Abstract
Clonal expansion of virus-specific naive T cells during an acute viral infection results in the formation of memory CD8 T cells that provide the host with long-term protective immunity against the pathogen. Memory CD8 T cells display enhanced effector functions compared with their naive precursors, allowing them to respond more rapidly and effectively to antigen re-encounter. The enhanced functions of memory CD8 T cells are mediated by heritable changes in gene regulation. Expression of select transcription factors along with locus-specific epigenetic modifications are coupled to and are essential in the formation of memory-specific gene expression patterns. Here, we will review the changes in gene expression that accompany development of memory CD8 T cells and discuss chromatin modifications as a potential means for heritable propagation of these changes during homeostatic cell division of self-renewing memory CD8 T cells. Also, we will discuss therapies that manipulate heritable gene regulation as a potential mechanism to restore function to non-functional memory CD8 T cells to combat chronic viral infection.
Keywords: CD8 T cell, epigenetic, immune memory, infection, T-cell differentiation
Introduction
CD8 T lymphocytes are a vital component of the adaptive immune response and are crucial to the control and clearance of intracellular pathogens. The CD8 T-cell response to an infection can be divided roughly into three stages: expansion, contraction and memory (Fig. 1A). In the expansion phase, antigen-specific naive T cells expand ∼105 fold, express select cytokines and homing markers and acquire what are known as ‘effector’ functions, the ability to kill infected cells, secrete anti-viral cytokines and activate other immune cells (1–11). Following viral clearance, the antigen-specific CD8 T-cell population contracts by 90–95% and the remaining 5–10% further differentiates into memory CD8 T cells that persist in the absence of antigen and provide long-term protection against re-infection.
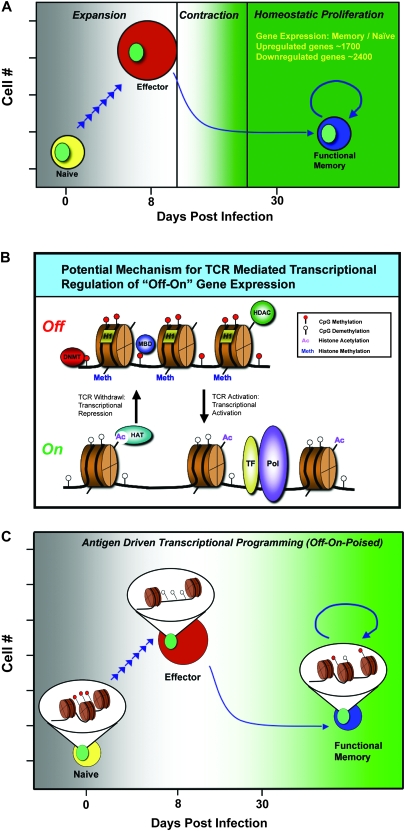
Fig. 1.
Mechanisms for acquired heritable changes in gene expression in the formation of functional memory CD8 T cells. (A) Cartoon depiction of antigen-driven clonal expansion (∼105 fold) of naive CD8 T cells differentiating into cytolytic effector cells. The effector cell population contracts 90–95% yielding a functional memory cell population that is capable of antigen-independent homeostatic proliferation. Gray = slow antigen-driven response, white = active antigen-driven response and green = poised for rapid antigen-driven response. The y-axis is a log scale. Differential gene expression values of memory versus naive cells were reported by Sarkar et al. (17). (B) Heritable chromatin accessibility is mediated by epigenetic modifications. Histone (brown cylinder) hypoacetylation and DNA (black line) hypomethylation correspond to transcriptional activation. Compact heterochromatin contains Mbd proteins and other to be determined proteins that block transcription factors (TF) and the RNA polymerase (Pol) from accessing the gene of interest. A list of known enzymes and proteins involved in the epigenetic process includes histone acetlyltransferases (HAT), DNA methyltransferases (Dnmt), histone deacetylases (HDAC) and proteins containing a methylated DNA binding domain (Mbd). Modifications to epigenetic marks are acquired for several different genes following TCR signaling of CD8 T cells. Genes programmed for downregulation obtain repressive marks correlated with chromatin condensation during the “off” stage of transcription, while activating marks are obtained by genes that become accessible to transcription factors and polymerase. (C) An emerging mechanism to explain the rapid recall potential of memory CD8 T cells suggests that genes that are rapidly expressed upon TCR-mediated activation have an acquired epigenetic program that either allows quick access by TF or is a result of the persistent presence of a TF.
Mouse models have shown that all memory CD8 T cells derive from effector cells that express cytolytic molecules such as granzyme B and perforin, key molecules involved in killing infected cells (12–14). As granzyme B expression is a hallmark feature of effector CD8 T cells, this indicates that memory cells are derived from effector cell precursors. However, not all effector CD8 T cells have equal capacities to give rise to memory cells. This is evident early during the expansion phase of the CD8 T-cell response. Antigen-specific CD8 T cells that express high levels of the IL-2 receptor CD25 a few days after an infection starts are predisposed to undergo contraction following viral clearance, while CD25low cells have the ability to give rise to memory cells. Later during the response, CD8 T cells expressing high levels of the IL-7 receptor CD127 and low levels of the activation marker Klrg1 show the highest capacity to generate memory cells, while their CD127low Klrg1high counterparts are more likely to be eliminated during the contraction phase (15–17).
Thus, it is clear that even at early time points after antigen exposure, CD8 T cells become committed to diverging pathways of differentiation. It has been proposed that the first such lineage commitment may occur during the asymmetric cell division that follows a naive T cell's initial encounter with antigen (18). An alternative hypothesis, which we favor, is that all antigen-specific T cells initially have the capacity to generate memory cell progeny but may lose this capacity and differentiate into ‘terminal effectors’ through a stochastic process. The duration of signals delivered through the T-cell receptor plays a key role in this process. An optimal length of antigen exposure promotes memory CD8 T-cell development while longer exposures drive CD8 T cells to terminal effector differentiation and the loss of memory precursor potential. In the extreme case, chronic persistent antigen exposure may lead a CD8 T-cell population to a state of reduced functionality known as ‘exhaustion’ (19).
The effector-to-memory transition results in a shift of the virus-specific CD8 T-cell immune response from active cytolytic functions to a state of preparedness. Upon antigen re-exposure, memory CD8 T cells express key effector molecules, such as the cytokines interferon-γ and interleukin 2, much more rapidly than their naive predecessors (1, 20, 21). Although they undergo similar rates of antigen-dependent proliferation compared with naive cells, memory cells divide more frequently in the absence of antigen, a process referred to as homeostatic proliferation. This allows their numbers to be maintained for the life of the host (22–24).
The life-long immunity provided by memory CD8 T cells facilitates the efficient control of previously encountered viruses and intracellular pathogens (25). In this review, we will focus on potential mechanisms that may allow for these heightened functional properties of memory CD8 T cells to be established during differentiation and propagated during homeostatic proliferation. We will also briefly discuss potential avenues for therapeutic intervention to repair non-functional memory CD8 T cells as a means to control chronic viral infections.
Changes in gene regulation define CD8 T-cell memory
Unlike B cells, whose heightened potential to clear infection arises through changes in the antigen-specific receptor, the B-cell receptor, the increased responsiveness of memory CD8 T cells occurs in the absence of change to TCR affinity (26). The functional qualities that define memory CD8 T cells result from stable changes in gene expression patterns that are propagated through cell division without changes to the DNA coding sequence, and without a requirement for continued antigenic stimulation. Resting pathogen-specific memory CD8 T cells have thousands of genes that are differentially expressed relative to naive CD8 T cells (17, 27–29). These genes may be up- or down-regulated relative to naive cells, while other genes are expressed at similar levels but are poised for rapid re-expression in memory cells upon antigen encounter (27, 28, 30) (Fig. 1A and C).
The dramatic difference in gene expression profiles between naive and memory CD8 T cells is potentially explained by the observed differential expression of lineage-specific transcription factors. It is well established that master regulatory transcription factor(s) control lineage fate decisions among many cell types, with the classic example being myogenic differentiation 1 expression promoting differentiation of muscle cells (31). Furthermore, expression of the transcription factor Forkhead box protein 3 (Foxp3) is essential and sufficient to induce CD4 regulatory T-lymphocyte development (32, 33).
On the other hand, memory formation in T cells is critically linked to the expression of several lymphocyte-specific transcription factors, yet a single ‘master regulatory’ transcription factor that is sufficient for inducing CD8 T-cell memory has not been found (34–38). Expression of inhibitor of DNA binding 2 (ID2), T-box expressed in T cells (Tbet) and eomesodermin is induced at the effector stage of CD8 T-cell differentiation. During the subsequent development of memory cells, levels of eomesodermin increase, Tbet expression decreases and ID2 expression remains constant (35, 36). Maintenance of these patterns of stage-specific up- and down-regulation for these genes is crucial in the establishment of functional memory T cells. The transcriptional regulators B lymphocyte-induced maturation protein 1 (Blimp-1) and B-cell CLL/lymphoma 6 (Bcl6) also play key roles in controlling memory cell development (39). Each transcription factor represses the expression of the other and promotes an opposing differentiation pathway; Bcl6 enhances CD8 T-cell memory formation (34), while Blimp-1 expression promotes formation of terminal effectors (37, 38). Furthermore, deletion of Blimp-1 results in a greater quantity of memory CD8 T cells relative to wild-type mice following acute viral infection (37, 38).
The mechanism for lineage commitment between terminal effectors and memory CD8 T cells has received a tremendous amount of attention recently. Interestingly, although quite distinct in their abilities to develop into either terminal effector or memory CD8 T cells, early precursors for each of these populations only exhibit minimal differences in transcript expression compared with the striking differences seen between bulk effector and memory virus-specific CD8 T cells (16, 17). Rather than having instantaneous differences in gene expression in the precursor subsets of effector cells, an alternative mechanism might be that heritable gene expression programs are applied in the precursor populations allowing for a delayed effect in gene regulation of the committed cells.
Taken together, the data described above clearly demonstrate that the commitment to a memory CD8 T-cell lineage is coupled with the expression of specific transcription factors. How then are these patterns of transcription factor expression maintained over time and through cycles of cell division? The answer is thought to lie in the structure of the chromatin encoding these genes.
Heritable access to chromatin
If the human genome were laid out in a straight line, its span would be roughly that of the height of an adult (∼2 m); thus, a mechanism has evolved that condenses our genome to fit within a nucleus with a diameter of ∼6 μm. Decades of research have revealed that this process of genomic condensation is quite dynamic and coupled to cell lineage-specific gene expression profiles (40, 41). In particular, naive T-cell chromatin accessibility is pliable and responsive to extracellular cues such as ligation of the TCR (signal 1) and CD28 (signal 2) (42, 43). Therefore, a description of antigen-induced adaptation to the gene expression profile should include modifications to the chromatin.
The mechanism for the heightened recall potential that a memory CD8 T cell has to antigen must be propagated during homeostatic proliferation to the daughter cell. As discussed above, much of the acquired function of a memory cell occurs in the form of differential gene regulation relative to naive cells. Heritable modifications to gene regulation without changes in the DNA sequence are referred to as epigenetic mechanisms. The major eukaryotic mechanisms for epigenetic programming include DNA methylation/demethylation, histone modifications and non-coding RNA-mediated transcriptional and post-transcriptional regulation (44, 45). Transcriptional regulation through these multiple types of epigenetic programs is thought to occur by blocking transcriptional initiation; however, our understanding of epigenetic mechanisms is incomplete thus the need for further exploration. A basic model for understanding epigenetic mechanisms is presented in Fig. 1(B). The formation of heterochromatin (i.e. tightly packed) versus euchromatin (i.e. loosely packed) is coupled to modifications of both histone (acetylation and methylation) and DNA (cytosine methylation). In general, nucleosomal modifications that lead to more compact heterochromatin result in transcriptional repression of that region of DNA, while modifications that are characteristic of euchromatin promote transcriptional activation (Fig. 1B) (45). The epigenetic enzymes associated with transcriptional repression include histone deacetylases (HDAC), histone methyltransferases, histone demethylases and cytosine-5 DNA methyltransferases (Dnmt). Interestingly, histone methylation patterns are multifaceted but allow faithful transmission of transcriptional regulation. It is now quite clear that different patterns of mono-, di- and tri-methylation on individual lysine residues of a histone are associated with repression, activation and poised states of transcription (41, 46). DNA methylation in mammals occurs predominantly at C–phosphate–G (CpG) dinucleotides (45, 47), while histone acetylation and methylation occur on many residues of histones, often referred to as the histone code (Fig. 1B) (45, 48, 49). Histone and DNA modifications appear to be coupled processes, that is, histone modifying enzymes are sensitive to the DNA methylation status and vice versa (49).
As a cell divides, the ensemble of epigenetic marks provides a program for the daughter cells to ‘remember’ the transcriptional status of the parental cell. Properly orchestrated epigenetic modifications are essential for the normal development of mammals (44, 45). Many examples exist where disruption of these epigenetic events results in human diseases including immunodeficiency, centromere instability and facial anomalies syndrome (50), Fragile X syndrome (50) and many different cancers (51, 52). Of particular relevance, hypomethylation of the DNA for select promoters is associated with aberrant T-cell development resulting in autoimmune diseases such as lupus (53, 54). The above described mechanisms of epigenetic gene regulation provide an attractive model for explaining how functional properties are acquired by memory CD8 T cells and propagated during the self-renewal process, but does experimental evidence support it?
In both human and mouse memory CD8 T cells, changes to histone modifications have been seen on genes that are up-regulated relative to naive CD8 T cells (55, 56). Treatment of memory CD8 T cells with curcumin, an inhibitor of histone lysine acetylation, blocks their ability to rapidly up-regulate granzyme B following antigen stimulation (57). An understanding of the mechanism for the propagation of histone modifications to the daughter cells following DNA synthesis is evolving but remains incomplete (58).
On the other hand, the mechanism for propagating CpG DNA methylation is better understood. The palindromic nature of the CpG substrate for methylation provides the template for copying the methylation program from parental to newly synthesized DNA daughter strand. The importance of this process for memory generation is highlighted by conditional deletion in CD8 T cells of Dnmt1, the enzyme that maintains the methylation pattern during DNA replication (47, 49, 59–62). Loss of Dnmt1 reduces the quantity and self-renewal capacity of memory CD8 T cells following acute viral infection (63, 64), and also leads to the aberrant expression of Foxp3 in CD8 T cells (65). Consistent with the conditional knockout studies of Dnmt1, deletion of Mbd2 (a DNA methylation binding transcriptional repressor) results in a normal effector CD8 T-cell response but a significant reduction in the quantity and quality of virus-specific memory CD8 T cells (63, 66). Thus, an essential feature in the formation of CD8 T-cell immunity is for DNA methylation programs to be maintained and properly interpreted in a proliferating memory CD8 T cell.
The level of DNA methylation has been observed to be inversely correlated with the expression of key effector molecules. Virus-specific CD8 T cells acquire a demethylated IL-2 promoter as they differentiate from naive to effector and the unmethylated status is maintained throughout memory development (20, 67). In contrast to this ‘off–on’ pattern of gene expression, where gene expression increases at the effector stage and stays elevated through memory, other genes display an ‘off-on-poised’ gene expression profile. Genes that follow this pattern of regulation increase at the effector stage and retract back to naive levels of expression, but are capable of rapid antigen induced re-expression (Fig. 1B and C) (17, 28, 68). This faster re-expression of many effector molecules, such as IFNγ and granzyme B is the defining feature of good functional memory.
How heritable programming facilitates memory responses
How are memory CD8 T cells heritably programmed for rapid re-expression of a gene? One possibility is that a transcription factor that was not present in the naive cell is now present in a memory CD8 T cell awaiting the signal from the TCR to initiate targeted gene expression. Another possibility is that the chromatin at the promoters of genes that are rapidly expressed in memory CD8 T cells has been heritably altered allowing for quicker access by a transcription factor relative to the naive counterpart (Fig. 1C). In the case of IFNγ gene regulation, it has been shown that the rapid cytokine production is a result of higher constitutive transcript expression along with an ability to produce new transcript more rapidly upon TCR-mediated stimulation of memory cells. Moreover, it was reported that specific epigenetic marks associated with a transcriptionally accessible state are observed in resting memory CD8 T cells (20, 21, 67).
Similarly, granzyme B, the cytolytic molecule of effector cells, is observed to be rapidly expressed only upon stimulation of memory CD8 T cells relative to naive CD8 T cells. More to the point, it was reported that the acquisition of memory in CD8 T cells was coincident with obtained heritable epigenetic programming at the chromatin of granzyme B, perforin and the memory-associated transcription factor eomesodermin (57, 69). Therefore, both epigenetic modifications and changes in transcription factor expression have been reported as underlying components in the formation of good memory functions. These mechanisms are not mutually exclusive and a temporal relationship between transcription factor binding and epigenetic modifications has yet to be established.
Further insights into the acquired transcriptional programs of memory CD8 T cells (Fig. 1A) may come from studies on the divergent pathway of CD4 T-cell differentiation (42, 70). Lymphocyte development in the periphery results in the formation of several specialized CD4 lymphocyte populations (71). Modifications of conserved non-coding sequence (CNS) elements are observed in the development of CD4 Th-cell subsets. The principal function used for defining Th1 versus Th2 CD4 T cells is the ability of the cell to express IFNγ or IL-4 respectively. Conditions for Th1 polarization along with TCR stimulation of naive CD4 T cells results in the demethylation of the IFNγ transcriptional regulatory regions, retention of an unmethylated proximal promoter of IFNγ and the retained DNA methylation of IL-4 transcriptional regulatory regions. In contrast, Th2 polarizing conditions resulted in demethylation of the IL-4 promoter and induced DNA methylation of the IFNγ promoter. Interpretation of these programs is also a vital aspect to formation of an efficient effector response. Studies on mice with Mbd2 knocked out revealed that the formation of naive CD4 T cells was not impaired, rather the lineage-specific cytokine production elicited post Th polarization was less restricted in Mbd2-deficient cells, with both Th1 and Th2 cells producing IFNγ and IL-4 (72). These and other studies suggest that Th polarization is mediated by a balance between the expression of cell-specific transcription factors along with restricted access to the IFNγ or IL-4 locus that is mediated by epigenetic marks and interpreters of those marks (64, 72–74).
A separate lineage of CD4 T cells, known as regulatory T cells (Tregs), is utilized to suppress overzealous immune responses. As mentioned earlier, development of regulatory CD4 T cells is dependent upon the expression of the master regulatory transcription factor Foxp3. It was recently demonstrated that a CNS upstream of the Foxp3 gene is dispensable for development of Tregs, but is essential for the retention of Foxp3 expression in dividing Tregs in the periphery (75). Further, it was demonstrated that this CNS element is demethylated, after which Foxp3 binds and promotes its own expression (75). This demonstrates that epigenetic mechanisms not only influence the expression of effector molecules, but are vital to the continued expression of lineage-specific transcription factors.
Conclusions
At present, it is difficult to say whether transcription factors provide the specificity for epigenetic modifications yielding the heritable memory gene expression profile or if some yet to be identified mechanism dictates the epigenetic program limiting the access that transcription factors have to chromatin, or even regulating the expression of the transcription factor directly. The answer may be all of the above. Understanding the mechanism for heritable functions of memory CD8 T cells will provide potential therapeutic avenues to rejuvenate non-functional CD8 T cells generated in response to chronic viral infections or tumors. Modifying transcriptional profiles by use of epigenetic targeting inhibitors is currently used to treat specific malignancies (76). Moreover, there is precedence that epigenetic therapies can modulate T-cell functions in vivo (77), providing motivation to pursue strategies of transcriptional manipulation as an approach to improve the antigen-specific response of exhausted CD8 T cells. Application of an epigenetic therapy requires a greater understanding of heritable gene regulation in functional virus-specific memory CD8 T cells. Such an understanding will provide insights into specific proteins or gene loci that may be manipulated to enhance memory-specific T-cell programs with the goal of improving specific transcriptional programs in non-functional memory CD8 T cells.
Funding
National Institutes of Health (AI030048 and N01-AI-50025) to R.A; American Cancer Society (PF-09-134-01-MPC) to B.Y; National Institutes of Health National Research Service Award (5-T32-AI007610) to C.W.D.
Acknowledgments
We thank Dr Rama Akondy for comments and critical reading of our manuscript.
References
- 1.Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78:5535. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 3.Bevan MJ, Goldrath AW. T-cell memory: you must remember this. Curr. Biol. 2000;10:R338. doi: 10.1016/s0960-9822(00)00461-9. [DOI] [PubMed] [Google Scholar]
- 4.Doherty PC, Topham DJ, Tripp RA. Establishment and persistence of virus-specific CD4+ and CD8+ T cell memory. Immunol. Rev. 1996;150:23. doi: 10.1111/j.1600-065x.1996.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 5.Lefrancois L, Masopust D. T cell immunity in lymphoid and non-lymphoid tissues. Curr. Opin. Immunol. 2002;14:503. doi: 10.1016/s0952-7915(02)00360-6. [DOI] [PubMed] [Google Scholar]
- 6.Parish IA, Kaech SM. Diversity in CD8(+) T cell differentiation. Curr. Opin. Immunol. 2009;21:291. doi: 10.1016/j.coi.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 2003;4:835. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 8.McKinstry KK, Strutt TM, Swain SL. The effector to memory transition of CD4 T cells. Immunol. Res. 2008;40:114. doi: 10.1007/s12026-007-8004-y. [DOI] [PubMed] [Google Scholar]
- 9.Sallusto F, Lanzavecchia A. Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur. J. Immunol. 2009;39:2076. doi: 10.1002/eji.200939722. [DOI] [PubMed] [Google Scholar]
- 10.Murali-Krishna K, Altman JD, Suresh M, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 11.Blattman JN, Antia R, Sourdive DJ, et al. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 2002;195:657. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opferman JT, Ober BT, Ashton-Rickardt PG. Linear differentiation of cytotoxic effectors into memory T lymphocytes. Science. 1999;283:1745. doi: 10.1126/science.283.5408.1745. [DOI] [PubMed] [Google Scholar]
- 13.Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399:593. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- 14.Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323:505. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 2003;4:1191. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 16.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 2008;205:625. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang JT, Palanivel VR, Kinjyo I, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 19.Wherry EJ, Teichgraber V, Becker TC, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 2003;4:225. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 20.Kersh EN, Fitzpatrick DR, Murali-Krishna K, et al. Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells. J. Immunol. 2006;176:4083. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- 21.Bachmann MF, Barner M, Viola A, Kopf M. Distinct kinetics of cytokine production and cytolysis in effector and memory T cells after viral infection. Eur. J. Immunol. 1999;29:291. doi: 10.1002/(SICI)1521-4141(199901)29:01<291::AID-IMMU291>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Zimmermann C, Prevost-Blondel A, Blaser C, Pircher H. Kinetics of the response of naive and memory CD8 T cells to antigen: similarities and differences. Eur. J. Immunol. 1999;29:284. doi: 10.1002/(SICI)1521-4141(199901)29:01<284::AID-IMMU284>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 23.Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat. Med. 2001;7:913. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- 24.Lau LL, Jamieson BD, Somasundaram T, Ahmed R. Cytotoxic T-cell memory without antigen. Nature. 1994;369:648. [PubMed] [Google Scholar]
- 25.Crotty S, Ahmed R. Immunological memory in humans. Semin. Immunol. 2004;16:197. doi: 10.1016/j.smim.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Slifka MK, Whitton JL. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat. Immunol. 2001;2:711. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- 27.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 28.Wherry EJ, Ha S, Kaech SM, et al. Molecular signature of CD8 T cell exhaustion during chronic viral infection. Immunity. 2007;27:670. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J. Immunol. 2005;175:5895. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 30.Fann M, Godlove JM, Catalfamo M, et al. Histone acetylation is associated with differential gene expression in the rapid and robust memory CD8(+) T-cell response. Blood. 2006;108:3363. doi: 10.1182/blood-2006-02-005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weintraub H, Davis R, Tapscott S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 32.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057. [PubMed] [Google Scholar]
- 33.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat. Immunol. 2007;8:457. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 34.Ichii H, Sakamoto A, Hatano M, et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat. Immunol. 2002;3:558. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 35.Intlekofer AM, Takemoto N, Wherry EJ, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 36.Cannarile MA, Lind NA, Rivera R, et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat. Immunol. 2006;7:1317. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 37.Rutishauser RL, Martins GA, Kalachikov S, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin H, Blackburn SD, Intlekofer AM, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 2010;11:114. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 41.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 42.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4:616. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 43.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 44.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 45.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome–components and functional correlates. Genes Dev. 2006;20:3215. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 46.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 47.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 48.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 49.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robertson KD. DNA methylation and chromatin—unraveling the tangled web. Oncogene. 2002;21:5361. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 51.Jones PA. Epigenetics in carcinogenesis and cancer prevention. Ann. N. Y. Acad. Sci. 2003;983:213. doi: 10.1111/j.1749-6632.2003.tb05976.x. [DOI] [PubMed] [Google Scholar]
- 52.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 53.Deng C, Yang J, Scott J, Hanash S, Richardson BC. Role of the ras-MAPK signaling pathway in the DNA methyltransferase response to DNA hypomethylation. Biol. Chem. 1998;379:1113. doi: 10.1515/bchm.1998.379.8-9.1113. [DOI] [PubMed] [Google Scholar]
- 54.Gorelik G, Richardson B. Key role of ERK pathway signaling in lupus. Autoimmunity. 2010;43:17. doi: 10.3109/08916930903374832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DiSpirito JR, Shen H. Quick to remember, slow to forget: rapid recall responses of memory CD8+ T cells. Cell Res. 2010;20:13. doi: 10.1038/cr.2009.140. [DOI] [PubMed] [Google Scholar]
- 56.Dispirito JR, Shen H. Histone acetylation at the single-cell level: a marker of memory CD8(+) T cell differentiation and functionality. J. Immunol. 2010;184:4631. doi: 10.4049/jimmunol.0903830. [DOI] [PubMed] [Google Scholar]
- 57.Araki Y, Fann M, Wersto R, Weng NP. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B) J. Immunol. 2008;180:8102. doi: 10.4049/jimmunol.180.12.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 59.Flynn J, Glickman JF, Reich NO. Murine DNA cytosine-C5 methyltransferase: pre-steady- and steady-state kinetic analysis with regulatory DNA sequences. Biochemistry. 1996;35:7308. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]
- 60.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009;10:805. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tollefsbol TO, Hutchison CA., 3rd Mammalian DNA (cytosine-5-)-methyltransferase expressed in Escherichia coli, purified and characterized. J. Biol. Chem. 1995;270:18543. doi: 10.1074/jbc.270.31.18543. [DOI] [PubMed] [Google Scholar]
- 62.Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev. Biol. 2000;225:294. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- 63.Chappell C, Beard C, Altman J, Jaenisch R, Jacob J. DNA methylation by DNA methyltransferase 1 is critical for effector CD8 T cell expansion. J. Immunol. 2006;176:4562. doi: 10.4049/jimmunol.176.8.4562. [DOI] [PubMed] [Google Scholar]
- 64.Lee PP, Fitzpatrick DR, Beard C, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 65.Josefowicz SZ, Wilson CB, Rudensky AY. Cutting edge: TCR stimulation is sufficient for induction of Foxp3 expression in the absence of DNA methyltransferase 1. J. Immunol. 2009;182:6648. doi: 10.4049/jimmunol.0803320. [DOI] [PubMed] [Google Scholar]
- 66.Kersh EN. Impaired memory CD8 T cell development in the absence of methyl-CpG-binding domain protein 2. J. Immunol. 2006;177:3821. doi: 10.4049/jimmunol.177.6.3821. [DOI] [PubMed] [Google Scholar]
- 67.Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J. Immunol. 2006;177:1062. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 68.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 69.Juelich T, Sutcliffe EL, Denton A, et al. Interplay between chromatin remodeling and epigenetic changes during lineage-specific commitment to granzyme B expression. J. Immunol. 2009;183:7063. doi: 10.4049/jimmunol.0901522. [DOI] [PubMed] [Google Scholar]
- 70.Wilson CB, Merkenschlager M. Chromatin structure and gene regulation in T cell development and function. Curr. Opin. Immunol. 2006;18:143. doi: 10.1016/j.coi.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu. Rev. Immunol. 2010;28:445. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hutchins AS, Mullen AC, Lee HW, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol. Cell. 2002;10:81. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 73.Schoenborn JR, Dorschner MO, Sekimata M, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat. Immunol. 2007;8:732. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 75.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karberg S. Switching on epigenetic therapy. Cell. 2009;139:1029. doi: 10.1016/j.cell.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 77.Tao R, de Zoeten EF, Ozkaynak E, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 2007;13:1299. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]