Abstract
Streptococcus mutans is considered a primary pathogen for human dental caries. Its ability to produce a variety of peptide antibiotics called mutacins may play an important role in its invasion and establishment in the dental biofilm. S. mutans strain UA140 produces two types of mutacins, the lantibiotic mutacin I and the non-lantibitoc mutacin IV. In a previous study, we constructed a random insertional-mutation library to screen for genes involved in regulating mutacin I production, and found 25 genes/operons that have a positive effect on mutacin I production. In this study, we continued our previous work to identify genes that are negatively involved in mutacin I production. By using a high phosphate BHI plate that inhibited mutacin I production of the wild-type, we isolated 77 clones that consistently produced mutacin I under repressive conditions. From the 34 clones that we were able to obtain a sequence, 17 unique genes were identified. These genes encompass a variety of functional groups including the central metabolism, surface binding, sugar transport, and unknown functions. Some of the 17 mutations were further characterized and shown to increase mutacin gene expression during growth when it is usually not expressed in the wild-type. These results further demonstrate an intimate and intricate connection between mutacin production and the overall cellular homeostasis.
Introduction
Dental caries is one of the most common infectious diseases afflicting humans. Streptococcus mutans is considered as a primary agent in cariogenesis (Loesche, 1986). Its abilities to adhere and form biofilms on the tooth surface, to produce acids from metabolizeable carbohydrates, and to survive low pH and other environmental insults are believed to be critical in its persistence and eventual dominance in the dental plaque. The dental plaque consists of a complex bacterial community of >700 different species living in an environment with constant cycles of feast and famine (Socransky et al., 1998). In addition to the previously mentioned virulence properties, S. mutans also possesses a tremendous ability to kill other competing species in the dental plaque by producing peptide antibiotics called mutacins (Hamada & Ooshima, 1975; Parrot et al., 1989; Parrot et al., 1990). Mutacin production may allow S. mutans to invade and establish itself within the dental biofilm community and to persist when nutrients become limited in the dental plaque.
S. mutans strain UA140 produces two types of mutacins, the lantibiotic mutacin I and the non-lantibiotic mutacin IV (Qi et al., 2001). The lantibiotics are extensively modified peptide antibiotics, containing dehydrated threonine and serine residues and thioether bridges (Sahl & Bierbaum, 1998), while the non-lantibiotics are unmodified peptides. Mutacin production is controlled by many genetic as well as environmental factors. While the non-lantibiotic mutacin IV is controlled by quorum sensing via the three component system comCDE (Kreth et al., 2005; Kreth et al., 2006), regulation for the lantibiotic mutacin I is much more complex and less understood. Mutacin I can only be produced under high cell densities such as in the pellet or in a colony on the plate (Qi et al., 2000; Tsang et al., 2005; Tsang et al., 2006). In order to obtain a global view of how mutacin I production is regulated, we previously constructed a random insertional mutagenesis library in strain UA140. Using this library, we identified 25 genes that are positive regulators of mutacin I production (Tsang et al., 2005). In this study, we used the same library to identify genes that are negative regulators of mutacin production. We identified 17 unique genes and several of these were further characterized.
Materials and Methods
Bacterial Strains and Culture Conditions
Escherichia coli strain DH5α was used for cloning as well as plasmid amplifications. E. coli cells were grown in Luria-Bertani (LB, Fisher) medium aerobically at 37 °C. E. coli strains carrying plasmids were grown in LB medium containing spectinomycin (150 μg/ml). S. mutans wild-type strain UA140 was cultured in brain heart infusion (BHI, Difco) medium or on BHI agar plates. For selection of antibiotic resistant colonies, BHI medium was supplemented with spectinomycin (800 μg/ml) or erythromycin (15 μg/ml). BHI medium supplemented with potassium phosphate buffer (5 mM) was used to screen for mutants with increased mutacin production.
Library Screening
A previously constructed random insertional mutation library was used (Tsang et al., 2005). The library was constructed by randomly cleaving the genomic DNA of strain UA159 by CviJI restriction enzyme, and ligation of the 300–500 bp gel-purified fragments into the pZero-2 vector at the EcoRV site. After transforming into E. coli, the plasmid was isolated from 11, 000 pooled E. coli clones and transformed into S. mutans UA140. Eleven thousands transformants were randomly selected, which were then grown on 96-well plates, and stored at −80°C as the library. To screen for mutants that produce mutacin I in the presence of inhibitory amounts of phosphate, the library clones were grown in 96-well plates overnight and 5 μl of the culture was spotted onto BHI plates supplemented with 5 mM potassium phosphate and grown anaerobically for 24 hours. Each plate was then overlaid with a thin layer of soft agar mixed with overnight cultures of S. sobrinus OMZ176, which was only sensitive to mutacin I. The zone of inhibition (halo) was inspected after an overnight incubation under anaerobic conditions. Since this condition inhibits mutacin I production in the wild-type, the presence of a halo around any of the mutant colonies was considered to be positive. Positive clones from the first round of screening were subject to a second and third round of screening using the same procedures as described above. Clones that produced mutacin after all three rounds of screening were used for gene identification.
Identification of Mutated Genes
Chromosomal DNA from each selected mutant was prepared from 10 ml of overnight cell culture as follows: Cell cultures were centrifuged, the cell pellet was suspended in 1 ml TE buffer with 15 mg/ml freshly prepared lysozyme, and incubated at 37°C for 2–3 h. 1/10th volume of 10% SDS and 0.7 volume of phenol/chloroform (pH 6.7) were added to the mixture and mixed by vortexing for 1 min. The mixture was centrifuged at 14,000 rpm for 5 min and the supernatant (~0.9 ml) was removed. DNA was precipitated from the supernatant by 0.7 vol. of isopropanol in the presence of 0.15 M NaCl, and washed with 70% ethanol. The DNA pellet was suspended in 50 μl TE buffer containing 0.1 mg/ml RNase.
To obtain the identity of the gene mutation, chromosomal DNA was digested by Cvi JI (CHIMERx, Madison, WI), the enzyme that was used to generate near-random DNA fragments for construction of the insertional library used in this study (Tsang et al., 2005). The digestion reaction (30 μl) was performed in a 96-well plate and incubated at room temperature overnight. 2 μl of the digested DNA was ligated overnight at room temperature with 1 μl of pKS (+) plasmid digested with Bam HI and Eco RV, in a total vol. of 11 μl, also in a 96-well plate. 1 μl of the ligation mixture was used as template in a 25 μl PCR reaction using primers M13-reverse and Zero-F (5'-GTGTGCTGGAATTCTGCAG). The PCR product was purified with the PCR Purification Kit (Qiagen) and used directly for sequencing by the DNA Sequencing Facility of the Oklahoma Medical Research Foundation. Since EcoRV was used for cloning the original library fragments into pZero, confirmed mutations were expected to contain S. mutans sequence terminating at an Eco RV half-site followed by pZero plasmid sequence (Tsang et al., 2005). The sequences obtained were compared to the genomic sequences of S. mutans UA159 available at the Los Alamos Oral Pathogen Sequence Databases (http://www.oralgen.lanl.gov) via BLAST. The insertion point was defined as the junction with the Eco RV cleavage site.
Mutacin reporter assay in mutant backgrounds
Chromosomal DNA from each mutant was isolated and transformed into the mutA-luc reporter strain, UA140mutA-luc (He et al., 2008). Transformants were selected on BHI + 800 μg/ml Spc plates, and the right clones were further confirmed by PCR. To measure mutA gene expression, wild-type and mutant strains were grown overnight in BHI and 10 μl from each was spotted on BHI plates with or without the supplementation of 5 mM sodium phosphate. Cells were grown anaerobically for 16 h, then scraped from the plates, and resuspended in BHI. Luciferase activity was measured immediately after resuspension using the methods described previously (Kreth et al., 2004). The same cell suspensions were also measured for absorbance at 600 nm and the luciferase activities were normalized against OD600.
Results
Identification of genes involved in repression of mutacin I production
Previous studies demonstrated that mutacin I gene expression is repressed during growth and activated under mild stress conditions, such as those encountered during limited nutrient availability. In an effort to identify environmental factors that repress mutacin I gene expression, we previously found that buffering BHI with 5 mM phosphate buffer inhibited mutacin I production (Qi, unpublished). Further studies revealed that the inhibition is temporal; mutacin I was produced by about 24 h on low phosphate plates (BHI), whereas high phosphate plates (BHI + 5 mM phosphate) required 48 h for mutacin production (data not shown). Based on this observation, we assumed that phosphate availability may be one of the environmental factors that inhibits mutacin I gene expression during growth. Thus, we reasoned that by selecting mutants able to produce mutacin within 24 h under high phosphate conditions (i.e. nutrient excess), we would be able to identify genes involved in the repression of mutacin I gene expression during growth.
To this end, we screened a previously constructed random insertional mutagenesis library of 11,000 clones (Tsang et al., 2005) to look for mutacin production after 24 h of growth on phosphate supplemented BHI plates. Positive clones from the first round of screening were re-tested and after 3 rounds of testing, 77 mutants were confirmed and stored at −80°C.
To identify the mutated gene, genomic DNA (gDNA) was isolated from all 77 clones. A high throughput protocol was developed to sequence all clones simultaneously. In this protocol, we took advantage of the near randomness of the CviJI restriction enzyme. CviJI recognizes 5'-Pu G C Py-3' and cuts in between G and C, generating blunt end fragments. Chromosomal DNA from each clone was digested with CviJI and ligated with pKS vector digested with EcoRV. To specifically amplify the chromosome region where the insertion took place, a forward primer was designed, which is immediately upstream of the EcoRV site in pZero (the restriction site in the vector used to construct the original library) (Tsang et al., 2005). The reverse primer was the M13 reverse primer, which is located ~100 bp downstream of the EcoRV site in the pKS vector. After PCR amplification, most clones generated a ~300 – 400 bp fragment. After purification, these PCR products were directly used for sequencing with the M13 reverse primer. From these clones, we were able to obtain unambiguous sequences from 34 clones, from which 17 unique genes were identified (Table 1). To further confirm that these genes are truly responsible for the observed phenotype, each gene was individually deleted by using a single-crossover mutagenesis strategy (see Table S1 and Fig. S2). These mutants were tested for mutacin I production on BHI plates supplemented with 5 mM phosphate. All transformants displayed the same phenotype as the original clones, indicating that the observed phenotype in these clones is indeed due to insertions in the identified genes.
Table 1.
Genes associated with repression of mutacin I production
| # | Oralgen ID/GenBank tag (# of hit) | Annotation | Functional class |
|---|---|---|---|
| Class 1 mutant | |||
| 1 | SMu0557/SMU.611 (1) | DeaD, ATP dependent RNA helicase | Transcription, degradation of RNA |
| 2 | SMu0797/SMU.874 (1) | metH, 5-methyltetrahydrofolate-homocysteine methyltransferase | Amino acid biosynthesis, central intermediate metabolism, one carbon metabolism |
| 3 | SMu859/SMU.947 (1) | Dfr, dhfr, dyr, dihydrofolate reductase | Biosynthesis of cofactors, prothetic groups and carriers, folic acid |
| 4 | SMu1034/SMU.1132 (8) | pepN, lysyl-aminopeptidase | Degradation of proteins, peptides |
| 5 | SMu1849/SMU2035 (2) | mccF, possible bacteriocin immunity protein | Toxin production and resistance |
| 6 | SMu1850/SMU.2036 (2) | pepO, endopeptidase O | Degradation of proteins |
| 7 | SMu1885/SMU.2077 (2) | conserved hypothetical protein | unknown |
| Class 2 mutant | |||
| 8 | SMu0432/SMU.478 (1) | kguA, guanylate kinase | Purine metabolism |
| 9 | SMu0617/SMU.675 (1) | ptsI, phosphoenolpyruvate-protein phosphotransferase (enzyme I) | Signal transduction, PTS |
| 10 | SMu0646/SMU.707 (2) | endolysin | Biosynthesis & degradation of murein sacculus and peptidoglycan |
| 11 | SMu0649/SMU.712 (1) | Ppc/pepC, phosphoenolpyruvate carboxylase | Central intermediate metabolism |
| Class 3 mutant | |||
| 12 | SMu0538/SMU.591 (1) | HTP, upstream is a transcription regulator sharing IGS with furR | unknown |
| 13 | SMu0727/SMU.801 (1) | Obg, GTP-binding protein, GTP1/Obg family | unassigned |
| 14 | SMu1319/SMU.1449 (1) | fbp, pavA, fibrinogen-binding protein-A | Cellular process, pathogenesis, transport and binding proteins |
| 15 | SMu1708/SMU.1878 (1) | manM, mannose PTS system component IIC, a membrane protein | Signal transduction, PTS |
| 16 | SMu1709/SMU.1879 (7) | manN, mannose PTS system component IID, a membrane protein | Signal transduction, PTS |
| 17 | SMu1848/SMU.2033 (1) | conserved hypothetical protein | unknown |
Since two of the genes (SMU.478, SMU.947) reside in an operon, the downstream genes were individually deleted by a double-crossover strategy (see Fig. S1 and Table S1). Overall, nine mutagenesis attempts were made, and 8 mutants were obtained. No mutant could be obtained for SMU.481 possibly because this gene encodes methionyl-tRNA formyltransferase, which may be essential. All eight mutants were tested for their effect on mutacin production on BHI plus phosphate plate. All displayed a wild-type phenotype, except for clpX (SMU.949), which could not produce mutacin even on regular BHI plate where the wild-type produced mutacin (data not shown). These results suggested that the observed phenotype for SMU.478 and SMU.947 is not due to a polar effect on the downstream genes.
Preliminary characterization of selected mutants
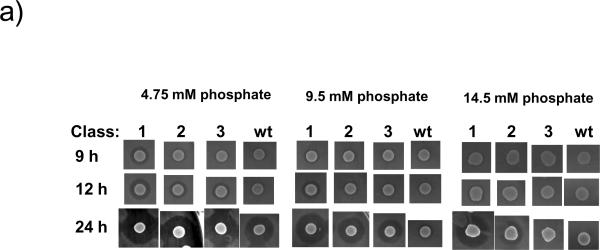
To further characterize the effects of these gene mutations on mutacin I production, all 17 mutants were further tested on BHI plates containing different concentrations of phosphate. Since the DIFCO BHI formulation contains 9.5 mM phosphate, we diluted BHI 1:1 with water and supplemented the 50% BHI with phosphate buffer to make the final concentrations of phosphate at 4.75 mM, 9.5 mM, and 14.5 mM, respectively. Mutants were spotted on the plates and the zone of inhibition was determined at 9 h, 12 h, and 24 h after inoculation. From the timing of mutacin production under different phosphate concentrations, three classes of mutants can be identified (Fig. 1A, and Table 1). Class 1 produced mutacin I at 9 h on the 4.75 mM phosphate plate, at 12 h on the 9.5 mM phosphate plate, and at >12 h on the 14.5 mM phosphate plate. At 24 h, these mutants produced the largest halo on all plates. Class 2 produced similar sized halos as the wild-type on the low phosphate plate (4.75 mM), but unlike the wild-type, it produced a strong halo on medium phosphate (9.5 mM) by 24 h. These mutants also produced halos on the highest phosphate plate, albeit smaller than the ones produced by the type 1 mutants. Class 3 produced more mutacin than the wild-type on the intermediate phosphate plate, but not on the highest phosphate plate.
Fig. 1.
Preliminary characterization of selected mutants.
a) A representative phenotype of the three mutant classes on BHI plate supplemented with different concentrations of phosphate. Mutants and the wild-type (wt) were grown overnight in BHI broth, and 5 μL of the overnight culture was spotted onto 50% BHI plates containing 4.75 mM, 9.5 mM, and 14.5 mM phosphate, respectively. The plates were grown anaerobically for 9 h, 12 h, and 24 h, respectively, and overlaid with the indicator strain S. sobrinus. Clear zones indicate the production of mutacin I. B.
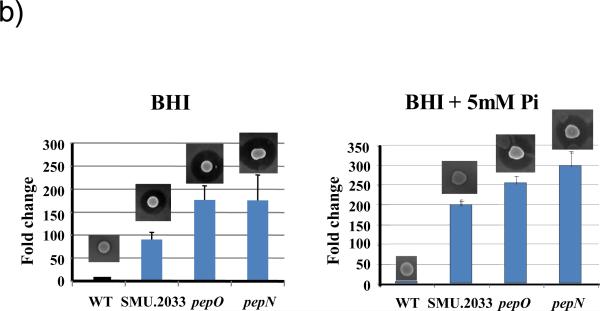
b) Mutacin production and mutA-luc gene expression in SMU. 2033, pepN, and pepO. Wild-type and mutant strains were grown overnight in BHI and 10 μ1 from each was spotted on BHI plates with or without the supplementation of 5 mM sodium phosphate. Cells were grown anaerobically for 16 h. One set was overlaid with the indicator strain for mutacin production as described above, and another set was used for luciferase assay. For luciferase assay, cells were scraped from the plates, and resuspended in BHI. Luciferase activity was measured immediately after resuspension. The same cell suspensions were also measured for absorbance at 600 nm and the luciferase activities were normalized against OD600. The luciferase activity of the wild-type was arbitrarily assigned as 1 (100%), and the level in the mutants was expressed as a ratio to the wild-type. The experiments were repeated at least 3 times with triplicate samples at each time. Presented here are representative results.
To investigate further whether the mutations affected mutacin I production at the transcriptional level, three mutations (pepN, pepO, and SMU.2033) were transformed into a mutA-luc reporter strain (He et al., 2008). The mutA-luc reporter activity was measured in cells grown on BHI plates as well as on BHI supplemented with 5 mM phosphate. As shown in Fig. 1B, all mutants displayed >100-fold increased mutA-luc reporter activity on both plates, indicating that all three genes affected mutacin production at the transcription level.
Discussion
This study is a continuation of our previous efforts to identify genes involved in regulating mutacin I production (Tsang et al., 2005). Using BHI plates supplemented with 5 mM phosphate to suppress mutacin I production in the wild-type, 77 mutant clones were originally identified, which produced mutacin in spite of abundant available phosphate. Of these, 34 sequences were obtained, and 17 unique genes were identified. We were not able to obtain clear sequence for the remaining clones, despite using additional approaches such as those used in our previous studies (Tsang et al., 2005). This suggests that these genes may have sequences with CviJI site too close to the cloning point (the original clone may have been generated from partial digestion), or they lack the restriction sites used in retrieving the inserted plasmid.
The effect of these genes on mutacin production was further analyzed under different phosphate concentrations. Based on their temporal pattern of mutacin production, these mutants were classified into three classes. Class 1 includes SMU.611, 797, 859, pepN, 2035, pepO, and 2077. These mutants exhibited the highest mutacin production on all plates tested. Although genes in this class are diverse, the two most prominent genes are pepN (SMU.1132) and pepO (SMU.2036). pepN was hit 8 times in the library screening, and pepO was hit 2 times (Table 1). Little has been reported regarding the function of pepN in streptococci; however, studies of pepN in other bacterial species may offer some indications. PepN belongs to a family of ATP-independent aminopeptidases that are involved in protein turnover inside the cytoplasm. In E. coli, pepN is a major aminopeptidase, and its mutation conferred resistance to sodium salicylate, indicating that it is a negative regulator for the sodium-salicylate induced stress response (Chandu et al., 2003; Chandu & Nandi, 2003). In addition, pepN gene expression was found to be induced by phosphate starvation in E. coli (Gharbi et al., 1985). Conspicuously, in S. mutans, pepN is located as the last gene in a 6-gene operon encoding proteins involved in phosphate uptake. Thus, based upon pepN gene expression in E. coli and its operon location in S. mutans, we speculate that the S. mutans pepN gene may be required for phosphate uptake or metabolism and its deletion may create a phosphate limitation signal responsible for inducing mutacin I production, even in the presence of an adequate supply of phosphate in the environment. PepO is a metalloendopeptidase, which belongs to the peptidase M13 family (Rawlings & Barrett, 1995). These peptidases are found in a wide range of organisms including mammals and bacteria. PepO is highly conserved among bacterial species, and among streptococci the similarity is ~90% at the amino acid level. However, the role of this peptidase in bacteria is not defined. A recent report by the Lamont laboratory suggested that pepO may play a role in the invasion of Porphyromonas gingivalis into host epithelial cells (Park et al., 2004). Here we demonstrate that both pepN and pepO mutations not only enhanced mutacin production on the high phosphate plate, but also during growth (Fig.1B). Furthermore, both gene mutations affected mutacin I production at the transcription level, increasing mutA gene expression >100 fold (Fig. 1B). The mechanism of how pepN and pepO mutations trigger mutacin production awaits further investigation.
Class 2 mutations include SMU.478, 675, 707, and 712. These mutants produced intermediate levels of mutacin on the highest phosphate plate tested. All genes in this class are annotated as genes in the central metabolism. The two interesting genes are ptsI (SMU.675) and pepC (SMU.712). pstI encodes phosphoenolpyruvate-protein phosphotransferase (enzyme I), which was first characterized in S. mutans by Boyd et al. as Enzyme I of the phosphoenolpyruvate-dependent phosphotransferase sugar transport system (Boyd et al., 1994). Subsequent studies by Cvitkovitch et al. demonstrated growth defects of ptsI mutant on glucose and raffinose (Cvitkovitch et al., 1995). In Streptococcus salivarius, a ptsI mutation strain was found to grow slower than the wild-type on glucose as well as possessing a much longer lag time. ptsI was also found to play a role in urease regulation (Weaver et al., 2000). It is likely that inactivation of ptsI reduced sugar transport, thus creating carbohydrate limitation in the cell, which triggered the stress response and mutacin I production. SMU.712 encodes a putative phosphoenolpyruvate carboxylase (PEPCase). PEPCase is an enzyme found in all multicellular plants, catalyzing the formation of oxaloacetate from phosphoenolpyruvate (PEP) and a hydrocarbonate ion (Sugimoto et al., 1992; Vazquez-Tello et al., 1993). In bacteria, PEPCase probably supplies oxaloacetate to the TCA cycle. Since S. mutans does not have a complete TCA cycle, it is likely that inactivation of the gene affected pyruvate metabolism, which is a downstream intermediate in oxaloacetate metabolism. Pyruvate is a central intermediate in energy metabolism as well as in amino acid biosynthesis. We speculate that disturbance in the intracellular metabolic balance may create a metabolic stress triggering mutacin I production.
The Class 3 mutations include SMU.591, 801, 1449, 1878, 1879, and 2033. These mutants produced mutacin on 9.5 mM phosphate, but could not produce mutacin on 14.5 mM phosphate. The two interesting genes in this class are manM (SMU.1878) and manN (SMU.1879) encoding mannose PTS system IIC and IID, respectively. Both genes are located in the same operon, and manN was hit 7 times in out library screening (Table 1). Although functions for manM and manN have not been studied, studies by Abranches et al. demonstrated that inactivation of manL, the first gene of the manLMN operon in S. mutans, resulted in impaired biofilm formation, decreased transformation efficiency, and altered expression of 62 genes (Abranches et al., 2006). Based on these results, it was suggested that EIIABMan (manL) plays a central regulatory role in the physiology and virulence of S. mutans by sensing the energy levels of the cell. The current finding that the inactivation of the EIICMan and EIIDMan components resulted in increased mutacin I production under repressive conditions is consistent with this notion.
In summary, we identified 17 genes whose mutation increased mutacin I production under repressive conditions. These genes are involved in a variety of cellular functions such as sugar transport, protein/peptide hydrolysis, amino acid and nucleotide synthesis, cell wall metabolism, and surface binding. The involvement of these genes in mutacin I production suggests an intimate and intricate connection between mutacin production and the overall cellular homeostasis.
Supplementary Material
Acknowledgements
This work was supported in part by NIH grants R01-DE014757 to F. Q., a COBRE P20-RR018741-05 grant to J. M., and a Delta Dental grant WDS78956 to W. S.
References
- Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. Journal of bacteriology. 2006;188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd DA, Cvitkovitch DG, Hamilton IR. Sequence and expression of the genes for HPr (ptsH) and enzyme I (ptsI) of the phosphoenolpyruvate-dependent phosphotransferase transport system from Streptococcus mutans. Infection and immunity. 1994;62:1156–1165. doi: 10.1128/iai.62.4.1156-1165.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandu D, Kumar A, Nandi D. PepN, the major Suc-LLVY-AMC-hydrolyzing enzyme in Escherichia coli, displays functional similarity with downstream processing enzymes in Archaea and eukarya. Implications in cytosolic protein degradation. The Journal of biological chemistry. 2003;278:5548–5556. doi: 10.1074/jbc.M207926200. [DOI] [PubMed] [Google Scholar]
- Chandu D, Nandi D. PepN is the major aminopeptidase in Escherichia coli: insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology (Reading, England) 2003;149:3437–3447. doi: 10.1099/mic.0.26518-0. [DOI] [PubMed] [Google Scholar]
- Cvitkovitch DG, Boyd DA, Hamilton IR. Regulation of sugar transport via the multiple sugar metabolism operon of Streptococcus mutans by the phosphoenolpyruvate phosphotransferase system. Journal of bacteriology. 1995;177:5704–5706. doi: 10.1128/jb.177.19.5704-5706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi S, Belaich A, Murgier M, Lazdunski A. Multiple controls exerted on in vivo expression of the pepN gene in Escherichia coli: studies with pepN-lacZ operon and protein fusion strains. Journal of bacteriology. 1985;163:1191–1195. doi: 10.1128/jb.163.3.1191-1195.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ooshima T. Inhibitory spectrum of a bacteriocinlike substance (mutacin) produced by some strains of Streptococcus mutans. Journal of dental research. 1975;54:140–145. doi: 10.1177/00220345750540010801. [DOI] [PubMed] [Google Scholar]
- He X, Wu C, Yarbrough D, Sim L, Niu G, Merritt J, Shi W, Qi F. The cia operon of Streptococcus mutans encodes a unique component required for calcium-mediated autoregulation. Molecular microbiology. 2008 Aug 8; doi: 10.1111/j.1365-2958.2008.06390.x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Bordador C, Shi W, Qi F. Transcriptional analysis of mutacin I (mutA) gene expression in planktonic and biofilm cells of Streptococcus mutans using fluorescent protein and glucuronidase reporters. Oral microbiology and immunology. 2004;19:252–256. doi: 10.1111/j.1399-302X.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Molecular microbiology. 2005;57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Zhu L, Shi W, Qi F. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS microbiology letters. 2006;265:11–17. doi: 10.1111/j.1574-6968.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiological reviews. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Yilmaz O, Jung IY, Lamont RJ. Identification of Porphyromonas gingivalis genes specifically expressed in human gingival epithelial cells by using differential display reverse transcription-PCR. Infection and immunity. 2004;72:3752–3758. doi: 10.1128/IAI.72.7.3752-3758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrot M, Charest M, Lavoie MC. Production of mutacin-like substances by Streptococcus mutans. Canadian journal of microbiology. 1989;35:366–372. doi: 10.1139/m89-056. [DOI] [PubMed] [Google Scholar]
- Parrot M, Drean MF, Trehan L, Lavoie MC. Incidence of bacteriocinogeny among fresh isolates of Streptococcus mutans. Canadian journal of microbiology. 1990;36:507–509. doi: 10.1139/m90-088. [DOI] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Applied and environmental microbiology. 2000;66:3221–3229. doi: 10.1128/aem.66.8.3221-3229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Applied and environmental microbiology. 2001;67:15–21. doi: 10.1128/AEM.67.1.15-21.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Evolutionary families of metallopeptidases. Methods in enzymology. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- Sahl HG, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annual review of microbiology. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. Microbial complexes in subgingival plaque. Journal of clinical periodontology. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Sugimoto T, Kawasaki T, Kato T, Whittier RF, Shibata D, Kawamura Y. cDNA sequence and expression of a phosphoenolpyruvate carboxylase gene from soybean. Plant molecular biology. 1992;20:743–747. doi: 10.1007/BF00046459. [DOI] [PubMed] [Google Scholar]
- Tsang P, Merritt J, Nguyen T, Shi W, Qi F. Identification of genes associated with mutacin I production in Streptococcus mutans using random insertional mutagenesis. Microbiology (Reading, England) 2005;151:3947–3955. doi: 10.1099/mic.0.28221-0. [DOI] [PubMed] [Google Scholar]
- Tsang P, Merritt J, Shi W, Qi F. IrvA-dependent and IrvA-independent pathways for mutacin gene regulation in Streptococcus mutans. FEMS microbiology letters. 2006;261:231–234. doi: 10.1111/j.1574-6968.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- Vazquez-Tello A, Whittier RF, Kawasaki T, Sugimoto T, Kawamura Y, Shibata D. Sequence of a soybean (Glycine max L.) phosphoenolpyruvate carboxylase cDNA. Plant physiology. 1993;103:1025–1026. doi: 10.1104/pp.103.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CA, Chen YY, Burne RA. Inactivation of the ptsI gene encoding enzyme I of the sugar phosphotransferase system of Streptococcus salivarius: effects on growth and urease expression. Microbiology (Reading, England) 2000;146(Pt 5):1179–1185. doi: 10.1099/00221287-146-5-1179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.