Abstract
Ubiquitin modification targets a protein for rapid degradation by the proteasome. However, poly-ubiquitination of proteins can result in multiple functions depending on the topology of the ubiquitin chain. Therefore ubiquitin signaling offers a more complex and versatile biology compared to many other post translational modifications. One area of potential for the application of this knowledge is the field of ischemia-induced brain damage, as occurs following a stroke. The ubiquitin proteasome system may exert a dual role on neuronal outcome following ischemia. Harmful ischemia results in an overload of the ubiquitin proteasome system, and blocking the proteasome reduces brain infarction following ischemia. However, the rapid and selective degradation of proteins following brief ischemia results in endogenous protection against ischemia. Therefore further understanding of the molecular signaling mechanisms which regulate the ubiquitin proteasome system may reveal novel therapeutic targets to reduce brain damage when ischemia is predicted, or to reduce the activation of the cell death mechanisms and the inflammatory response following stroke. The aim of this review is to discuss some of the recent advances in the understanding of protein ubiquitination and its implications for novel stroke therapies.
Keywords: rapid ischemic tolerance, preconditioning, bcl-2, synapse, neuroprotection
Ubiquitin was discovered in the 1970’s as a small (9 kDa) covalent post translational modification of proteins. A role for ubiquitination in the proteasomal degradation of short lived proteins was soon established (Ciechanover and others, 1984, Finley and others, 1984). The discovery of the role of the ubiquitin proteasome system in the anaphase promoting complex regulation of the cell cycle awakened many scientists to the potential for this system to regulate many biological processes (Pickart 2004). Although ubiquitin is best known as a prelude to proteasomal protein degradation, it also regulates protein-protein interactions and therefore numerous biological processes including protein translocation, signal transduction, gene transcription apoptosis and autophagic processes (Mukhopadhyay and Riezman 2007).
What makes ubiquitin particularly versatile as a post translational modification is the variety of modifications which it can form. Ubiquitin is conjugated to a lysine residue on the substrate protein (although N terminal linkage has also been reported). The availability of 7 lysine residues within the ubiquitin protein sequence enables the formation of poly-ubiquitin chains, each of which appears to encode different biological functions (Fig 1). The most common linkage is the Lys48 linkage, which is associated with protein degradation by the proteasome. In contrast, Lys63 linkages appear to play a role in protein-protein interactions mediating signal transduction, translocation and DNA repair (although see below). Ubiquitin also regulates lysosomal as well as proteasomal protein degradation. Lys63 linked poly-ubiquitin modified proteins are targeted to the lysosome for processing (Ikeda and Kerppola 2008). The role of other poly-ubiquitin lysine linkages is not yet clear, and still subject to investigation (Ikeda and Dikic 2008). With so many functions available to different poly-ubiquitin additions, it is clear that the process of poly-ubiquitination must be tightly regulated at a number of points.
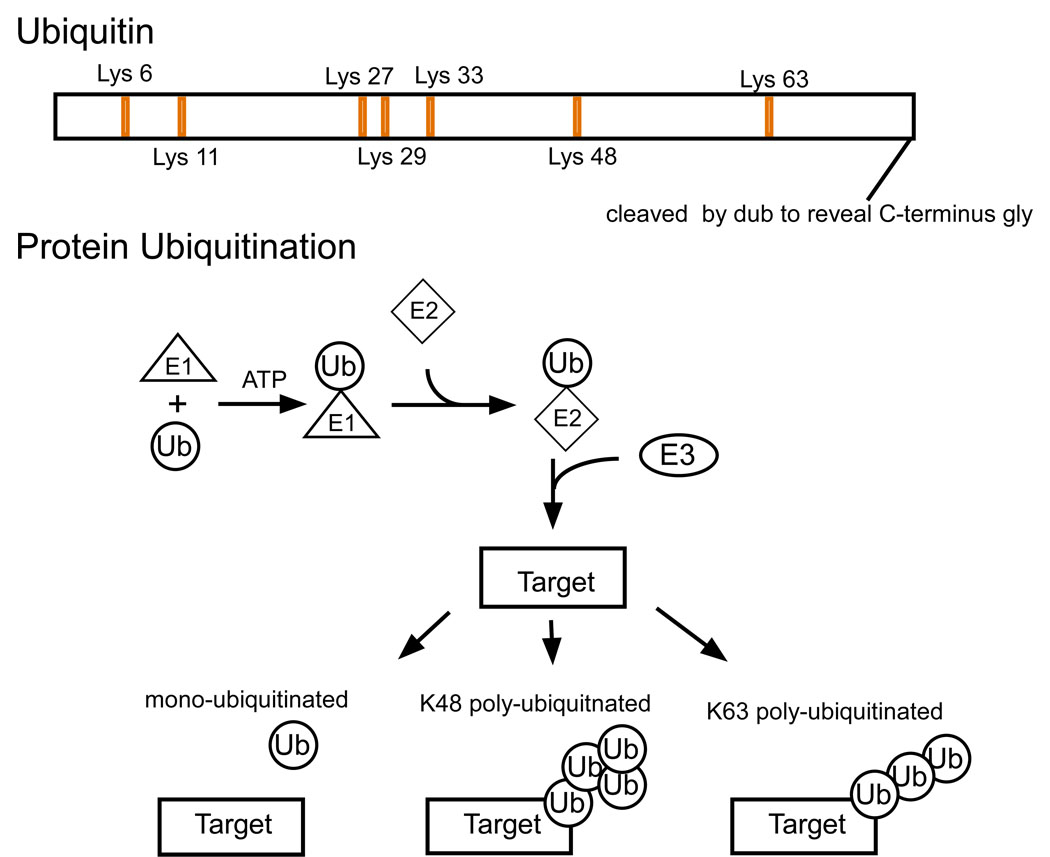
Figure 1.
Structure of ubiquitin. Upper) Ubiquitin is synthesized as a precursor protein, whose c terminus is cleaved by a de-ubiquitinating enzyme to reveal a C-terminus glycine residue. Seven lysine residues are present in the primary amino acid structure of ubiquitin, which allows multiple potential poly ubiquitin chain linkages. Lower) Ubiquitin is added to a substrate protein by the sequential action of an E1-ligase, E2-ligase and an E3- ligase. The different poly-ubiquitin chains have different topologies, which are associated with their function. Ubiquitin chains with a Lys 48 linkage usually result in proteasomal degradation of the protein. Lys 63 linked chains can target proteins to the proteasome, lysosome or play a role in protein-protein interactions.
Ubiquitin is added and removed from a target protein by E3-ligase and de-ubiquitinating enzymes respectively, which function similarly to protein kinases and phosphatases regulating protein phosphorylation. The number of potential E3-ligases encoded in the genome is similar to the potential number of protein kinases, suggesting that ubiquitination may play a critical role in regulating as many biological processes as protein phosphorylation. The ubiquitin system requires a sequential process of E1-, E2- and E3-ligases to conjugate ubiquitin to a substrate (Hershko 1983) (Fig 1). Ubiquitin is synthesized as a precursor, and the C-terminus tail of the pro-protein is removed by a de-ubiquitinylating enzyme (DUB) to expose a C-terminal glycine residue (Lund and others, 1985). Ubiquitin forms a high energy thioester bond with an E1-ligase, in a reaction requiring ATP (Pickart 2001). This step is the only step of the conjugation process which requires ATP, and may account for the observation that ubiquitination may occur under conditions of low ATP levels such at ischemia (see later).
The ubiquitin is transferred from the E1-ligase to the E2-ligase (Fig 1). There are approximately 20 E2-ligases encoded by the human genome, in contrast to only one E1-ligase. Some E2-ligases are used for one ubiquitin family member, for example Ubc9 is an E2-ligase unique to sumoylation (Johnson and Blobel 1997). Different E2-ligases have specificity for different steps of the ubiquitination process; UbcH5a catalyzes the non-specific lysine linkage of ubiquitin, where as Ubc13 formed only Lys63 linked poly-ubiquitin chains (Windheim and others, 2008). In another recent report the rapid mono-ubiquitination of cell cycle regulatory proteins by the APC (anaphase promoting complex) was identified as being mediated by Ubc4, where as Lys 48 poly-ubiquitination by the APC was mediated by Ubc1 (Rodrigo-Brenni and Morgan 2007). Interestingly the E2-ligases interact with different domains on the E3-ligase (Windheim and others, 2008). The choice of E2-ligase may determine the topology of the substrate ubiquitination modification, and poly-ubiquitination of a target may require multiple E2-/ E3-ligase combinations.
In the final step of the process, the ubiquitin is transferred from the E2-ligase to the substrate by an E3-ligase. This reaction defines the target specificity of ubiquitination. There are two major E3-ligase families, the RING (really interesting new gene) and HECT (Homologous to the E6-AP Carboxyl Terminus) families. Additional E3-ligases have recently been identified including a novel zinc finger protein containing E3 ligase and U box domain containing proteins (Jiang and others, 2001, Wertz and others, 2004). However, U-box proteins may be involved with poly-ubiquitin chain extension / E4 ligase activity (see below). Furthermore E3-ligase independent mono-ubiquitination has been reported; however the significance of this observation is not yet clear (Hoeller and others, 2007). The RING and HECT E3-ligases have quite different mechanisms for transferring the ubiquitin to the target protein. RING E3-ligases mediate the direct transfer of the ubiquitin from the E2-ligase directly to the target protein. This may involve a conformational change in the E2-ligase once it has bound to the E3-ligase, to enable the access of the ubiquitin for transfer. In contrast, the HECT E3-ligases form an intermediate E3-ligase-ubiquitin complex prior to the transfer of the ubiquitin to the target protein.
Whether all E3-ligases mono-ubiquitinate or poly-ubiquitinate target proteins is not yet clear. A fourth enzyme class, the E4-ligase was recently described and shown to regulate poly-ubiquitin chain extension. Poly-ubiquitin chains in excess of four ubiquitin residues are required for proteasomal degradation of proteins (Thrower and others, 2000). Ubiquitin fusion degradation protein 2 (UFD2) functions with E1, E2 and E3-ligases to ubiquitinate a UFD substrate (UFD4). Omitting UFD2 from the reaction resulted in termination of the poly-ubiquitin chain with only three ubiquitins (Koegl and others, 1999). In addition the first three ubiquitins were added via Lys 29 linkages whereas chain extension was Lys48 mediated linkages (as shown by incubation with K29R and K48R ubiquitin mutants respectively) (Koegl and others, 1999). This study also showed that the E4-ligase contains a poly-ubiquitin binding domain, although it is not clear whether the binding of the E4 changes the conformation of the already attached ubiquitin. The authors suggest that E4-ligases may promote the degradation of post-translationally activated protein, by extending the oligo-ubiquitin chain (Koegl and others, 1999).
UFD2 contains a U box domain, which may identify potential new E4-ligase members. An interesting member of this family is the E4-ligase CHIP (C terminus of Hsc70-interacting protein), which functions with the E3-ligase mutated in a familial form of Parkinson’s disease, PARKIN, to regulate Pael receptor degradation (Hoppe 2005, Imai and others, 2002). However, other studies suggest that CHIP also acts as an E3 ligase (Windheim and others, 2008). Whether all proteins which contain a U-box are E4-ligases remains to be determined, but E4-ligase activitiy has been reported in proteins that do not contain a U-box domain (Hoppe 2005). The nuclear protein P300 recognizes MDM2 (Murine double minute 2) mediated mono-ubiquitination of p53 promoting further ubiquitin chain extension (Grossman and others, 2003). P300 does not contain a U-box domain in its sequence, suggesting that another domain on this protein may function as the E4-ligase or the P300 recruits another E4-ligase. The target specificity of E4-ligases is not yet clear, or whether this function is mediated by E2/ E3 ligase combinations (Windheim and others, 2008). The elucidation of proteins with E3- and E4-ligase activity for specific proteins may reveal additional targets for drug manipulation of this system.
One of the more perplexing questions which is currently being investigated by a number of groups is the mechanism(s) by which the ubiquitin chain on a protein substrate is extended. A number of complex models have been proposed (Fig 2), and yet a clear answer is not apparent (see review by Hochstrasser (Hochstrasser 2006)). Following addition of one or more ubiquitins to the substrate protein the E3 ligase, which interacts with the substrate protein will be too far from the end of the chain to mediate the conjugation of the next ubiquitin molecule. A number of models have been proposed to explain this issue. One suggested mechanism is that the poly-ubiquitin chains are synthesized prior to addition to the substrate, either by the E2 or E3-ligase. For example the Ube2g2/ gp78 mediated poly-ubiquitination involves the assembly of poly-ubiquitin chains on the E2-ligase, prior to transfer to the substrate (Li and others, 2007). This may be the reason a number of studies have reported auto-ubiquitination of E2- and E3-ligases. Alternatively, the initial E3-ligase mono-ubiquitinates the substrate and then either different E2 ligases regulate chain extension, or an additional factor (the E4 ligase) regulates extension (Hoppe 2005). Currently different E3-ligases are being studied in different systems and under different physiological conditions, which may account for the unclear picture from these recent studies.
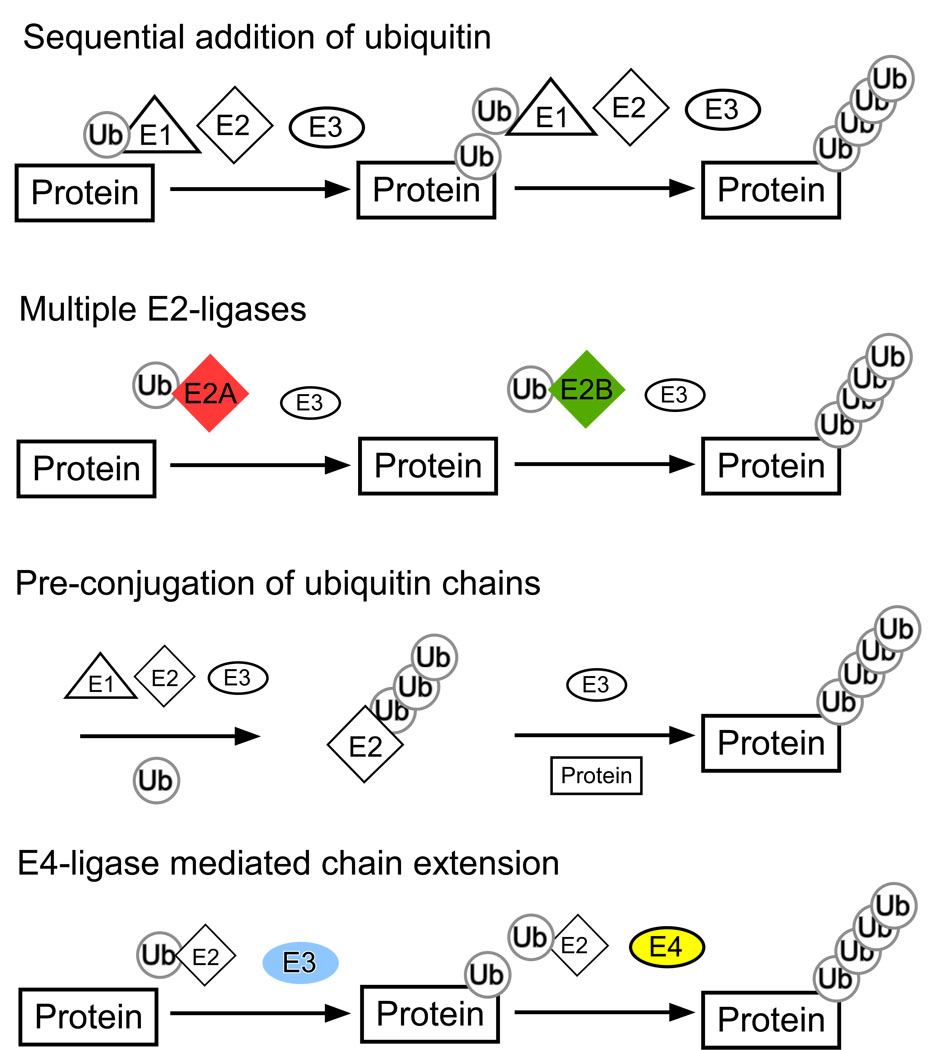
Figure 2.
Potential models of poly-ubiquitination. Sequential addition of ubiquitin refers to the process of ubiquitin transferring from the E1, to the E2 and then E3 ligase onto the substrate, this process would then be repeated for each additional ubiquitin added to the chain. However this model fails to describe how the E3-ligase which interacts with the substrate protein can physically reach the end of the ubiquitin chain. Some protein ubiquitination is mediated by multiple E3-ligases acting with the E3-ligase. The first E2-ligase mediates mono-ubiquitination, and the second E2-ligase promotes poly-ubiquitination. Some E2-ligases can assist in the transfer of pre-synthesized poly-ubiquitin onto the substrate. Finally, some proteins have E4 ligase activity. When added to the reaction, E4 ligases promote poly-ubiquitin chain extension.
Ubiquitin chains can be remodeled to change the function of the ubiquitin modification to regulate degradation (see Fig 3). Wertz and others showed how following Tumor Necrosis Factor (TNF) receptor activation, the signaling protein receptor interacting protein (RIP-1) is bound to the TNF receptor complex via a Lys63 linked poly-ubiquitin modification (Wertz and others, 2004). To turn down TNF receptor signaling, RIP-1 is deubiquitinated by the co-factor A20 (via its ovarian tumor domain (a DUB family domain)). Following de-ubiquitination, RIP-1 is Lys48 poly-ubiquitinated by A20 targeting it for proteasomal degradation and thereby turning off TNFR signaling. Interestingly, A20 contains a novel E3 ligase domain. The A20 zinc finger domain is structurally divergent from the RING zinc finger domain common to many E3 ligases (Wertz and others, 2004).
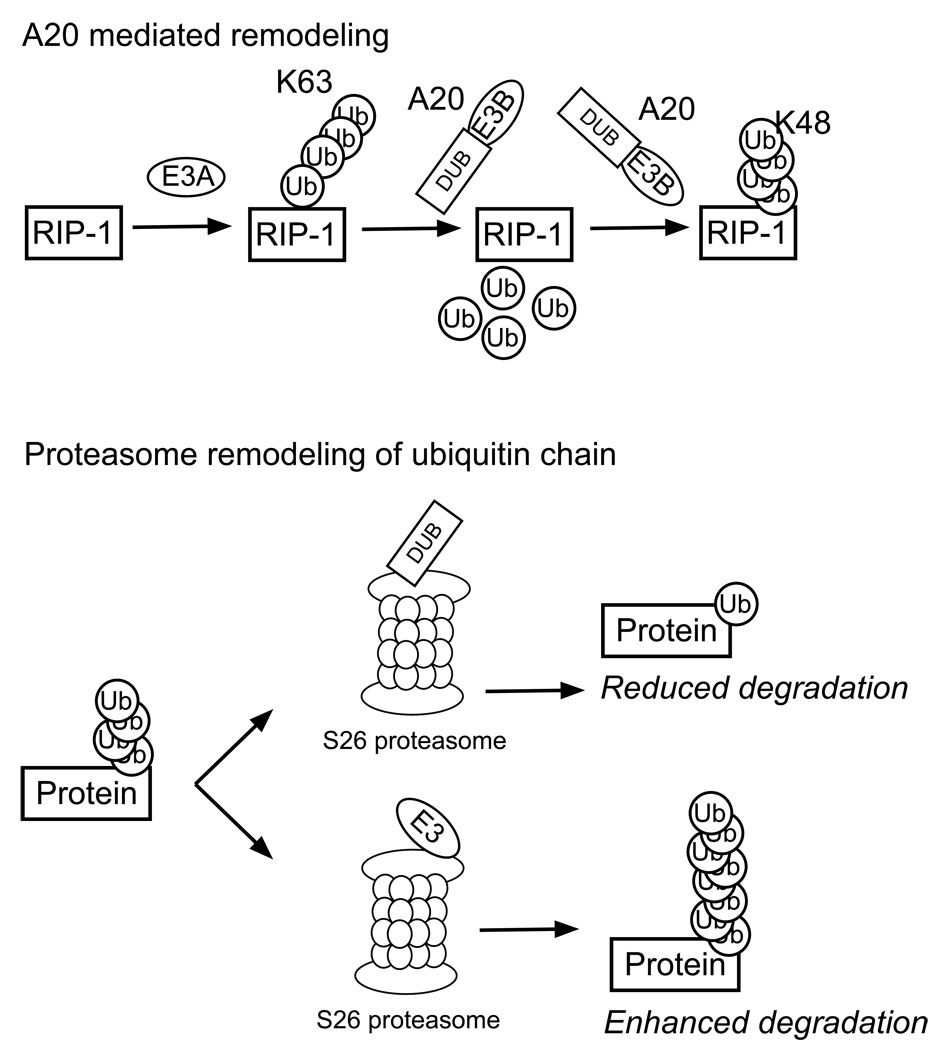
Figure 3.
Examples of ubiquitin chain remodeling. A20 mediates de-ubiquitination of Lys63 poly-ubiquitinated RIP-1. A20 can also catalyze the Lys48 poly-ubiquitination of RIP-1. At the proteasome the actions of de-ubiquitinating enzymes and E3-ligases can prevent a substrate from proteasome degradation, or extend the poly-ubiquitin chain and therefore enhance degradation.
In addition to modification of the poly-ubiquitin chain prior to proteasomal trafficking, proteins at the proteasome are also subject to chain editing (Fig 3). The S26 proteasome contains multiple E3 ligases and DUB domains. Recent studies of the interplay between the ligase Hul5 and the DUB Ubp6, show that these enzymes can regulate substrate degradation by extending their ubiquitin chains and enhancing proteasome activity, or removing their ubiquitin modifications to reduce degradation (Crosas and others, 2006).
Taken together these studies show that the process of protein ubiquitination is a tightly regulated process. The degradation of proteins by the ubiquitin proteasome system and subsequent loss of function determines the consequence for the cell (i.e. degradation of cell death proteins promotes protective phenotypes (Meller and others, 2006). However, ubiquitin has multiple functions, not just protein degradation. Consistent with a role in poly-ubiquitin chains mediating protein-protein interactions, a family of ubiquitin-like modifier proteins has been identified.
Ubiquitin binding domains, protein interactions by ubiquitination
The binding of ubiquitin chains, which are associated with non-proteasome targeting, is accomplished by ubiquitin binding domains (UBD) on the acceptor proteins. There are a number (>30) of different ubiquitin binding domain motifs that can bind to mono- and poly-ubiquitin. Interestingly the affinity of a UBD and ubiquitin is low, typically in the low micromolar range (Ikeda and Dikic 2008). Since ubiquitin levels in cells are relatively high, this suggests that the regulation of binding is due to accumulation of UBD containing proteins in cellular compartments and the use of multiple UBDs within a protein/complex (Ikeda and Dikic 2008).
Further analysis of this issue is ongoing, and it was recently shown that the 3D topology of Lys63 and Lys48 linked poly-ubiquitin may offer some ideas as to how the binding domains specific to each are accomplished. X-ray crystallography of Lys48 linked di-ubiquitin show a compact arrangement of the ubiquitin moieties, in contrast Lys63 ubiquitin is more open and linear in shape (Varadan and others, 2004, Varadan and others, 2002). Therefore the structure of the Lys48 and Lys63 linkages are quite different. In a recent study of the binding affinities of different UBDs four classes were described, those which bound mono-ubiquitin, linkage selective and nonselective domains, and one class which did not bind any ubiquitin ligand (Raasi and others, 2005). Interestingly, the protein homologs containing UBDs show a remarkable conservation of ubiquitin chains specificity between species (Raasi and others, 2005).
Ubiquitin-like modifiers
Ubiquitin is a member of an ever growing family of ubiquitin-like modifiers, which include SUMO, NEDD8 and Atg. For more detailed information on these proteins the reader is referred to some recent reviews (Geiss-Friedlander and Melchior 2007, Kerscher and others, 2006). These proteins have a similar tertiary structure to ubiquitin, but have a lower amino acid homology to ubiquitin. A similar series of sequential reactions regulate the modification of proteins by ubiquitin-like modifiers, for example SUMO-ylation requires an E1, E2 and E3-ligase mediated reaction. Modification by ubiquitin-like modifiers results in the regulation of protein function and localization, rather than direct protein degradation. These additional protein modifiers also interact with the ubiquitin system to regulate protein function, as discussed below.
Mono-ubiquitination
Most research has focused on how poly-ubiquitination (Lys48) results in protein degradation by the proteasome. Mono-ubiquitination has also been shown to regulate a number of divergent cellular processes (Hicke 2001). One of the more commonly studied functions of mono-ubiquitination is the regulation of gene expression. Mono-ubiquitination of histone H2A and H2B is an important event for the elongation step of transcription. In addition, mono-ubiquitination functions as a signaling molecule for the assembly of transcription enzymes and unraveling of chromatin (see (Weake and Workman 2008) for a review of histone ubiquitination and gene expression).
Mono-ubiquitination can also affect protein function outside of the nucleus. One such example is the regulation of receptor endocytosis (Hicke 2001). Mono-ubiquitination of a receptor or ion channel promotes endocytosis and internalization. These proteins are either de-ubiquitinated and recycled, or subject to lysosomal degradation. In addition, the tumor suppressor protein p53 is exported from the nucleus to the mitochondria when it is mono-ubiquitinated (Marchenko and others, 2007), where as poly-ubiquitination results in its proteasomal degradation. p53 is then de-ubiquitinated by HAUSP at the mitochondria which enables its interaction with Bcl-2 family members (Marchenko and others, 2007).
Crosstalk between ubiquitin, phosphorylation and other ubiquitin-like modifiers
Recent studies suggest that post translational modifiers do not work alone, but rather form a complex system of signaling crosstalk to regulate biological processes(Hunter 2007). The ubiquitination of a substrate can be regulated by its phosphorylation, for example the phosphorylation of Bim on Ser 65 by ERK1/2 enables its interaction with its E3-ligase, resulting in ubiquitination and proteasomal degradation (Ley and others, 2003) (Fig 4). Interestingly the ubiquitination of multiple Bcl-2 family proteins are regulated by the phosphorylation of critical residues (Breitschopf and others, 2000a). In addition nitrosylation of Bcl-2 regulates its degradation by the proteasome (Chanvorachote and others, 2006). Some kinases are also regulated by ubiquitination, which can turn on or off a biological signal, for example RIP-1 is regulated by A20, which controls IKKβ function following TNF receptor signaling (Wertz and others, 2004).
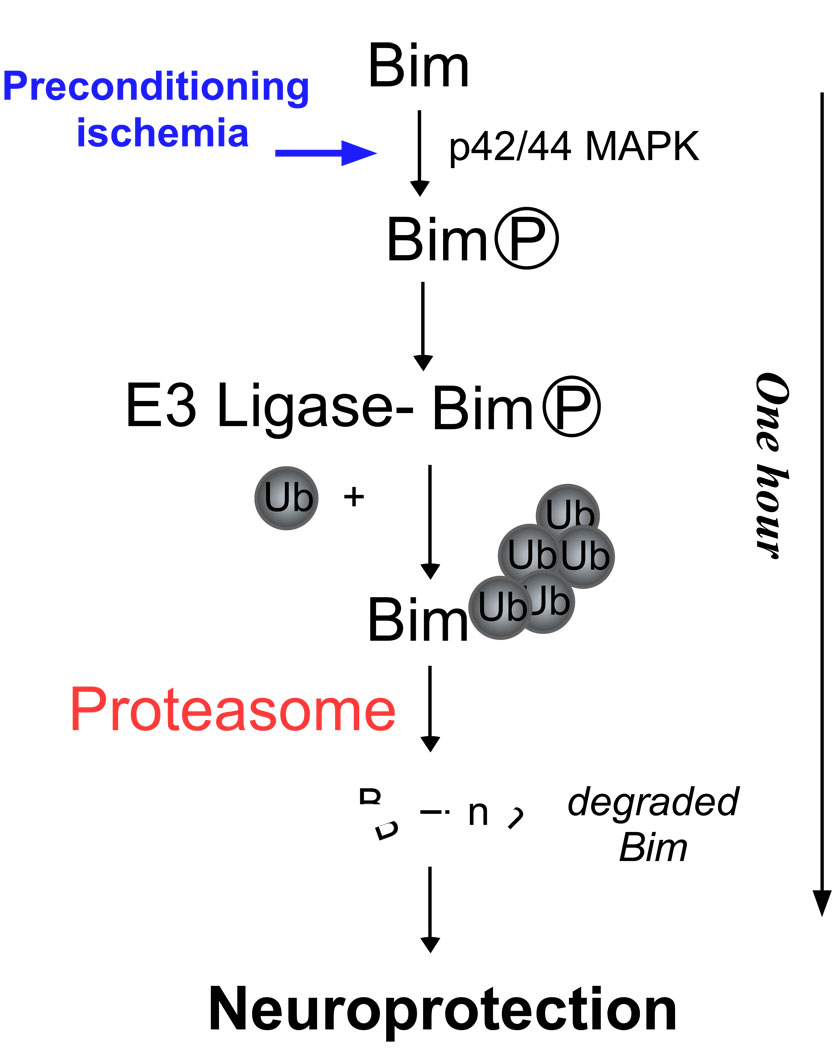
Figure 4.
Crosstalk between phosphorylation and ubiquitination promotes the degradation of Bim following brief preconditioning ischemia. Following ischemia, activation of p42/p44 MAPK promotes Bim phosphorylation. Phosphorylated Bim is ubiquitinated by its E3-ligase and degraded by the proteasome. The loss of Bim results in protection against further ischemia. What is unclear, is which E3-ligase mediates Bim ubiquitination following preconditioning ischemia.
The crosstalk between ubiquitin and the ubiquitin like modifier SUMO has been the focus of many studies. Under normoxic conditions, hypoxia induced factor (HIF) 1 is hydroxylated promoting its ubiquitination by the E3-ligase VHL(von-Hippel Lindau tumor factor) resulting in protein degradation. Hypoxia induced the sumoylation of HIF1α, which also enables its ubiquitination by VHL. De-sumoylation of HIF1α results in stabilization of the protein by preventing it’s ubiquitination and promoting the expression of HIF1-regulated genes (Cheng and others, 2007). These data are in contrast to other reports which suggest that sumoylation promotes HIF1α gene expression (Bae and others, 2004), and suggest that either sumoylation/ desumoylation may be involved in HIF1 degradation, perhaps similar to A20 mediated regulation of RIP-1, or additional signaling mediators may regulate HIF1α in response to hypoxia.
The tumor suppressor p53 is subject to complex regulation as signaling crosstalk, including phosphorylation, acetylation, nitrosylation, sumoylation, mono-ubiquitination and poly-ubiquitination (Lavin and Gueven 2006). The stability of the E3-ligase that regulates p53 ubiquitination MDM2, is regulated by the tumor suppressor p14ARF and other regulatory proteins (Zhang and others, 1998). In addition Arf can regulate protein sumoylation (Gallagher and others, 2006). Hence, one can determine that ubiquitination (and other ubiquitin-like modifications of proteins) does not happen in isolation, but rather as part of a highly complex, intricately regulated signaling system. Given the specificity within the system and the remarkable variety of target proteins whose function and stability are regulated by the ubiquitin proteasome system, it is clear that further study of this system may reveal targets for therapeutic manipulation.
The proteasome
In order to be targeted to the proteasome, a protein requires a poly-ubiquitin tail of at least four residues, resulting in a high affinity for the proteasome and subsequent processing. The proteasome complex allows the rapid complete and selective degradation of substrates due to its structure. The physical tube like structure of the proteasome ensures that only targeted proteins are processed. The core is narrow, which requires the unfolding of a protein into a linear peptide sequence (Pickart and Cohen 2004). The importance of the proteasome is underlined by the fact that many knockouts of proteasome subunits result in lethal phenotypes (Heinemeyer and others, 1991, Orlowski 1999).
The S26 proteasome is made up of an S20 core and an S19 cap (the S denotes sedimentary co-efficient, the molecular weight of the core and cap structures are approximately 760 and 1 Mda respectively). The proteasome is structurally similar to the HsIV and CIsP proteases found in prokaryotic cells, however on an evolutionary basis these proteases are unrelated. The S20 proteasome shows little protease activity on its own and requires the S19 cap proteins for full function (Pickart and Cohen 2004). The catalytic core of the proteasome is formed by four 7-membered rings of alpha and beta subunits (Baumeister and others, 1998). The catalytic sites are on the β subunits, where as the α sub units control the passage of substrates to the catalytic sites.
The proteasome S19 cap is made up of approximately 17 subunits (designated RPN in C. elegans or yeast). The cap is the site of protein unfolding, a process which requires ATP. Indeed many of the cap subunits possess ATPase activity. The cap forms a ring and a lid type feature to regulate entry of the protein to the protease (Baumeister and others, 1998, Pickart and Cohen 2004). As well as unfolding the protein, the cap is the site for removal of the ubiquitin chain prior to degradation of the protein allowing ubiquitin to be recycled by the cell. Two types of de-ubiquitinating enzymes act on substrates, USP14 removes the proximal ubiquitin from a protein, where as UCH (ubiquitin C Terminal hydrolases) removes distal ubiquitins from the substrate. Recently, it was shown that the cap also contains E3-ligase activity. The role of this is not clear, but the E3-ligase Hul5 functions with the de-ubiquitinating enzyme USP14 to regulate protein degradation (Crosas and others, 2006).
The role of the ubiquitin-proteasome system in ischemia and ischemic tolerance
The ubiquitin proteasome system has been implicated in a number of pathologies which effect neuronal structure and function. Proteasome inhibitors when administered for long durations or at high concentrations induce neuronal cell death (Qiu and others, 2000). Blocking proteasome function delays axonal degeneration following cell injury or axon cutting (Wallerian degeneration) (Zhai and others, 2003). Interestingly, the wlds mutant mouse shows a slowing of the degeneration, and express a mutant form of the UFD2 E4-ligase lacking catalytic activity fused to nicotinamide mononucleotide transferase (Mack and others, 2001).
Ubiquitin-rich inclusions are a common feature of certain neurodegenerative diseases, such as Alzheimer’s disease and Parkinson’s disease. These plaques tend to be enriched in ubiquitin, and it has been suggested that they are deposits of mis-folded proteins. Accumulation of the proteins into aggregates may overwhelm the proteasome system resulting in cell stress and neuronal death. For more details on the role of the ubiquitin proteasome system in these diseases the reader is referred to the following reviews ((Lim 2007, Oddo 2008)).
A number of studies have investigated protein ubiquitination and proteasome activity following ischemia (Fig 5). These studies suggest detrimental effects of the ubiquitin proteasome system following ischemia, resulting in damage to cell components, or mediating inflammatory responses and leukocyte infiltration to the brain. Ischemia in the brain is modeled by either a local reduction in blood flow to a discrete brain area (focal ischemia) or a complete reduction in blood flow to the entire brain (global ischemia) (Traystman 2003). Following global ischemia the ubiquitination of proteins which form aggregates has been reported (Liu and others, 2005b). These protein aggregates contain poly-ribosomes, translation associated proteins and the E3/ E4-ligase CHIP (Liu and others, 2005b). Following global ischemia, the prolonged accumulation of poly ubiquitinated proteins in the post synaptic density has been reported in the selectively vulnerable hippocampal regions, but were briefly found in the ischemia resilient dentate gyrus cells (Liu and others, 2005b, Liu and others, 2004). However, these studies did not identify which type of poly-ubiquitin linkage was added to the proteins. The proteasome is also affected by harmful ischemia resulting in cap disassembly and the trafficking of these cap subunits to protein aggregates and a reduction in proteasome function. The formation of aggregates by ubiquitinated proteins, due to impaired proteasome function, may contribute to cell stress following ischemia thereby amplifying the neuronal damage to the neurons.
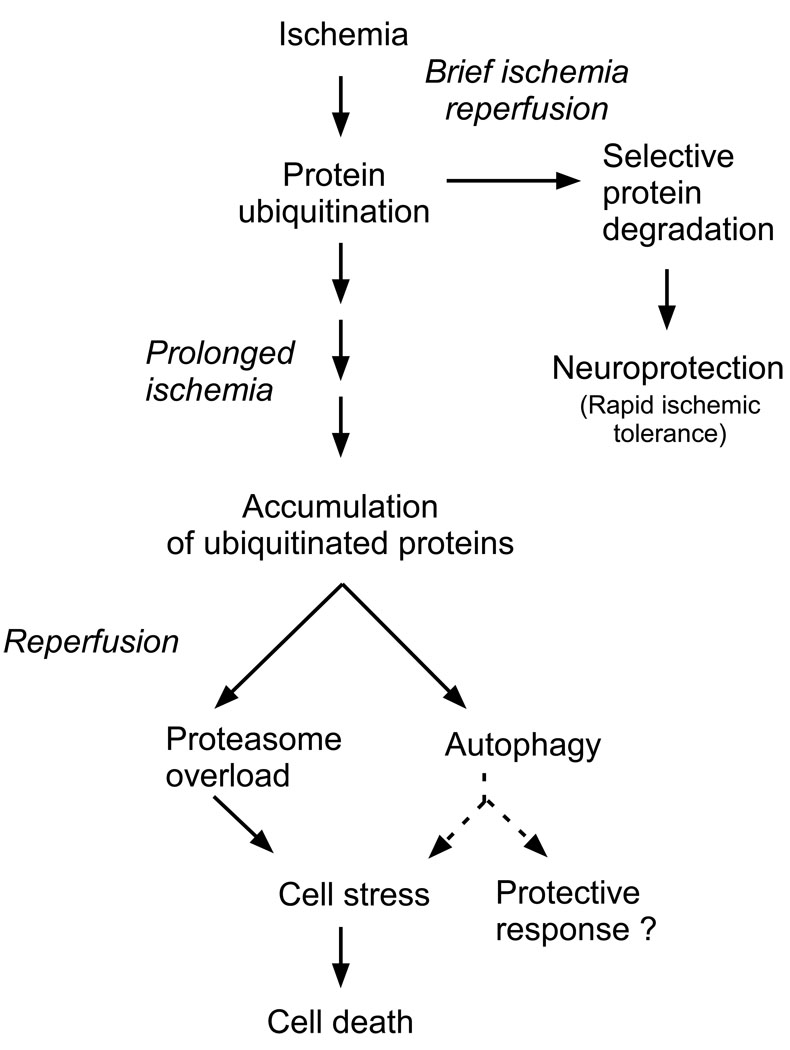
Figure 5.
Overview of potential role of ubiquitin proteasome system in mediating the cellular response to ischemia. Brief ischemia can result in the rapid degradation of proteins by the proteasome and tolerance to ischemia. In contrast following harmful ischemia the accumulation of ubiquitinated proteins may contribute to cell stress. Harmful ischemia reduces proteasome function. The ubiquitinated proteins form aggregates which are subject to autophagic processing. Whether autophagy is a protective response or promotes further cell stress following ischemia is unclear.
Preconditioning neurons with brief ischemia resulted in a reduction in protein aggregation following ischemia (Liu and others, 2005a). Both the formation of triton-X100 insoluble ubiquitinated proteins and the depletion of free ubiquitin following harmful ischemia were reversed in ischemic tolerant (delayed) neurons. However, it is not clear whether the proteins are less ubiquitinated because tolerant cells are not damaged by the ischemia or whether the tolerance reduces protein aggregation, thereby protecting the cells. Interestingly, ischemic tolerance is associated with raised heat shock protein expression, and raised heat shock protein levels result in less ubiquitin redistribution in neurons following ischemia (Ouyang and others, 2005).
A number of protein substrates have been identified as being regulated by the proteasome following ischemia. Ubiquitination of survival proteins following ischemia has been reported in the retina. Analysis of a gracile axonal dystrophy mouse reveals an exon deletion of ubiquitin C-terminal hydrolase-L1. These mice show less ubiquitination of Bcl-2 and XIAP in response to ischemia, compared to wild type mice (Harada and others, 2004). In addition PKC gamma, type 2 iodinase and AKAP121 are degraded by the proteasome following ischemia (Carlucci and others, 2008, Lamirand and others, 2007, Matsumoto and others, 2004). Hence a number of proteins are subject to ubiquitination following ischemia, which may regulate cell fate.
Ubiquitination may regulate autophagic processing of proteins following ischemia as well as proteasomal protein degradation. Autophagy is a lysosomal-dependent mechanism of degrading proteins, and organelles. Autophagosomes form by the enveloping of the cytosol by a phagophore (a membrane sac) in a highly regulated process, which then fuses with a lysosome containing proteases that degrade the contents. Autophagy has divergent effects, which are relevant to ischemia and other neuropathological conditions. Autophagic neuronal death has been reported in response to ischemia (Adhami and others, 2006, Carloni and others, 2008, Koike and others, 2008, Qin and others, 2008, Uchiyama and others, 2008). However, whether autophagy is a survival or damaging response is not yet clear. While autophagic cell death has been reported since the 1960’s, and is a mechanism of cell death which can proceed without the apparent biochemical hallmarks of programmed cell death (i.e. caspase cleavage and activation), some studies suggest a protective role (Carloni and others, 2008). Hence the activation of autophagic processes following ischemia/ neuronal damage could be a double-edged sword (for a review on ischemia and autophagy see (Rami and Kogel 2008)). Therefore it is not yet clear whether the autophagic clearance of ubiquitinated proteins is a reserve for the proteasome, or a separate system for protein clearance which is activated in response to an overwhelming of the proteasome following ischemia.
Proteasome inhibitors reduce ischemia-induced brain injury
Further evidence implicating a role of the ubiquitin proteasome system in ischemia induced cell damage come from studies showing that proteasome inhibitors reduce brain infarction following ischemia in animal models. There are two classes of proteasome inhibitors. The first such as clasto-lactacystin-β-lactone (usually administered as a pro drug lactacystin) are derived from naturally occurring compounds from bacteria/ fungi. The second class of inhibitors are short modified peptides, such as the classical proteasome inhibitor MG132 (carbobenzoxy-L-leucyl-L-leucyl-L-leucinal) and bortezomib. It is noteworthy that some of these inhibitors can block other proteases such as caspases, calpains and cathepsin. For structures and pharmacology of proteasome inhibitors the reader is referred to the review of Wojcik and Napoli (Wojcik and Di Napoli 2004).
The proteasome inhibitor MLN519, which is structurally similar to lactacystin(Wojcik 1999), provides effective neuroprotection in models of focal ischemia (2 hours middle cerebral artery occlusion) when given up to 4 hours post ischemia (Williams and others, 2003). MLN519 treated rats show less neuronal damage, astrocytic damage, leukocyte infiltration and infarction compared to untreated rats subjected to injurious ischemia. The dose of MLN519 reduced blood proteasome activity by 80% and also decreased NFKB activation. Protection was not evident in this study at 8 hours post ischemia (Williams and others, 2003).
In follow up studies, MLN519 shows neuroprotective effects in behavioural assessment when administered up to 10–48 hours following ischemia (Williams and others, 2004, Williams and others, 2005). MLN519 increased survival and reduced ischemia induced weight loss. In addition animals subjected to ischemia and MLN519 also performed better in some neurological tests than ischemia treated controls. This exciting study suggests that proteasome inhibitors may be a useful strategy to reduce delayed post ischemic brain injury. Indeed there is a similar temporal profile of post ischemic brain injury and neuroinflamation (Dirnagl and others, 1999). This may be a more feasible therapeutic time point to investigate, given the failure of previous neuroprotective therapeutic strategies which targeted apical events following ischemia and hence had a narrow therapeutic time window (NMDA receptor antagonists and calcium channel blockers).
Bortezomib (Velcade), a di-peptide boronate derivative approved for multiple myeloma, also reduces ischemia induced brain injury. In a study of embolic focal ischemia, bortezomib induced protection up to 4 hours post ischemia, but enhanced the therapeutic time widow for Tissue plasminogen activator therapy to 6 h (Zhang and others, 2006b). In a separate study, bortezomib reduced brain injury in a permanent MCAO model and a second embolitic model (Henninger and others, 2006). These studies show that bortezomib reduces blood proteasome activity by 77% and also reduces NFKB and eNOS (nitric oxide synthase) expression following harmful ischemia (Henninger and others, 2006).
These studies suggest that as a post-stoke therapy, proteasome inhibitors may have therapeutic efficacy in reducing neuro-inflammation associated delayed cell death in the brain. This view is supported by the fact proteasome inhibitors do not protect neurons from in vitro modeled ischemia or glutamate toxicity (Meller and others, 2008, Waataja and others, 2008). The global blockade of the proteasome, however, may result in many unwanted side effects and as such the future target for this research will be to identify critical molecular mechanisms that are blocked by the proteasome inhibitors to develop more specific therapeutics. In addition, the proteasome may also mediate protective events, which may be promoted to induce protection against ischemia. This mechanism is slowly being revealed by the study of the endogenous protective phenomenon of ischemic tolerance.
Protein ubiquitination and ischemic tolerance
Tolerance, induced by a preconditioning event, is a universal phenomenon and has been observed in many species and organs. A preconditioning agent is defined as an agent or stimuli, which at a low dose is not toxic but induces protection against a larger toxic dose (tolerance) (Dirnagl and others, 2003, Janoff 1964). The protection induced by preconditioning can render the organ/ system protected from other harmful stimuli (cross-tolerance), for example endotoxin (lipo-poly saccharide (LPS)), seizures or spreading depression all induce tolerance to ischemia (Ahmed and others, 2000, Kobayashi and others, 1995, Rosenzweig and others, 2007). Many early studies of ischemic tolerance focused on the protection of the myocardium following the induction of ischemic tolerance, however, ischemic tolerance was soon reported in the brain (Kitagawa and others, 1990). While many of the earlier heart studies have guided experiments in the brain, differences exist between the two systems (Tauskela and others, 1999).
Ischemic tolerance has been observed in murine, gerbil and rat models of ischemia, and following focal or global ischemia (Dirnagl and others, 2003). Ischemic tolerance has been modeled in vitro using oxygen glucose deprivation (OGD), hypoxia (low oxygen) chemical ischemia with metabolic inhibitors and glutamate/ N-Methyl D aspartate (NMDA) agonists, suggesting that the ischemic tolerance phenomenon is not just a blood reperfusion effect (Chen and Simon 1997, Meller and others, 2005). However, the ability of preconditioning in one body region to induce ischemic tolerance in another (remote preconditioning) suggests that in the whole body situation a vascular preconditioning mediator is released, which can induce tolerance in a different tissue (Ren and others, 2008).
Evidence from prospective and retrospective studies suggests a potential role of ischemic tolerance in humans (Schaller 2005, Schaller and Graf 2002, Wegener and others, 2004, Weih and others, 1999). Patients with prior transient ischemic attacks to the same territory of the brain affected by a subsequent stroke, have a reduced infarct volume and severity of symptoms. Hence, endogenous ischemic tolerance in the human brain exists.
There are two accepted time windows for ischemic tolerance following the preconditioning stimuli. A rapid time window has been shown to exist 30 minutes-1 hour following preconditioning. In contrast delayed tolerance is induced 24 hours following preconditioning, and can last up to 3 days. Although intermediate time windows have been reported (Ren and others, 2008). The difference in time windows of the two types of tolerance gives clues as to the biological mechanism of each. Delayed tolerance is mediated via new protein synthesis and a change in the genomic response to ischemia. The protein synthesis inhibitor cycloheximide blocks delayed tolerance in both in vivo and in vitro models (Barone and others, 1998, Meller and others, 2006, Meller and others, 2005). In contrast, cycloheximide did not block an in vitro rapid ischemic tolerance model in rat cortical neurons (Meller and others, 2006). Many studies of ischemic tolerance have focused on the gene based, de-novo protein synthesis dependent long-term/ delayed ischemic tolerance, for more detailed reviews (Chen and Simon 1997, Dirnagl and others, 2003, Obrenovitch 2008, Steiger and Hanggi 2007).
Studies of the molecular mechanism of delayed ischemic tolerance have identified a number of important genes whose neuroprotective effects may mediate tolerance to harmful ischemia, including Bcl-2, Bcl-w, heat shock proteins, Proliferating Cell Nuclear Antigen, glutamate transport proteins EAAT2/ 3 and erythropoietin (Bossenmeyer-Pourie and Daval 1998, Currie and others, 2000, McLaughlin and others, 2003, Minami and others, 2000, Ruscher and others, 2002). More recently it has been proposed that a preconditioning event results in a re-programming of the genomic response of the brain to subsequent harmful ischemia, and has similarities to the natural protective state of hibernation (Stenzel-Poore and others, 2007, Stenzel-Poore and others, 2004, Stenzel-Poore and others, 2003). Gene-based and metabolic-based mechanisms of endogenous tolerance to ischemia are observed in a number of divergent species and scenarios including mammal hibernation, deep sea turtles, arctic tree squirrels, fish and beetle larvae (Frerichs and Hallenbeck 1998, Frerichs and others, 1994, Hoback and others, 2000, Lee and others, 2006, Podrabsky and others, 2007, Storey 2003, Storey 2007).
Rapid ischemic tolerance in animal and cell culture models of ischemia
In contrast to delayed ischemic tolerance, rapid ischemic tolerance has been less extensively studied. Rapid ischemic tolerance has been described in the heart (Eisen and others, 2004) and also occurs in brain. However, less is known about the molecular mechanisms by which rapid ischemic tolerance induces its neuroprotective effects in brain, compared to delayed tolerance. Indeed approximately 20 studies directly investigate the molecular mechanisms of rapid tolerance induced neuroprotection.
The first report of rapid tolerance to ischemia in the brain was by Schurr and others in 1986. They showed that prior exposure of rat hippocampal slices to anoxia 30 minutes prior to harmful ischemia (anoxia) maintained electrical responsiveness of the cells (Schurr and others, 1986). Rapid ischemic tolerance has been observed in primary cortical neuronal cultures, organotypic preparations or slices of hippocampus using anoxia, oxygen and glucose deprivation, NMDA and pharmacological preconditioning stimuli (Bandyopadhyay and others, 2002, Meller and others, 2006, Perez-Pinzon and Born 1999, Reshef and others, 1996, Schurr and others, 1986). Typically most studies use a one hour interval between preconditioning and harmful ischemia, although protection is also observed within a 30 min window (Perez-Pinzon and others, 1997). Rapid ischemic tolerance in vitro protects against oxygen glucose deprivation, chemical induced ischemia (iodoacetate), anoxia and NMDA excitotoxicity (Meller and others, 2008, Reshef and others, 1996)
Rapid ischemic tolerance has also been described in animal models of ischemia. The protection one observes depends on the duration of reperfusion following the second, harmful ischemia and the severity of the second ischemic insult. For example, rapid ischemic tolerance, using 3 × 5 minute periods of middle cerebral artery (MCA) occlusion in mice, 30 min prior to 60 min MCA occlusion (MCAO) or permanent MCAO, reduced infarction 24 hours following the final ischemic insult (Stagliano and others, 1999). In a global model of ischemia in rats, protection induced by 2 minutes bi-lateral carotid occlusion followed by 30 minutes of reperfusion resulted in protection to a 10 minute occlusion when assessed 3 but not 7 days later (Perez-Pinzon and others, 1997). Other studies have reported rapid ischemic tolerance in focal models of ischemia (Atochin and others, 2003, Nakamura and others, 2002). Preconditioning animals with 30 min MCAO resulted in protection from 180 min MCAO one hour later, and the protection was evident 7 day following the last ischemic insult. We observed similar protection in a focal model of rapid ischemic tolerance when we paired 30 min MCAO with a 60 min reperfusion followed by a 100 min harmful MCAO (Minami and others, 2003).
The concept of remote preconditioning to induce ischemic tolerance has been investigated (Ren and others, 2008). The authors showed that three cycles of occlusion of the femoral artery (left) resulted in protection from ischemia induced by permanent occlusion of the left distal MCA combined with a 30 min occlusion of the bilateral common carotid arteries. This suggests that rapid ischemic tolerance can be induced remotely, which has interesting clinical implications. Furthermore this suggests that endogenous rapid ischemic tolerance in the whole animal is induced by the release of a mediator into the blood stream, as well as occurring in the tissue.
A number of mechanisms have been investigated with respect to rapid ischemic tolerance. Many studies have described a role of adenosine receptors in mediating tolerance induced by anoxia, isoflurane and adenosine receptor agonists (Liu and others, 2006, Perez-Pinzon and others, 1996, Reshef and others, 1996). In addition in vitro and in vivo rapid ischemic tolerance is blocked by the adenosine A1 receptor antagonists 8-Cyclopentyl-1,3-dipropylxanthine (DCPCX) (Nakamura and others, 2002, Perez-Pinzon and others, 1996). The source of the adenosine which is mediating these effects following the brief ischemic event is not yet clear, and potential mechanisms could include excytotoxic release of adenosine, transporter-mediated release or metabolism of ATP into adenosine (Parkinson and others, 2002). Other neurotransmitter receptors which may regulate rapid ischemic tolerance include the delta opioid receptor. Rapid ischemic tolerance to glutamate excitoitoxicity following hypoxic preconditioning is blocked by the delta opioid receptor antagonist naltrindole (Zhang and others, 2006a). The mechanisms which regulate delta opioid induced tolerance are not yet clear.
Rapid ischemic tolerance induced by preconditioning with anoxia and the metabolic poison iodo-acetate can be blocked by the inhibitor of the ATP-sensitive potassium (KATP) channel glibenclamide. The effect of adenosine in mediating protection against iodoacetate is also blocked by glibenclamide. A rapid tolerant state in neurons can be induced by incubation for 30–60 min with the KATP channel openers chromakalin, diazoxide and pinacidil (Perez-Pinzon and Born 1999, Reshef and others, 1998). Taken together, these data suggest that the activation of kATP channels following preconditioning via the increased adenosine A1 receptor activation plays a role in rapid ischemic tolerance.
The ubiquitin proteasome system and rapid ischemic tolerance
The most obvious difference between delayed and rapid ischemic tolerance is the time window for effect. As the name implies, rapid ischemic tolerance occurs quickly following the preconditioning stimuli and is not blocked by the protein synthesis inhibitor cycloheximide (1 µM) (Meller and others, 2006). This suggests that the neuroprotective mechanism of rapid tolerance occurs independently of new protein synthesis. Therefore, the protective mechanism of rapid ischemic tolerance would be predicted to result in the rapid post-translational modification of insitu proteins to mediate the neuroprotective event.
Consistent with this hypothesis, potential role for the ubiquitin proteasome system in regulating rapid ischemic tolerance both in brain and in heart was recently described (Cai and others, 2008, Meller and others, 2006, Meller and others, 2008). Rapid ischemic tolerance is blocked by multiple proteasome inhibitors (clasto-lactacystin β-lactone, MG132 (Z-Leu-Leu-Leu-CHO) and MG115 (Z-Leu-Leu-Nva-H), suggesting that following preconditioning ischemia proteins are degraded resulting in a neuroprotective phenotype (Meller and others, 2006, Meller and others, 2008). This concept of down-regulation or depletion of proteins to generate a protective phenotype has been suggested by others; gene transcriptional down-regulation is a common phenotype of delayed ischemic tolerance (Stenzel-Poore and others, 2003). In addition ischemic tolerance in a cardiac protection model was recently shown to be blocked in a transgenic mouse deficient for low molecular mass polypeptide 2 (LMP-2) (a subunit of the immunoproteasome). This mechanism of protection was attributed to activation of AKT, due to degradation of the repressor PTEN (phosphatase and tensin homologue deleted on chromosome 10) (Cai and others, 2008).
Ubiquitination of the pro-cell death protein Bim following preconditioning ischemia
The first protein shown to be ubiquitinated in a model of rapid ischemic tolerance was the pro-cell death member of the Bcl-2 protein family Bim. The balance between pro-survival and pro-apoptotic Bcl-2 family proteins regulate the initiation of intrinsic cell death signaling upstream of the mitochondria. Multiple Bcl-2 family proteins are regulated by the ubiquitin-proteasome system (Basu and Haldar 2002, Breitschopf and others, 2000a, Breitschopf and others, 2000b, Li and Dou 2000). Bim, a pro-cell death member of the bcl-2 family is a member of the BH3 only subgroup of proteins (O'Connor and others, 1998). Bim is commonly expressed as BimEL (a 23–26 kDa protein) but splice variants result in a truncated BimL or BimS, which are also potent cell death inducers (O'Connor and others, 1998). Additional splice variants have been identified in humans, but their function is not yet clear (Miyashita and others, 2001). Bim can induce apoptosis via the direct activation of Bax/ Bak, where as other BH3 only containing Bcl-2 proteins result in sensitization alone (Kuwana and others, 2005). Bim induction has been shown to be essential for neuronal cell death (Putcha and others, 2001), whereas reducing Bim protein levels reduces cell death in neurons (Strasser and others, 2000) and renders them resilient to harmful ischemia (Meller and others, 2006).
Our studies show that only Bim, but not Bax or Bid protein levels are rapidly decreased following preconditioning ischemia, and that this decrease is associated with an increase in ubiquitination of Bim (Meller and others, 2006). The effect on Bim was transient; ischemic tolerance is lost 4 hours following preconditioning coincident with Bim levels returning to normal (Meller and others, 2006). The decrease in Bim was blocked by the proteasome inhibitor MG132, which is consistent with preconditioning ischemia inducing Bim ubiquitination and proteasomal degradation that renders neurons protected against the harmful effects of ischemia.
Previous studies in MEF cells show that Bim ubiquitination is dependent on its phosphorylation of a Ser65 residue by p42/p44 MAPK (Ley and others, 2003, Ley and others, 2005). Furthermore, blocking p42/p44 MAPK activation blocks ischemic tolerance (Meller and others, 2006). Consistent with Bim ubiquitination playing a role in ischemic tolerance, blocking p42/p44 MAPK also prevents Bim ubiquitination and degradation (Fig 4). Given the promiscuous nature of p42/p44 MAPK being involved in many cell functions, it is reasonable to assume that this is not the only target for MAPK mediated phosphorylation following preconditioning ischemia.
What has yet to be identified is the E3-ligase responsible for ubiquitinating phosphorylated Bim following preconditioning ischemia. It has been suggested that the tyrosine kinase c-Cbl is a Bim E3-ligase in osteoclasts, (Akiyama and others, 2003). However, other studies failed to show an increase in Bim protein levels in the testis of c-Cbl knockout mice, which would be expected if the E3-ligase is absent (El Chami and others, 2005). Further evidence against a role of c-Cbl come from studies which show the biology of the interaction between c-Cbl and Bim does not match the known biology of Bim ubiquitination (Wiggins and others, 2007). For example, Bim ubiquitination is not affected by mutation of tyrosine residues on Bim, whereas mutation of serine residues (Ser 69) prevents Bim ubiquitination. Bim failed to interact with c-Cbl under conditions where Bim ubiquitination increased, Bim stability was not affected by knockout of c-Cbl in MEF cells, and finally p42/p44 MAPK-mediated Bim ubiquitination occurs in fibroblasts obtained from c-Cbl knockout mice. Recently the E3 ligase CIS has been suggested as a Bim E3-ligase (Zhang and others, 2008). The authors show how CIS is associated with the microtubule scaffold in cells, and that microtubule de-polymerizing agents release Bim and enable its association with CIS, resulting in its ubiquitination. However the authors did not investigate the regulation of this system by p42/p44 MAPK. This leads to an intriguing situation: what else is ubiquitinated following preconditioning ischemia? Is Bim ubiquitinated by multiple E3-ligases? And do different E3-ligases interact with Bim under different biological conditions?
Proteomic analysis of ubiquitinated proteins in rapid ischemic tolerance
We recently employed a non-biased proteomic approach to screen for ubiquitinated proteins in rapid ischemic tolerance (Meller and others, 2008). The agarose bound UBD of p62 was used to enrich ubiquitinated proteins from preconditioned tissue, which were subsequently identified by mass spectrometry. Our results show a higher pulldown/ identification of proteins following preconditioning ischemia compared to control. Some proteins were identified in both control and preconditioned tissue, and unfortunately the absolute levels could not be determined. It was not determined whether these proteins are regulated by the same or different E3-ligases. Whether these proteins are Lys63 or Lys48 liked is not clear. However, further analysis showed that the degradation of two candidate proteins (Marcks and fascin) was inhibited by the proteasome inhibitor MG132. This data would suggest that the ubiquitination of these proteins resulted in their proteasomal degradation.
Bioinformatic analysis of the proteins identified by our proteomic approach revealed an interesting function of the identified proteins, 16 out of 25 of the proteins had been identified to interact either directly or indirectly with proteins associated with the post synaptic density. This strongly suggested that the ubiquitin proteasome could be changing synaptic function, which may be associated with protection.
Actin is a major cytoskeletal component of the post synaptic density, so we focused our investigations on two actin binding proteins, fascin and MARCKS (Meller and others, 2008). In order to maintain the shape and function of the spine, actin forms filaments which are cross-linked by other proteins to provide strength, and one such protein is fascin. Fascin regulates shape and trajectory of neurites in drosophila neurons (Kraft and others, 2006). Actin does not directly anchor to the plasma membrane, but contacts intermediate proteins such as myristoylated, alanine-rich C-kinase substrate (MARCKS) (Hartwig and others, 1992, Sundaram and others, 2004). Interestingly, NMDA induces degradation of MARCKS but via the proteases cathepsin and calpain rather than the proteasome (Graber and others, 2004). Following preconditioning ischemia, proteasome-mediated degradation of MARCKS and fascin occurs, resulting in a decrease in the interaction of these proteins with actin (Meller and others, 2008).
In our study we show the proteasome dependent degradation of two actin binding proteins following preconditioning ischemia, which is associated with the solublization of actin filaments, and the loss of dendritic spines. The loss of spines was blocked by jasplakinolide and the proteasome inhibitor MG132, suggesting that the ubiquitination of proteins and actin depolymerization following preconditioning results in structural reorganization of the neuron (Meller and others, 2008). Since ischemic tolerance induced protection was also blocked by jasplakinolide and MG132, this suggests that these structural changes are critical for rapid ischemic tolerance (Meller and others, 2008).
The concept that ischemia can remodel the synapse is not new. Indeed in a series of studies Mark Goldberg’s group showed that brief ischemia results in dendritic spine loss, where as longer ischemia results in permanent spine loss (Faddis and others, 1997, Hasbani and others, 2001a, Hasbani and others, 2001b). Dendritic spine loss following ischemia has also been reported following focal ischemia in vivo (Zhang and others, 2005). Dendritic spine loss is recoverable if the exposure to ischemia is transient and non-harmful, but is persistent when toxic levels of ischemia occur (Park and others, 1996, Zhang and others, 2005). Spine numbers recover approximately 2–4 hours following ischemia (Meller and others, 2008, Park and others, 1996). Interestingly, the recovery of the spines occurs at the same location on the dendrites that the spines were originally lost, both in vitro (Hasbani and others, 2001a) and in vivo (Zhang and others, 2005), suggesting a post synaptic remodeling event.
The ubiquitin proteasome system has also been implicated in synaptic reorganization in response to neuronal activity and in LTP (Dong and others, 2008, Ehlers 2003, Fonseca and others, 2006). The dendritic spine is the major site of post synaptic input to a neuron. The post synaptic density, a region of dense cellular material adjacent to the synapse, is rich in proteasome subunits and ubiquitin (Chapman and others, 1992). The development of synapses in a number of species is regulated by the ubiquitin proteasome system (Yi and Ehlers 2007). In addition, proteasome inhibitors can induce neurite outgrowth in PC12 cells treated with NGF (Maufroid and others, 1996), but inhibition of N-end ubiquitination result in reduced neurite outgrowth (Obin and others, 1999).
When neurons are activated, proteasome sub units enter the dendritic spine and are associated with the actin cytoskeleton (Bingol and Schuman 2006). Hence the proteasome is positioned to regulate (degrade) cytoskeleton proteins as reported following both ischemia and neuronal activation (Ehlers 2003, Meller and others, 2008). Activation of neurons resulted in remodeling of the synapse by the enhanced and decrease degradation of post synaptic associated proteins. The proteins regulated included neurotransmitter receptors and structural proteins (Ehlers 2003). These structural changes resulted in long lasting enhancement of NMDA mediated CREB signaling. Phosphorylation of the transcription factor CREB is associated with protection and survival in neurons following ischemia (Walton and others, 1999).
Stimulation of neurons can also change the receptor composition of the post synaptic density. PSD-95 levels decrease reducing AMPA receptor subunit levels at the membrane surface and endocytosis of GluR1 subunits (Bingol and Schuman 2004, Colledge and others, 2003). Further studies show that PSD-95 is ubiquitinated by MDM2 (which is normally associated with p53 regulation) (Colledge and others, 2003). In contrast activation of a neuron reduces GABA receptor ubiquitination enhancing their numbers at the membrane surface (Saliba and others, 2007) (in unpublished studies we have observed enhance GABA currents following preconditioning ischemia). Interestingly it has recently been shown that nicotine can inhibit proteasome function, resulting in the accumulation of ubiquitinated proteins (Rezvani and others, 2007). Nicotine had a direct binding ability with the proteasome, which may have many implications given the high social use of this drug.
These studies would suggest that, perhaps not surprisingly, one of the major consequences of ischemia induced protein ubiquitination and degradation is a change in synaptic function (Fig 6). Our studies have confirmed this hypothesis, in that we observe a reduction in NMDA currents and toxic NMDA signaling following preconditioning in a rapid ischemic tolerance model (Meller and others, 2008). The reduction in NMDA excitotoxicity following preconditioning was blocked by jasplakinolide and MG132, which suggests that the same mechanisms which regulate tolerance to ischemia, also result in tolerance to toxic levels of NMDA.
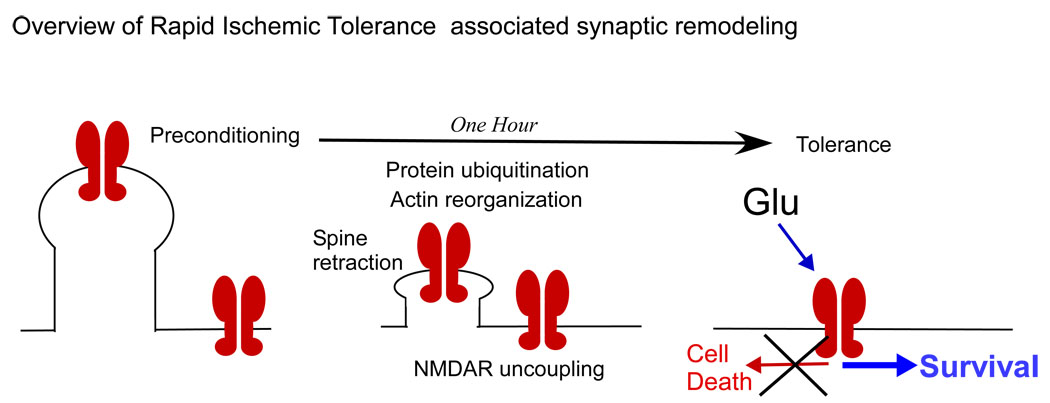
Figure 6.
Overview of preconditioning ischemia mediated morphological re-organization in dendrite and alterations in NMDA receptor mediated signaling. Following preconditioning ischemia, protein degradation and actin re-organization result in uncoupling of the NMDA receptor complexes from the actin cytoskeleton. The morphological and cytoskeleton changes results in an uncoupling of the toxic NMDA receptor mediated signaling but not physiological NMDA receptor function. As a result NMDA mediated excitotoxicity is reduced following preconditioning ischemia. This result may help identify how to switch off harmful NMDA receptor mediated signaling.
A loss of synaptic NMDA receptors has been shown to result in tolerance to OGD (Sattler and others, 2000) and disrupting post synaptic NMDA receptor-PSD-95 interactions also result in reduced toxicity following ischemia (Sattler and others, 1999). Waataja recently reported a study which correlates with our own observations, in that NMDA receptor activation resulted in PSD-95 degradation by the proteasome, and a loss in dendritic spines (Waataja and others, 2008). These changes were blocked by the proteasome inhibitor MG132 and over expression of p14ARF (which regulates MDM2 stability). Furthermore a loss of spines following NMDA exposure resulted in a loss of EPSPs (excitatory post synaptic potentials), which suggests that synaptic NMDA receptors were blocked or absent (Waataja and others, 2008). Glutamate toxicity in these cells was not mediated by the proteasome, similar to OGD mediated cell death in our experiments (Meller and others, 2008). However, toxicity to glutamate was enhanced in ARF overexpressing cells (Waataja and others, 2008). Furthermore the glutamate induced loss of spines was similar in time course to the ischemia induced loss of spines, complete by one hour and recovering to control levels by 3–4 hours (Hasbani and others, 2001a, Meller and others, 2008). The study by Waataja did not study whether the glutamate response or excitoxicity was reduced in cells pretreated with glutamate (Waataja and others, 2008). Taken together, these two studies suggest that proteasome dependent removal of dendritic spines occurs following a preconditioning ischemia and that this is a protective event.
Translating the promise:- future targets for research
Following ischemia the pattern of activation of intracellular mechanisms denotes the potential therapeutic time windows to a given strategy. Ischemia induced cell death is a progressive event and requires 24–48 hours to fully develop (Dirnagl and others, 1999) (Fig 7). Studies on the role of the ubiquitin proteasome system in ischemia and rapid ischemic tolerance suggest two potential therapeutic strategies (Fig 7). Based on studies of rapid ischemic tolerance, acute neuroprotection may be an obtainable therapeutic goal. By further understanding the mechanism by which cell death mediating proteins are rapidly degraded by the ubiquitin proteasome system, one could devise therapies to stimulate their degradation. Bim degradation is rapid, occuring one hour following preconditioning. Given the established role of the Bcl-2 family of proteins in mediating cell death following ischemia and other brain injuries, a reduction in Bim may result in neuroprotection (Meller and others, 2006, Shinoda and others, 2004).
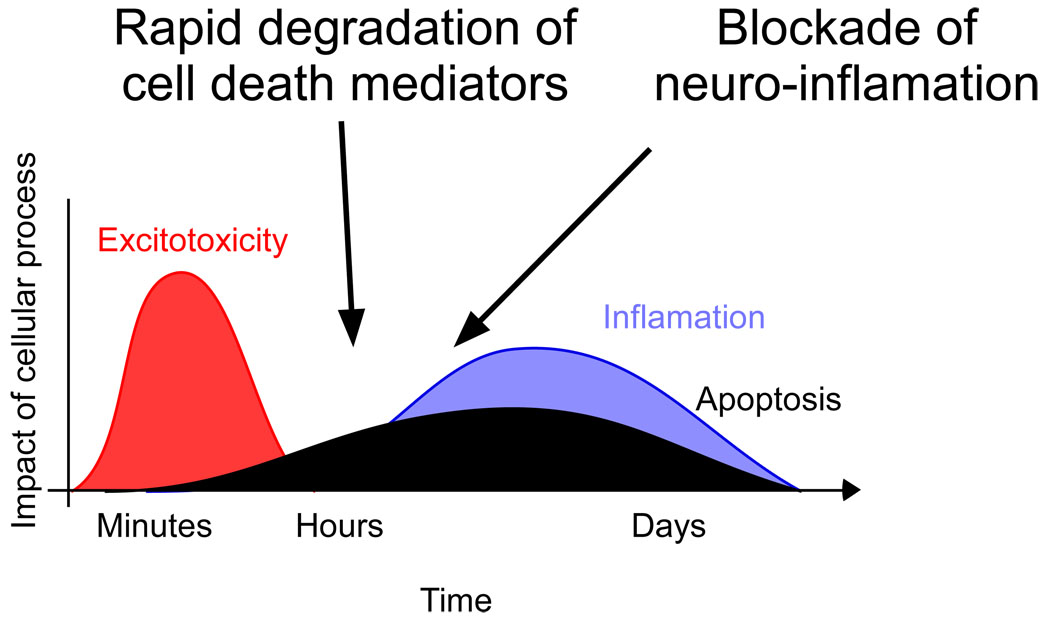
Figure 7.
Potential therapeutic sites of intervention based on ubiquitin proteasome mediated protection. Initial therapeutic strategies may target proteins whose rapid degradation is identified as being neuroprotective in rapid ischemic tolerance models. Delayed cell death has also been attributed to neuro-inflammation processes following ischemia. Proteasome inhibitors have shown to provide delayed therapeutic effects in reducing infarction and behavioral deficits by reducing neuro-inflammation. A combined therapeutic strategy targeting multiple processes which contribute to cell death following ischemia may effectively reduce brain injury following stroke. Time course figure adapted from (Dirnagl and others, 1999).
A second phase of therapy would involve the reduction in secondary brain injury and neuro-inflammation. These have been shown to be reduced by proteasome inhibitors. While acute therapy with these agents may be tolerated, proteasome inhibitors may aggravate part of the response to ischemia, by inducing accumulation of ubiquitinated proteins that must then be cleared by autophagic processes. By further understanding the molecular mechanisms that regulate neuro-inflammation a more specific target therapy may be devised. Indeed, the ubiquitin proteasome system contains many elements, not only the E1, E2 and E3 ligases, but additional modifying factors each of which are potential targets for molecular intervention. Furthermore knowledge of the mechanisms which regulate these processes may be transferable to other neurological diseases of which cell death is a feature. This remains an exciting time for ubiquitin research and therapies based on this versatile signaling system are no doubt being developed.
Acknowledgements
We wish to thank the NIH/ NINDS (NS050669 & NS054023 Meller, NS024728 & NS039016 Simon), the American Heart Association (0465430Z Meller) and the Legacy Good Samaritan Foundation for funding. The author would like to acknowledge the assistance of Simon Thompson PhD, Theresa Lusardi PhD, Liam Loftus PhD, Manabu Minami MD, Tao Yang, Andrea Ordonez, Veronica Jessick, Michelle Ashley, Daniel Torrey, Corrin Clayton and Jennifer Cameron with these studies. The author would like to thank Drs R P Simon, D. Henshall, Z. Xiong and J. Saugstad for their assistance pertaining to this project.
References
- Adhami F, Liao G, Morozov YM, Schloemer A, Schmithorst VJ, Lorenz JN, Dunn RS, Vorhees CV, Wills-Karp M, Degen JL, Davis RJ, Mizushima N, Rakic P, Dardzinski BJ, Holland SK, Sharp FR, Kuan CY. Cerebral ischemia-hypoxia induces intravascular coagulation and autophagy. Am J Pathol. 2006;169:566–583. doi: 10.2353/ajpath.2006.051066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed SH, He YY, Nassief A, Xu J, Xu XM, Hsu CY, Faraci FM. Effects of lipopolysaccharide priming on acute ischemic brain injury. Stroke. 2000;31:193–199. doi: 10.1161/01.str.31.1.193. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI, Fukuda A, Hikita A, Seto H, Okada T, Inaba T, Sanjay A, Baron R, Kawaguchi H, Oda H, Nakamura K, Strasser A, Tanaka S. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. Embo J. 2003;22:6653–6664. doi: 10.1093/emboj/cdg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atochin DN, Clark J, Demchenko IT, Moskowitz MA, Huang PL. Rapid cerebral ischemic preconditioning in mice deficient in endothelial and neuronal nitric oxide synthases. Stroke. 2003;34:1299–1303. doi: 10.1161/01.STR.0000066870.70976.57. [DOI] [PubMed] [Google Scholar]
- Bae SH, Jeong JW, Park JA, Kim SH, Bae MK, Choi SJ, Kim KW. Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochemical and Biophysical Research Communications. 2004;324:394–400. doi: 10.1016/j.bbrc.2004.09.068. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Johnson L, Chung W, Thakor NV. Protection by rapid chemical preconditioning of stressed hippocampal slice: a study of cellular swelling using optical scatter imaging. Brain Res. 2002;945:79–87. doi: 10.1016/s0006-8993(02)02693-8. [DOI] [PubMed] [Google Scholar]
- Barone FC, White RF, Spera PA, Ellison J, Currie RW, Wang X, Feuerstein GZ. Ischemic preconditioning and brain tolerance: temporal histological and functional outcomes, protein synthesis requirement, and interleukin-1 receptor antagonist and early gene expression. Stroke. 1998;29:1937–1950. doi: 10.1161/01.str.29.9.1937. discussion 1950-1. [DOI] [PubMed] [Google Scholar]
- Basu A, Haldar S. Signal-induced site specific phosphorylation targets Bcl2 to the proteasome pathway. Int J Oncol. 2002;21:597–601. [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. A proteasome-sensitive connection between PSD-95 and GluR1 endocytosis. Neuropharmacology. 2004;47:755–763. doi: 10.1016/j.neuropharm.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- Bossenmeyer-Pourie C, Daval JL. Prevention from hypoxia-induced apoptosis by pre-conditioning: a mechanistic approach in cultured neurons from fetal rat forebrain. Brain Res Mol Brain Res. 1998;58:237–239. doi: 10.1016/s0169-328x(98)00123-5. [DOI] [PubMed] [Google Scholar]
- Breitschopf K, Haendeler J, Malchow P, Zeiher AM, Dimmeler S. Posttranslational modification of Bcl-2 facilitates its proteasome- dependent degradation: molecular characterization of the involved signaling pathway. Mol Cell Biol. 2000a;20:1886–1896. doi: 10.1128/mcb.20.5.1886-1896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschopf K, Zeiher AM, Dimmeler S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J Biol Chem. 2000b;275:21648–21652. doi: 10.1074/jbc.M001083200. [DOI] [PubMed] [Google Scholar]
- Cai ZP, Shen Z, Van Kaer L, Becker LC. Ischemic preconditioning-induced cardioprotection is lost in mice with immunoproteasome subunit low molecular mass polypeptide-2 deficiency. Faseb J. 2008 doi: 10.1096/fj.08-105940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carloni S, Buonocore G, Balduini W. Protective role of autophagy in neonatal hypoxia-ischemia induced brain injury. Neurobiol Dis. 2008 doi: 10.1016/j.nbd.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Carlucci A, Adornetto A, Scorziello A, Viggiano D, Foca M, Cuomo O, Annunziato L, Gottesman M, Feliciello A. Proteolysis of AKAP121 regulates mitochondrial activity during cellular hypoxia and brain ischaemia. Embo J. 2008;27:1073–1084. doi: 10.1038/emboj.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvorachote P, Nimmannit U, Stehlik C, Wang L, Jiang BH, Ongpipatanakul B, Rojanasakul Y. Nitric oxide regulates cell sensitivity to cisplatin-induced apoptosis through S-nitrosylation and inhibition of Bcl-2 ubiquitination. Cancer Research. 2006;66:6353–6360. doi: 10.1158/0008-5472.CAN-05-4533. [DOI] [PubMed] [Google Scholar]
- Chapman AP, Courtney SC, Smith SJ, Rider CC, Beesley PW. Ubiquitin immunoreactivity of multiple polypeptides in rat brain synaptic membranes. Biochem Soc Trans. 1992;20:155S. doi: 10.1042/bst020155s. [DOI] [PubMed] [Google Scholar]
- Chen J, Simon R. Ischemic tolerance in the brain. Neurology. 1997;48:306–311. doi: 10.1212/wnl.48.2.306. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- Currie RW, Ellison JA, White RF, Feuerstein GZ, Wang X, Barone FC. Benign focal ischemic preconditioning induces neuronal Hsp70 and prolonged astrogliosis with expression of Hsp27. Brain Res. 2000;863:169–181. doi: 10.1016/s0006-8993(00)02133-8. [DOI] [PubMed] [Google Scholar]
- Dirnagl U, Iadecola C, Moskowitz MA. Pathobiology of ischaemic stroke: an integrated view. Trends Neurosci. 1999;22:391–397. doi: 10.1016/s0166-2236(99)01401-0. [DOI] [PubMed] [Google Scholar]
- Dirnagl U, Simon RP, Hallenbeck JM. Ischemic tolerance and endogenous neuroprotection. Trends Neurosci. 2003;26:248–254. doi: 10.1016/S0166-2236(03)00071-7. [DOI] [PubMed] [Google Scholar]
- Dong C, Upadhya SC, Ding L, Smith TK, Hegde AN. Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn Mem. 2008;15:335–347. doi: 10.1101/lm.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Eisen A, Fisman EZ, Rubenfire M, Freimark D, McKechnie R, Tenenbaum A, Motro M, Adler Y. Ischemic preconditioning: nearly two decades of research. A comprehensive review. Atherosclerosis. 2004;172:201–210. doi: 10.1016/S0021-9150(03)00238-7. [DOI] [PubMed] [Google Scholar]
- El Chami N, Ikhlef F, Kaszas K, Yakoub S, Tabone E, Siddeek B, Cunha S, Beaudoin C, Morel L, Benahmed M, Regnier DC. Androgen-dependent apoptosis in male germ cells is regulated through the proto-oncoprotein Cbl. J Cell Biol. 2005;171:651–661. doi: 10.1083/jcb.200507076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faddis BT, Hasbani MJ, Goldberg MP. Calpain activation contributes to dendritic remodeling after brief excitotoxic injury in vitro. J Neurosci. 1997;17:951–959. doi: 10.1523/JNEUROSCI.17-03-00951.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Ciechanover A, Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984;37:43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52:239–245. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Frerichs KU, Hallenbeck JM. Hibernation in ground squirrels induces state and species-specific tolerance to hypoxia and aglycemia: an in vitro study in hippocampal slices. J Cereb Blood Flow Metab. 1998;18:168–175. doi: 10.1097/00004647-199802000-00007. [DOI] [PubMed] [Google Scholar]
- Frerichs KU, Kennedy C, Sokoloff L, Hallenbeck JM. Local cerebral blood flow during hibernation, a model of natural tolerance to "cerebral ischemia". J Cereb Blood Flow Metab. 1994;14:193–205. doi: 10.1038/jcbfm.1994.26. [DOI] [PubMed] [Google Scholar]
- Gallagher SJ, Kefford RF, Rizos H. The ARF tumour suppressor. Int J Biochem Cell Biol. 2006;38:1637–1641. doi: 10.1016/j.biocel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Graber S, Maiti S, Halpain S. Cathepsin B-like proteolysis and MARCKS degradation in sub-lethal NMDA-induced collapse of dendritic spines. Neuropharmacology. 2004;47:706–713. doi: 10.1016/j.neuropharm.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- Harada T, Harada C, Wang YL, Osaka H, Amanai K, Tanaka K, Takizawa S, Setsuie R, Sakurai M, Sato Y, Noda M, Wada K. Role of ubiquitin carboxy terminal hydrolase-L1 in neural cell apoptosis induced by ischemic retinal injury in vivo. Am J Pathol. 2004;164:59–64. doi: 10.1016/S0002-9440(10)63096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig JH, Thelen M, Rosen A, Janmey PA, Nairn AC, Aderem A. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature. 1992;356:618–622. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- Hasbani MJ, Schlief ML, Fisher DA, Goldberg MP. Dendritic spines lost during glutamate receptor activation reemerge at original sites of synaptic contact. J Neurosci. 2001a;21:2393–2403. doi: 10.1523/JNEUROSCI.21-07-02393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasbani MJ, Viquez NM, Goldberg MP. NMDA receptors mediate hypoxic spine loss in cultured neurons. Neuroreport. 2001b;12:2731–2735. doi: 10.1097/00001756-200108280-00028. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. Embo J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninger N, Sicard KM, Bouley J, Fisher M, Stagliano NE. The proteasome inhibitor VELCADE reduces infarction in rat models of focal cerebral ischemia. Neurosci Lett. 2006;398:300–305. doi: 10.1016/j.neulet.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Hershko A. Ubiquitin: roles in protein modification and breakdown. Cell. 1983;34:11–12. doi: 10.1016/0092-8674(83)90131-9. [DOI] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hoback WW, Podrabsky JE, Higley LG, Stanley DW, Hand SC. Anoxia tolerance of con-familial tiger beetle larvae is associated with differences in energy flow and anaerobiosis. J Comp Physiol [B] 2000;170:307–314. doi: 10.1007/s003600000104. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Hoppe T. Multiubiquitylation by E4 enzymes: 'one size' doesn't fit all. Trends Biochem Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Kerppola TK. Lysosomal Localization of Ubiquitinated Jun Requires Multiple Determinants in a Lysine-27 Linked Polyubiquitin Conjugate. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-05-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Janoff A. Alterations in Lysosomes (Intracellular Enzymes) During Shock; Effects of Preconditioning (Tolerance) and Protective Drugs. Int Anesthesiol Clin. 1964;2:251–269. doi: 10.1097/00004311-196402000-00008. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Matsumoto M, Tagaya M, Hata R, Ueda H, Niinobe M, Handa N, Fukunaga R, Kimura K, Mikoshiba K, et al. 'Ischemic tolerance' phenomenon found in the brain. Brain Res. 1990;528:21–24. doi: 10.1016/0006-8993(90)90189-i. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Harris VA, Welsh FA. Spreading depression induces tolerance of cortical neurons to ischemia in rat brain. J Cereb Blood Flow Metab. 1995;15:721–727. doi: 10.1038/jcbfm.1995.92. [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Koike M, Shibata M, Tadakoshi M, Gotoh K, Komatsu M, Waguri S, Kawahara N, Kuida K, Nagata S, Kominami E, Tanaka K, Uchiyama Y. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am J Pathol. 2008;172:454–469. doi: 10.2353/ajpath.2008.070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R, Escobar MM, Narro ML, Kurtis JL, Efrat A, Barnard K, Restifo LL. Phenotypes of Drosophila brain neurons in primary culture reveal a role for fascin in neurite shape and trajectory. J Neurosci. 2006;26:8734–8747. doi: 10.1523/JNEUROSCI.2106-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Lamirand A, Mercier G, Ramauge M, Pierre M, Courtin F. Hypoxia stabilizes type 2 deiodinase activity in rat astrocytes. Endocrinology. 2007;148:4745–4753. doi: 10.1210/en.2007-0625. [DOI] [PubMed] [Google Scholar]
- Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death and Differentiation. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Miyake SI, Wakita H, McMullen DC, Azuma Y, Auh S, Hallenbeck JM. Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J Cereb Blood Flow Metab. 2006 doi: 10.1038/sj.jcbfm.9600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 Signaling Pathway Promotes Phosphorylation and Proteasome-dependent Degradation of the BH3-only Protein, Bim. J Biol Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005;12:1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci U S A. 2000;97:3850–3855. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- Lim KL. Ubiquitin-proteasome system dysfunction in Parkinson's disease: current evidence and controversies. Expert Rev Proteomics. 2007;4:769–781. doi: 10.1586/14789450.4.6.769. [DOI] [PubMed] [Google Scholar]
- Liu C, Chen S, Kamme F, Hu BR. Ischemic preconditioning prevents protein aggregation after transient cerebral ischemia. Neuroscience. 2005a;134:69–80. doi: 10.1016/j.neuroscience.2005.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Ge P, Zhang F, Hu BR. Co-translational protein aggregation after transient cerebral ischemia. Neuroscience. 2005b;134:1273–1284. doi: 10.1016/j.neuroscience.2005.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Martone ME, Hu BR. Protein ubiquitination in postsynaptic densities after transient cerebral ischemia. Journal of Cerebral Blood Flow and Metabolism. 2004;24:1219–1225. doi: 10.1097/01.WCB.0000136706.77918.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xiong L, Chen S, Wang Q. Isoflurane tolerance against focal cerebral ischemia is attenuated by adenosine A1 receptor antagonists. Can J Anaesth. 2006;53:194–201. doi: 10.1007/BF03021827. [DOI] [PubMed] [Google Scholar]
- Lund PK, Moats-Staats BM, Simmons JG, Hoyt E, D’Ercole AJ, Martin F, Van Wyk JJ. Nucleotide sequence analysis of a cDNA encoding human ubiquitin reveals that ubiquitin is synthesized as a precursor. J Biol Chem. 1985;260:7609–7613. [PubMed] [Google Scholar]
- Mack TG, Reiner M, Beirowski B, Mi W, Emanuelli M, Wagner D, Thomson D, Gillingwater T, Court F, Conforti L, Fernando FS, Tarlton A, Andressen C, Addicks K, Magni G, Ribchester RR, Perry VH, Coleman MP. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat Neurosci. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO Journal. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Shamloo M, Matsumoto E, Isshiki A, Wieloch T. Protein kinase C-gamma and calcium/calmodulin-dependent protein kinase II-alpha are persistently translocated to cell membranes of the rat brain during and after middle cerebral artery occlusion. J Cereb Blood Flow Metab. 2004;24:54–61. doi: 10.1097/01.WCB.0000095920.70924.F5. [DOI] [PubMed] [Google Scholar]
- Maufroid JP, Bradshaw RA, Boilly B, Hondermarck H. Nerve growth factor induced neurite outgrowth from amphibian neuroepithelial precursor cells is prevented by dipeptides inhibiting ubiquitin-mediated proteolysis. Int J Dev Biol. 1996;40:609–611. [PubMed] [Google Scholar]
- McLaughlin B, Hartnett KA, Erhardt JA, Legos JJ, White RF, Barone FC, Aizenman E. Caspase 3 activation is essential for neuroprotection in preconditioning. Proc Natl Acad Sci U S A. 2003;100:715–720. doi: 10.1073/pnas.0232966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller R, Cameron JA, Torrey DJ, Clayton CE, Ordonez AN, Henshall DC, Minami M, Schindler CK, Saugstad JA, Simon RP. Rapid degradation of Bim by the ubiquitin-proteasome pathway mediates short-term ischemic tolerance in cultured neurons. J Biol Chem. 2006;281:7429–7436. doi: 10.1074/jbc.M512138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller R, Minami M, Cameron JA, Impey S, Chen D, Lan JQ, Henshall DC, Simon RP. CREB-mediated Bcl-2 protein expression after ischemic preconditioning. J Cereb Blood Flow Metab. 2005;25:234–246. doi: 10.1038/sj.jcbfm.9600024. [DOI] [PubMed] [Google Scholar]
- Meller R, Thompson SJ, Lusardi TA, Ordonez AN, Ashley MD, Jessick V, Wang W, Torrey DJ, Henshall DC, Gafken PR, Saugstad JA, Xiong ZG, Simon RP. Ubiquitin proteasome-mediated synaptic reorganization: a novel mechanism underlying rapid ischemic tolerance. J Neurosci. 2008;28:50–59. doi: 10.1523/JNEUROSCI.3474-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami M, Jin KL, Li W, Nagayama T, Henshall DC, Simon RP. Bcl-w expression is increased in brain regions affected by focal cerebral ischemia in the rat. Neurosci Lett. 2000;279:193–195. doi: 10.1016/s0304-3940(99)00987-8. [DOI] [PubMed] [Google Scholar]
- Minami M, Meller R, Henshall DC, Simon RP. Society for Neuroscience. New Orleans: 2003. The temporal profile of rapid and delayed tolerance to focal cerebral ischemia in mouse.. Program. [Google Scholar]
- Miyashita T, Shikama Y, Tadokoro K, Yamada M. Molecular cloning and characterization of six novel isoforms of human Bim, a member of the proapoptotic Bcl-2 family. FEBS Lett. 2001;509:135–141. doi: 10.1016/s0014-5793(01)03145-3. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Nakakimura K, Matsumoto M, Sakabe T. Rapid tolerance to focal cerebral ischemia in rats is attenuated by adenosine A1 receptor antagonist. J Cereb Blood Flow Metab. 2002;22:161–170. doi: 10.1097/00004647-200202000-00004. [DOI] [PubMed] [Google Scholar]
- O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, Huang DC. Bim: a novel member of the Bcl-2 family that promotes apoptosis. Embo J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obin M, Mesco E, Gong X, Haas AL, Joseph J, Taylor A. Neurite outgrowth in PC12 cells. Distinguishing the roles of ubiquitylation and ubiquitin-dependent proteolysis. J Biol Chem. 1999;274:11789–11795. doi: 10.1074/jbc.274.17.11789. [DOI] [PubMed] [Google Scholar]
- Obrenovitch TP. Molecular physiology of preconditioning-induced brain tolerance to ischemia. Physiol Rev. 2008;88:211–247. doi: 10.1152/physrev.00039.2006. [DOI] [PubMed] [Google Scholar]
- Oddo S. The ubiquitin-proteasome system in Alzheimer's disease. J Cell Mol Med. 2008;12:363–373. doi: 10.1111/j.1582-4934.2008.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski RZ. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 1999;6:303–313. doi: 10.1038/sj.cdd.4400505. [DOI] [PubMed] [Google Scholar]
- Ouyang YB, Xu L, Giffard RG. Geldanamycin treatment reduces delayed CA1 damage in mouse hippocampal organotypic cultures subjected to oxygen glucose deprivation. Neurosci Lett. 2005;380:229–233. doi: 10.1016/j.neulet.2005.01.055. [DOI] [PubMed] [Google Scholar]
- Park JS, Bateman MC, Goldberg MP. Rapid alterations in dendrite morphology during sublethal hypoxia or glutamate receptor activation. Neurobiol Dis. 1996;3:215–227. doi: 10.1006/nbdi.1996.0022. [DOI] [PubMed] [Google Scholar]
- Parkinson FE, Sinclair CJ, Othman T, Haughey NJ, Geiger JD. Differences between rat primary cortical neurons and astrocytes in purine release evoked by ischemic conditions. Neuropharmacology. 2002;43:836–846. doi: 10.1016/s0028-3908(02)00083-7. [DOI] [PubMed] [Google Scholar]
- Perez-Pinzon MA, Born JG. Rapid preconditioning neuroprotection following anoxia in hippocampal slices: role of the K+ ATP channel and protein kinase C. Neuroscience. 1999;89:453–459. doi: 10.1016/s0306-4522(98)00560-0. [DOI] [PubMed] [Google Scholar]
- Perez-Pinzon MA, Mumford PL, Rosenthal M, Sick TJ. Anoxic preconditioning in hippocampal slices: role of adenosine. Neuroscience. 1996;75:687–694. doi: 10.1016/0306-4522(96)00311-9. [DOI] [PubMed] [Google Scholar]
- Perez-Pinzon MA, Xu GP, Dietrich WD, Rosenthal M, Sick TJ. Rapid preconditioning protects rats against ischemic neuronal damage after 3 but not 7 days of reperfusion following global cerebral ischemia. J Cereb Blood Flow Metab. 1997;17:175–182. doi: 10.1097/00004647-199702000-00007. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Podrabsky JE, Lopez JP, Fan TW, Higashi R, Somero GN. Extreme anoxia tolerance in embryos of the annual killifish Austrofundulus limnaeus: insights from a metabolomics analysis. J Exp Biol. 2007;210:2253–2266. doi: 10.1242/jeb.005116. [DOI] [PubMed] [Google Scholar]
- Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A, Johnson EM. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron. 2001;29:615–628. doi: 10.1016/s0896-6273(01)00238-0. [DOI] [PubMed] [Google Scholar]
- Qin AP, Zhang HL, Qin ZH. Mechanisms of lysosomal proteases participating in cerebral ischemia-induced neuronal death. Neurosci Bull. 2008;24:117–123. doi: 10.1007/s12264-008-0117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JH, Asai A, Chi S, Saito N, Hamada H, Kirino T. Proteasome inhibitors induce cytochrome c-caspase-3-like protease-mediated apoptosis in cultured cortical neurons. J Neurosci. 2000;20:259–265. doi: 10.1523/JNEUROSCI.20-01-00259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Rami A, Kogel D. Apoptosis meets autophagy-like cell death in the ischemic penumbra: Two sides of the same coin? Autophagy. 2008;4:422–426. doi: 10.4161/auto.5778. [DOI] [PubMed] [Google Scholar]
- Ren C, Gao X, Steinberg GK, Zhao H. Limb remote-preconditioning protects against focal ischemia in rats and contradicts the dogma of therapeutic time windows for preconditioning. Neuroscience. 2008;151:1099–1103. doi: 10.1016/j.neuroscience.2007.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef A, Sperling O, Zoref-Shani E. Preconditioning of primary rat neuronal cultures against ischemic injury: characterization of the "time window of protection'. Brain Res. 1996;741:252–257. doi: 10.1016/s0006-8993(96)00939-0. [DOI] [PubMed] [Google Scholar]
- Reshef A, Sperling O, Zoref-Shani E. Opening of ATP-sensitive potassium channels by cromakalim confers tolerance against chemical ischemia in rat neuronal cultures. Neurosci Lett. 1998;250:111–114. doi: 10.1016/s0304-3940(98)00458-3. [DOI] [PubMed] [Google Scholar]
- Rezvani K, Teng Y, Shim D, De Biasi M. Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J Neurosci. 2007;27:10508–10519. doi: 10.1523/JNEUROSCI.3353-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Rosenzweig HL, Minami M, Lessov NS, Coste SC, Stevens SL, Henshall DC, Meller R, Simon RP, Stenzel-Poore MP. Endotoxin preconditioning protects against the cytotoxic effects of TNFalpha after stroke: a novel role for TNFalpha in LPS-ischemic tolerance. J Cereb Blood Flow Metab. 2007;27:1663–1674. doi: 10.1038/sj.jcbfm.9600464. [DOI] [PubMed] [Google Scholar]
- Ruscher K, Freyer D, Karsch M, Isaev N, Megow D, Sawitzki B, Priller J, Dirnagl U, Meisel A. Erythropoietin is a paracrine mediator of ischemic tolerance in the brain: evidence from an in vitro model. J Neurosci. 2002;22:10291–10301. doi: 10.1523/JNEUROSCI.22-23-10291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba RS, Michels G, Jacob TC, Pangalos MN, Moss SJ. Activity-dependent ubiquitination of GABA(A) receptors regulates their accumulation at synaptic sites. J Neurosci. 2007;27:13341–13351. doi: 10.1523/JNEUROSCI.3277-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler R, Xiong Z, Lu WY, Hafner M, MacDonald JF, Tymianski M. Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein. Science. 1999;284:1845–1848. doi: 10.1126/science.284.5421.1845. [DOI] [PubMed] [Google Scholar]
- Sattler R, Xiong Z, Lu WY, MacDonald JF, Tymianski M. Distinct roles of synaptic and extrasynaptic NMDA receptors in excitotoxicity. J Neurosci. 2000;20:22–33. doi: 10.1523/JNEUROSCI.20-01-00022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller B. Ischemic preconditioning as induction of ischemic tolerance after transient ischemic attacks in human brain: its clinical relevance. Neurosci Lett. 2005;377:206–211. doi: 10.1016/j.neulet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Schaller B, Graf R. Cerebral ischemic preconditioning. An experimental phenomenon or a clinical important entity of stroke prevention? J Neurol. 2002;249:1503–1511. doi: 10.1007/s00415-002-0933-8. [DOI] [PubMed] [Google Scholar]
- Schurr A, Reid KH, Tseng MT, West C, Rigor BM. Adaptation of adult brain tissue to anoxia and hypoxia in vitro. Brain Research. 1986;374:244–248. doi: 10.1016/0006-8993(86)90418-x. [DOI] [PubMed] [Google Scholar]
- Shinoda S, Schindler CK, Meller R, So NK, Araki T, Yamamoto A, Lan JQ, Taki W, Simon RP, Henshall DC. Bim regulation may determine hippocampal vulnerability after injurious seizures and in temporal lobe epilepsy. J Clin Invest. 2004;113:1059–1068. doi: 10.1172/JCI19971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagliano NE, Perez-Pinzon MA, Moskowitz MA, Huang PL. Focal ischemic preconditioning induces rapid tolerance to middle cerebral artery occlusion in mice. J Cereb Blood Flow Metab. 1999;19:757–761. doi: 10.1097/00004647-199907000-00005. [DOI] [PubMed] [Google Scholar]
- Steiger HJ, Hanggi D. Ischaemic preconditioning of the brain, mechanisms and applications. Acta Neurochir (Wien) 2007;149:1–10. doi: 10.1007/s00701-006-1057-1. [DOI] [PubMed] [Google Scholar]
- Stenzel-Poore MP, Stevens SL, King JS, Simon RP. Preconditioning reprograms the response to ischemic injury and primes the emergence of unique endogenous neuroprotective phenotypes: a speculative synthesis. Stroke. 2007;38:680–685. doi: 10.1161/01.STR.0000251444.56487.4c. [DOI] [PubMed] [Google Scholar]
- Stenzel-Poore MP, Stevens SL, Simon RP. Genomics of preconditioning. Stroke. 2004;35:2683–2686. doi: 10.1161/01.STR.0000143735.89281.bb. [DOI] [PubMed] [Google Scholar]
- Stenzel-Poore MP, Stevens SL, Xiong Z, Lessov NS, Harrington CA, Mori M, Meller R, Rosenzweig HL, Tobar E, Shaw TE, Chu X, Simon RP. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- Storey KB. Mammalian hibernation. Transcriptional and translational controls. Adv Exp Med Biol. 2003;543:21–38. [PubMed] [Google Scholar]
- Storey KB. Anoxia tolerance in turtles: metabolic regulation and gene expression. Comp Biochem Physiol A Mol Integr Physiol. 2007;147:263–276. doi: 10.1016/j.cbpa.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Strasser A, Puthalakath H, Bouillet P, Huang DC, O'Connor L, O'Reilly LA, Cullen L, Cory S, Adams JM. The role of bim, a proapoptotic BH3-only member of the Bcl-2 family in cell-death control. Ann N Y Acad Sci. 2000;917:541–548. doi: 10.1111/j.1749-6632.2000.tb05419.x. [DOI] [PubMed] [Google Scholar]
- Sundaram M, Cook HW, Byers DM. The MARCKS family of phospholipid binding proteins: regulation of phospholipase D and other cellular components. Biochem Cell Biol. 2004;82:191–200. doi: 10.1139/o03-087. [DOI] [PubMed] [Google Scholar]
- Tauskela JS, chakravarthy BR, Murray CL, Wang Y, Comas T, Hogan M, Hakim A, Morley P. Evidence from cultured rat cortical neurons of differences in the mechanism of ischemic preconditioning of brain and heart. Brain Res. 1999;827:143–151. doi: 10.1016/s0006-8993(99)01322-0. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traystman RJ. Animal models of focal and global cerebral ischemia. Ilar J. 2003;44:85–95. doi: 10.1093/ilar.44.2.85. [DOI] [PubMed] [Google Scholar]
- Uchiyama Y, Koike M, Shibata M. Autophagic neuron death in neonatal brain ischemia/hypoxia. Autophagy. 2008;4:404–408. doi: 10.4161/auto.5598. [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- Waataja JJ, Kim HJ, Roloff AM, Thayer SA. Excitotoxic loss of post-synaptic sites is distinct temporally and mechanistically from neuronal death. J Neurochem. 2008;104:364–375. doi: 10.1111/j.1471-4159.2007.04973.x. [DOI] [PubMed] [Google Scholar]
- Walton M, Woodgate AM, Muravlev A, Xu R, During MJ, Dragunow M. CREB phosphorylation promotes nerve cell survival. J Neurochem. 1999;73:1836–1842. [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wegener S, Gottschalk B, Jovanovic V, Knab R, Fiebach JB, Schellinger PD, Kucinski T, Jungehulsing GJ, Brunecker P, Muller B, Banasik A, Amberger N, Wernecke KD, Siebler M, Rother J, Villringer A, Weih M. Transient ischemic attacks before ischemic stroke: preconditioning the human brain? A multicenter magnetic resonance imaging study. Stroke. 2004;35:616–621. doi: 10.1161/01.STR.0000115767.17923.6A. [DOI] [PubMed] [Google Scholar]
- Weih M, Kallenberg K, Bergk A, Dirnagl U, Harms L, Wernecke KD, Einhaupl KM. Attenuated stroke severity after prodromal TIA: a role for ischemic tolerance in the brain? Stroke. 1999;30:1851–1854. doi: 10.1161/01.str.30.9.1851. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Wiggins CM, Band H, Cook SJ. c-Cbl is not required for ERK1/2-dependent degradation of Bim(EL) Cell Signal. 2007;19:2605–2611. doi: 10.1016/j.cellsig.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Berti R, Dave JR, Elliot PJ, Adams J, Tortella FC. Delayed treatment of ischemia/reperfusion brain injury: extended therapeutic window with the proteosome inhibitor MLN519. Stroke. 2004;35:1186–1191. doi: 10.1161/01.STR.0000125721.10606.dc. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Hale SL, Moffett JR, Dave JR, Elliott PJ, Adams J, Tortella FC. Delayed treatment with MLN519 reduces infarction and associated neurologic deficit caused by focal ischemic brain injury in rats via antiinflammatory mechanisms involving nuclear factor-kappaB activation, gliosis, and leukocyte infiltration. J Cereb Blood Flow Metab. 2003;23:75–87. doi: 10.1097/01.WCB.0000039285.37737.C2. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Myers TM, Cohn SI, Sharrow KM, Lu XC, Tortella FC. Recovery from ischemic brain injury in the rat following a 10 h delayed injection with MLN519. Pharmacol Biochem Behav. 2005;81:182–189. doi: 10.1016/j.pbb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Windheim M, Peggie M, Cohen P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. 2008;409:723–729. doi: 10.1042/BJ20071338. [DOI] [PubMed] [Google Scholar]
- Wojcik C. Proteasomes in apoptosis: villains or guardians? Cell Mol Life Sci. 1999;56:908–917. doi: 10.1007/s000180050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C, Di Napoli M. Ubiquitin-proteasome system and proteasome inhibition: new strategies in stroke therapy. Stroke. 2004;35:1506–1518. doi: 10.1161/01.STR.0000126891.93919.4e. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol Rev. 2007;59:14–39. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Wang J, Kim A, Liu Q, Watts R, Hoopfer E, Mitchison T, Luo L, He Z. Involvement of the ubiquitin-proteasome system in the early stages of wallerian degeneration. Neuron. 2003;39:217–225. doi: 10.1016/s0896-6273(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Qian H, Zhao P, Hong SS, Xia Y. Rapid hypoxia preconditioning protects cortical neurons from glutamate toxicity through delta-opioid receptor. Stroke. 2006a;37:1094–1099. doi: 10.1161/01.STR.0000206444.29930.18. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang ZG, Liu X, Hozeska A, Stagliano N, Riordan W, Lu M, Chopp M. Treatment of embolic stroke in rats with bortezomib and recombinant human tissue plasminogen activator. Thromb Haemost. 2006b;95:166–173. [PubMed] [Google Scholar]
- Zhang S, Boyd J, Delaney K, Murphy TH. Rapid reversible changes in dendritic spine structure in vivo gated by the degree of ischemia. J Neurosci. 2005;25:5333–5338. doi: 10.1523/JNEUROSCI.1085-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Cheng GZ, Gong J, Hermanto U, Zong CS, Chan J, Cheng JQ, Wang LH. RACK1 and CIS mediate the degradation of BIMEL in cancer cells. J Biol Chem. 2008 doi: 10.1074/jbc.M802360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]