointed out that C/EBPs could act as a transcriptional enhancer and/ or repressor through a mechanism that does not rely on binding to a C/EBP site. Here, in the present study, we observed that the C/EBP deletion construct not only has a very low basal transcription activity, compared to wt IGFBP5 promoter construct, but also is non-responsive to 4HPR treatment, in functional studies. Using deletion and point mutation analysis of the IGFBP5 promoter, it was established that mutations in the C/EBP site decreased basal promoter activity and also eliminated the stimulatory effect on luciferase expression (Ji et al., 1999). On the other hand, mutation of CCAAT/enhancer element in an IGFBP5-promoter reporter construct has been shown to increase the transcription of transiently transfected plasmid in human neuroblastoma cell line (Cesi et al., 2005). Thus, the regulation of IGFBP5 expression during the neuronal differentiation of RPE cells by these factors reflects their cell specific effects, especially during cell growth and differentiation.
In summary, we have identified IGFBP5 as a gene differentially regulated by 4HPR. IGFBP5 is expressed in human RPE cells, and its expression, mRNA as well as protein, are greatly decreased during the neuronal differentiation of RPE cells induced by 4HPR. However, this decrease in IGFBP5 expression and the neuronal differentiation appears to occur through different mechanisms, since ectopic expression of IGFBP5 does not block the neuronal differentiation induced by 4HPR. U0126, an inhibitor of MEK1/2, inhibits the 4HPR-induced down regulation of IGFBP5 expression, indicating that MAPK pathway plays a central role in these processes as is the case with 4HPR-induced neuronal differentiation. Further, the inhibition of 4HPR-induced neuronal differentiation and the blockade of IGFBP5 down-regulation by C/EBPβ suggest that C/EBPs function downstream of MAPK pathway in determining the cellular differentiation of human RPE cells. The human IGFBP5 promoter showed a decrease in transcriptional activation in the presence of C/EBP response element after 4HPR treatment. Deletion of the C/EBP responsive element markedly decreased the basal promoter activity and abolished its responsiveness to 4HPR treatment, suggesting that the expression of IGFBP5 appears to be regulated by C/EBP. Thus, our results identify C/EBPβ as a potent regulator of neurite outgrowth in human RPE cells, and suggest an important role for this transcription factor in modulating the expression of IGFBP5 during 4HPR-induced neuronal differentiation of RPE cells.
Fig.6.
Acknowledgements
This research work was supported by the Intramural Research Program of the National Eye Institute, National Institutes of Health.
Abbreviations used are
- RPE
retinal pigment epithelium
- 4HPR
N-(4-Hydroxyphenyl)-retinamide
- IGF
Insulin-like growth factor
- IGFBP
Insulin-like growth factor binding protein
- C/EBP
CCAAT/enhancer binding protein
- MAPK
mitogen-activated protein kinase
- MEK1/2
mitogen activated protein kinase/extracellular signal-regulated kinase 1 and 2
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
References
- Allander SV, Larsson C, Ehrenborg E, Suwanichkul A, Weber G, Morris SL, Bajalica S, Kiefer MC, Luthman H, Powell DR. Characterization of the chromosomal gene and promoter for human insulin-like growth factor binding protein-5. The Journal of biological chemistry. 1994;269:10891–10898. [PubMed] [Google Scholar]
- Beattie J, Allan GJ, Lochrie JD, Flint DJ. Insulin-like growth factor-binding protein-5 (IGFBP-5): a critical member of the IGF axis. The Biochemical journal. 2006;395:1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D. The retinal pigment epithelium: a versatile partner in vision. J Cell Sci. 1993;17 Suppl:189–195. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- Cesi V, Giuffrida ML, Vitali R, Tanno B, Mancini C, Calabretta B, Raschella G. C/EBP alpha and beta mimic retinoic acid activation of IGFBP-5 in neuroblastoma cells by a mechanism independent from binding to their site. Experimental cell research. 2005;305:179–189. doi: 10.1016/j.yexcr.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes & development. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- Chen S, Samuel W, Fariss RN, Duncan T, Kutty RK, Wiggert B. Differentiation of human retinal pigment epithelial cells into neuronal phenotype by N-(4-hydroxyphenyl)retinamide. Journal of neurochemistry. 2003;84:972–981. doi: 10.1046/j.1471-4159.2003.01608.x. [DOI] [PubMed] [Google Scholar]
- Cobb LJ, Salih DA, Gonzalez I, Tripathi G, Carter EJ, Lovett F, Holding C, Pell JM. Partitioning of IGFBP-5 actions in myogenesis: IGF-independent anti-apoptotic function. Journal of cell science. 2004;117:1737–1746. doi: 10.1242/jcs.01028. [DOI] [PubMed] [Google Scholar]
- Darlington GJ, Ross SE, MacDougald OA. The role of C/EBP genes in adipocyte differentiation. The Journal of biological chemistry. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- de Jong PT. Age-related macular degeneration. The New England journal of medicine. 2006;355:1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Hayashi A, Nakae K, Naka H, Ohji M, Tano Y. Cytokine effects on phagocytosis of rod outer segments by retinal pigment epithelial cells of normal and dystrophic rats. Current eye research. 1996;15:487–499. doi: 10.3109/02713689609000760. [DOI] [PubMed] [Google Scholar]
- Hicks D, Bugra K, Faucheux B, Jeanny JC, Laurent M, Malecaze F, Mascareli F, Raulais D, Cohen Y, Courtois Y. In: Progress in Retinal Research. Osborne NN, Chader GJ, editors. Oxford: Pergamon Press; 1991. pp. 333–374. [Google Scholar]
- Ji C, Chen Y, Centrella M, McCarthy TL. Activation of the insulin-like growth factor-binding protein-5 promoter in osteoblasts by cooperative E box, CCAAT enhancer-binding protein, and nuclear factor-1 deoxyribonucleic acid-binding sequences. Endocrinology. 1999;140:4564–4572. doi: 10.1210/endo.140.10.7061. [DOI] [PubMed] [Google Scholar]
- Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. Journal of cell science. 2005;118:2545–2555. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- Kuemmerle JF, Zhou H. Insulin-like growth factor-binding protein-5 (IGFBP-5) stimulates growth and IGF-I secretion in human intestinal smooth muscle by Ras-dependent activation of p38 MAP kinase and Erk1/2 pathways. The Journal of biological chemistry. 2002;277:20563–20571. doi: 10.1074/jbc.M200885200. [DOI] [PubMed] [Google Scholar]
- Lin SC, Wang CP, Chen YM, Lu SY, Fann MJ, Liu CJ, Kao SY, Chang KW. Regulation of IGFBP-5 expression during tumourigenesis and differentiation of oral keratinocytes. The Journal of pathology. 2002;198:317–325. doi: 10.1002/path.1220. [DOI] [PubMed] [Google Scholar]
- McKnight SL. McBindall--a better name for CCAAT/enhancer binding proteins? Cell. 2001;107:259–261. doi: 10.1016/s0092-8674(01)00543-8. [DOI] [PubMed] [Google Scholar]
- Miyakoshi N, Richman C, Kasukawa Y, Linkhart TA, Baylink DJ, Mohan S. Evidence that IGF-binding protein-5 functions as a growth factor. The Journal of clinical investigation. 2001;107:73–81. doi: 10.1172/JCI10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Wilson EM, Rotwein P. Molecular endocrinology (Baltimore, Md. Vol. 22. 2008. Insulin-like growth factor (IGF) binding protein-5 blocks skeletal muscle differentiation by inhibiting IGF actions; pp. 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag TC, Wadhwa S. Developmental expression of calretinin immunoreactivity in the human retina and a comparison with two other EF-hand calcium binding proteins. Neuroscience. 1999;91:41–50. doi: 10.1016/s0306-4522(98)00654-x. [DOI] [PubMed] [Google Scholar]
- Otteson DC, Cirenza PF, Hitchcock PF. Persistent neurogenesis in the teleost retina: evidence for regulation by the growth-hormone/insulin-like growth factor-I axis. Mechanisms of development. 2002;117:137–149. doi: 10.1016/s0925-4773(02)00188-0. [DOI] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Pittack C, Jones M, Reh TA. Basic fibroblast growth factor induces retinal pigment epithelium to generate neural retina in vitro. Development. 1991;113:577–588. doi: 10.1242/dev.113.2.577. [DOI] [PubMed] [Google Scholar]
- Pochet R, Blachier F, Malaisse W, Parmentier M, Pasteels B, Pohl V, Resibois A, Rogers J, Roman A. Calbindin-D28 in mammalian brain, retina, and endocrine pancreas: immunohistochemical comparison with calretinin. Adv Exp Med Biol. 1989;255:435–443. doi: 10.1007/978-1-4684-5679-0_46. [DOI] [PubMed] [Google Scholar]
- Punglia RS, Lu M, Hsu J, Kuroki M, Tolentino MJ, Keough K, Levy AP, Levy NS, Goldberg MA, D'Amato RJ, Adamis AP. Regulation of vascular endothelial growth factor expression by insulin-like growth factor I. Diabetes. 1997;46:1619–1626. doi: 10.2337/diacare.46.10.1619. [DOI] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. The Biochemical journal. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Yin P, Duan C. IGFBP-5 regulates muscle cell differentiation by binding to IGF-II and switching on the IGF-II auto-regulation loop. The Journal of cell biology. 2008;182:979–991. doi: 10.1083/jcb.200712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi DS, Janick LM, Reh TA. Basic fibroblast growth factor (FGF-2) induced transdifferentiation of retinal pigment epithelium: generation of retinal neurons and glia. Dev Dyn. 1997;209:387–398. doi: 10.1002/(SICI)1097-0177(199708)209:4<387::AID-AJA6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Salih DA, Tripathi G, Holding C, Szestak TA, Gonzalez MI, Carter EJ, Cobb LJ, Eisemann JE, Pell JM. Insulin-like growth factor-binding protein 5 (Igfbp5) compromises survival, growth, muscle development, and fertility in mice. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4314–4319. doi: 10.1073/pnas.0400230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel W, Kutty RK, Nagineni S, Gordon JS, Prouty SM, Chandraratna RA, Wiggert B. Regulation of stearoyl coenzyme A desaturase expression in human retinal pigment epithelial cells by retinoic acid. The Journal of biological chemistry. 2001;276:28744–28750. doi: 10.1074/jbc.M103587200. [DOI] [PubMed] [Google Scholar]
- Samuel W, Kutty RK, Sekhar S, Vijayasarathy C, Wiggert B, Redmond TM. Mitogen-activated protein kinase pathway mediates N-(4-hydroxyphenyl)retinamide-induced neuronal differentiation in the ARPE-19 human retinal pigment epithelial cell line. Journal of neurochemistry. 2008;106:591–602. doi: 10.1111/j.1471-4159.2008.05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MR, Lahm H, Wu M, Hoeflich A, Wolf E. Transgenic mouse models for studying the functions of insulin-like growth factor-binding proteins. Faseb J. 2000;14:629–640. doi: 10.1096/fasebj.14.5.629. [DOI] [PubMed] [Google Scholar]
- Stewart CE, Rotwein P. Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiological reviews. 1996;76:1005–1026. doi: 10.1152/physrev.1996.76.4.1005. [DOI] [PubMed] [Google Scholar]
- Tanno B, Cesi V, Vitali R, Sesti F, Giuffrida ML, Mancini C, Calabretta B, Raschella G. Silencing of endogenous IGFBP-5 by micro RNA interference affects proliferation, apoptosis and differentiation of neuroblastoma cells. Cell death and differentiation. 2005;12:213–223. doi: 10.1038/sj.cdd.4401546. [DOI] [PubMed] [Google Scholar]
- Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- Thrailkill KM, Siddhanti SR, Fowlkes JL, Quarles LD. Differentiation of MC3T3-E1 osteoblasts is associated with temporal changes in the expression of IGF-I and IGFBPs. Bone. 1995;17:307–313. doi: 10.1016/8756-3282(95)00223-z. [DOI] [PubMed] [Google Scholar]
- Umayahara Y, Billiard J, Ji C, Centrella M, McCarthy TL, Rotwein P. CCAAT/enhancer-binding protein delta is a critical regulator of insulin-like growth factor-I gene transcription in osteoblasts. The Journal of biological chemistry. 1999;274:10609–10617. doi: 10.1074/jbc.274.15.10609. [DOI] [PubMed] [Google Scholar]
- Umek RM, Friedman AD, McKnight SL. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- Williams SC, Baer M, Dillner AJ, Johnson PF. CRP2 (C/EBP beta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. The EMBO journal. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X, Hou YT, Li L, Schmiedlin-Ren P, Christman GM, Cheng HL, Bitar KN, Zimmermann EM. IGF-I increases IGFBP-5 and collagen alpha1(I) mRNAs by the MAPK pathway in rat intestinal smooth muscle cells. American journal of physiology. 2004;286:G777–G783. doi: 10.1152/ajpgi.00293.2003. [DOI] [PubMed] [Google Scholar]
- Yan RT, Ma WX, Wang SZ. neurogenin2 elicits the genesis of retinal neurons from cultures of nonneural cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:15014–15019. doi: 10.1073/pnas.261455698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chaum E. A reassessment of insulin-like growth factor binding protein gene expression in the human retinal pigment epithelium. Journal of cellular biochemistry. 2003;89:933–943. doi: 10.1002/jcb.10570. [DOI] [PubMed] [Google Scholar]
- Yeh LC, Adamo ML, Duan C, Lee JC. Osteogenic protein-1 regulates insulin-like growth factor-I (IGF-I), IGF-II, and IGF-binding protein-5 (IGFBP-5) gene expression in fetal rat calvaria cells by different mechanisms. Journal of cellular physiology. 1998;175:78–88. doi: 10.1002/(SICI)1097-4652(199804)175:1<78::AID-JCP9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Zhao S, Rizzolo LJ, Barnstable CJ. Differentiation and transdifferentiation of the retinal pigment epithelium. International review of cytology. 1997;171:225–266. doi: 10.1016/s0074-7696(08)62589-9. [DOI] [PubMed] [Google Scholar]