Summary
Purified DNA translocases Rdh54 and Rad54 can dissociate complexes formed by eukaryotic RecA-like recombinases on double-stranded DNA. Here we show Rad51 complexes are dissociated by these translocases in mitotic cells. Rad51 overexpression blocked growth of cells deficient in Rdh54 activity. This toxicity was associated with accumulation of Rad51 foci on undamaged chromatin. At normal Rad51 levels, rdh54 deficiency resulted in slight elevation of Rad51 foci. A triple mutant lacking Rdh54, Rad54, and a third Swi2/Snf2 homologue Uls1, accumulated Rad51 foci, grew slowly, and suffered chromosome loss. Thus, Uls1 and Rad54 can partially substitute for Rdh54 in the removal of toxic, non–damage-associated Rad51-DNA complexes. Additional data suggest that the function of Rdh54 and Rad54 in removal of Rad51 foci is significantly specialized; Rad54 predominates for removal of damage-associated foci and Rdh54 predominates for removal of non-damage-associated foci.
Introduction
Homologous recombination (HR) is an essential process that functions in diverse aspects of chromosome metabolism. Multiple HR pathways (reviewed in Branzei and Foiani, 2008; Sung and Klein, 2006) repair spontaneous and programmed double-stranded DNA breaks (DSBs). In addition, HR repairs the single-stranded DNA (ssDNA) gaps and DNA ends that form at damaged replication forks, and is further involved in telomere maintenance, meiotic recombination, and chromosome segregation. DNA strand exchange proteins or “recombinases” promote the central process of HR (Eggleston and Kowalczykowski, 1991). Saccharomyces cerevisiae, like most other eukaryotes, encodes two RecA-like recombinases, Rad51 and Dmc1 (Bishop et al., 1992; Shinohara et al., 1992). Rad51 functions in mitotic and meiotic recombination, while Dmc1 functions only in meiosis. Both proteins promote DNA strand exchange between ssDNA and homologous dsDNA in vitro. To catalyze this reaction, recombinases must first form helical recombinase-DNA filaments on ssDNA (Eggleston and Kowalczykowski, 1991; Sehorn et al., 2004). The resulting nucleoprotein filaments then invade homologous double-stranded DNA (dsDNA) to form regions of heteroduplex DNA that are bound by the recombinases, making it possible for the homologous region to be used as template for high fidelity repair.
Like RecA, Rad51 and Dmc1 require ATP, but not ATP hydrolysis for strand exchange (Hong et al., 2001; Menetski and Kowalczykowski, 1990; Sung and Stratton, 1996).
Although able to promote strand exchange in vitro, the yeast recombinases differ from RecA by exhibiting little or no binding preference for ssDNA versus dsDNA (Hong et al., 2001; Ogawa et al., 1993). In cells, the lack of specificity of yeast recombinases for ssDNA may be compensated for by protein-protein interactions between Rad51, the ssDNA binding protein RPA, and accessory proteins referred to as Rad51 assembly “mediators”. A cytological manifestation of mediated Rad51 assembly is the colocalization of Rad51 immunostaining foci with RPA foci during DNA repair (Gasior et al., 2001, and references therein). In addition, yeast recombinases require additional accessory proteins for optimal activity in vitro, including the Swi2/Snf2-like translocase proteins Rdh54 and Rad54 (Petukhova et al., 1998; Petukhova et al., 2000; Petukhova et al., 1999). These translocases may also contribute indirectly to the assembly of Rad51 at sites of ssDNA by promoting dissociation of non-recombinogenic complexes on dsDNA (as discussed below).
Rad54 has been studied more extensively than Rdh54, and several biochemical activities have been described for Rad54, including Rad51 filament stabilization, DNA unwinding, nucleosome displacement, Holliday junction displacement, and dissociation of filaments from dsDNA (San Filippo et al., 2008). Rdh54 shares the ability to promote Rad51 filament stabilization, DNA unwinding, and dissociation of recombinase from dsDNA, and may share additional activities.
Given this plethora of biochemical properties, an important challenge is determining which properties contribute to the function of Rad54 and Rdh54 in living cells. A related question is to understand how many of the Swi2/Snf2-related translocases function in HR and can remove Rad51 from DNA. This is particularly relevant, given that there are 17 proteins in this family in yeast, seven of which have been shown to function in some aspect of HR or DNA repair (Heyer et al., 2006).
One biochemical activity that is important in meiotic cells is the ability of translocases to promote dissociation of strand exchange proteins from dsDNA. This dissociation activity contributes to meiotic recombination in two distinct ways. First, Dmc1 protein was shown to bind randomly to meiotic chromatin forming complexes that are not associated with recombination (Holzen et al., 2006). In the absence of both Rdh54 and Rad54, meiotic recombination fails. This failure appears to be the consequence of sequestration of Dmc1 at random dsDNA sites on chromatin. Secondly, C. elegans homologs of the translocases were recently shown to promote dissociation of Rad51 from sites of DSBs during meiotic recombination (Ward et al., 2010). Thus two roles for translocase-dependent dissociation of strand exchange proteins have been described in meiotic cells; similar mitotic functions have not been previously demonstrated.
Although partially redundant, the roles of Rad54 and Rdh54 in living cells do not appear to be identical; Rad54 is more important than Rdh54 for mitotic DNA repair and homology-mediated mating-type switching (Heyer et al., 2006; San Filippo et al., 2008). (Petukhova et al., 1999; Schmuckli-Maurer et al., 2003) and Rdh54 is more important for Dmc1 dynamics and interhomolog recombination during meiosis (Holzen et al., 2006; Shinohara et al., 2000; Shinohara et al., 1997).
The role for Rdh54 in mitotic recombination is not as clearly defined. Rdh54 is required for recovery from the arrest caused by an induced DSB during mitosis, and such arrest depends on RAD51 (Lee et al., 2001). rdh54 mutants have reduced mitotic interhomolog recombination, but the reduction is less than ten-fold, much less than the reduction seen in other HR mutants (Klein, 1997). Additionally, while rad54 mutants are highly sensitive to DNA damaging agents, rdh54 mutants are only slightly sensitive. The defects of a rad54 rdh54 double mutant are greater than the respective single mutants, indicating some overlap in function (Klein, 1997; Shinohara et al., 1997). Biochemical experiments raise the possibility that this shared function could involve the dissociation of Rad51 from dsDNA (Chi et al., 2006; Solinger et al., 2002).
In this study we show that Rad51 can bind undamaged chromatin in living cells. When overexpressed, Rad51 forms complexes on undamaged chromatin that can be visualized by immunostaining. Overexpression is detrimental when Rad51 is not removed from undamaged chromatin. We examined the ability of Rdh54 and Rad54 to promote dissociation of Rad51 from chromatin in cells overexpressing Rad51. The results demonstrate a new in vivo function for these translocases. We also examined a third Swi2/Snf2-related factor, Uls1/Ris1/Tid4 (hereafter Uls1) (Uzunova et al., 2007), and find that it too can contribute to Rad51 turnover from undamaged chromatin.
Importantly, we show that in the absence of the Rad54, Rdh54 and Uls1 translocases, Rad51 accumulates spontaneously on chromatin. The Rad51 accumulation in the triple translocase mutant occurs without Rad51 overexpression and results in genome instability and chromosome loss. The results indicate that, similar to Dmc1 regulation during meiosis, Rdh54 is a major translocase involved in Rad51 dissociation from sites of nonspecific dsDNA binding during mitosis. Importantly, Rad51 association with chromatin occurs in the absence of induced DNA damage or DSBs, blocks cell growth, and increases genomic instability
Results
RAD51 overexpression can inhibit the growth of cells
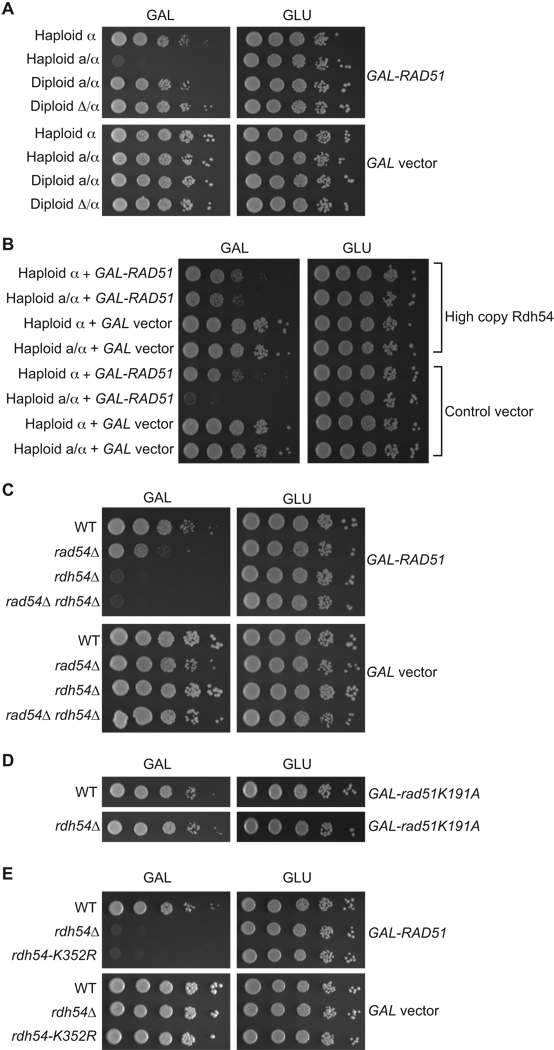
We used Rad51 overexpression as a probe for factors that could counteract possible deleterious consequences of Rad51 bound inappropriately to chromatin. RAD51 was overexpressed using a plasmid with the coding region of RAD51 fused to the GAL1 promoter resulting in a fifteen-fold increase in protein levels (Figure S1). The GAL1 promoter is strongly induced by growth on medium containing galactose and repressed on medium containing glucose. The GAL1-RAD51 plasmid or a GAL1 empty vector control was introduced into both haploid and diploid wild type strains. After growth to mid log phase in glucose medium, equivalent numbers of cells were spotted by serial dilutions on medium containing galactose to induce RAD51 expression. The plates were monitored for any growth defects associated with altered RAD51 expression.
Although all control cells showed equivalent growth, a MATa/MATα diploid strain showed a modest, but reproducible, defect in growth on galactose medium (Figure 1A). In contrast, the growth of a MATα haploid strain was only slightly slower on galactose as compared to glucose. The difference in growth for the MATa/MATα diploid compared to the MATα haploid was greater in liquid cultures than on plates; Rad51 overexpression increased the doubling time 1.9 fold in diploids and 1.1 fold in haploids.
Figure 1.
Cell type sensitivity to RAD51 overexpression is due to limiting Rdh54 protein. A. Haploid and diploid strains with the indicated mating types were transformed with GAL-RAD51 or GAL empty vector plasmids. The resulting transformants were grown as described in the Experimental Procedures and then serially diluted and plated onto galactose or glucose plates. Plates were incubated at 30°C for three days. B. The haploid strains from panel A were additionally transformed with a high copy plasmid carrying the RDH54 gene or the control vector. The resulting transformants were grown as described and then serially diluted and plated onto galactose or glucose plates and incubated at 30°C for three days. C. Haploid cells of the indicated genotypes were transformed with the GAL-RAD51 or GAL empty vector plasmids and the resulting transformants were grown as described and then serially diluted and plated onto galactose or glucose plates and incubated at 30°C for three days. WT is wild type. D. Haploid cells were transformed with the GAL-rad51K191A plasmid and the resulting transformants were grown as described and then serially diluted and plated onto galactose or glucose plates and incubated at 30°C for three days. E. Haploid cells of the indicated genotypes were transformed with the GAL-RAD51 or GAL empty vector plasmids and the resulting transformants were grown as described and then serially diluted and plated onto galactose or glucose plates and incubated at 30°C for three days.
Cell type-specific transcriptional regulation, not ploidy, controls cellular sensitivity to Rad51 overexpression
Differences in the phenotype of isogenic haploid versus diploid cells can be a consequence of either the ploidy difference between the two strains or the fact that the two strains express distinct sets of genes owing to expression of different cell type-specific transcription factors encoded by the MAT locus. To determine which of these two possibilities accounted for the observed sensitivity to RAD51 overexpression in wild type diploids, we examined a haploid wild type strain that expresses both MATa and MATα (MATa/MATα) and a diploid strain expressing only MATα (matΔ/MATα). Growth of the strains was assayed following induction of high RAD51 expression on galactose plates. The MATa/MATα haploid is even more sensitive to RAD51 overexpression than the MATa/MATα diploid (Figure 1A). This is in contrast to the matΔ/MATα diploid, which shows almost no sensitivity to RAD51 overexpression and grows similarly to a MATα haploid.
These results indicate that one or more differences between the transcriptome of MATα cells and MATa/MATα cells is responsible for the difference in the sensitivity of haploid and diploid strains to RAD51 overexpression. When strains that express both Mata and MATα are compared, the relative sensitivity of haploids compared to diploids suggests that diploidy has a protective effect against RAD51 overexpression.
In Silico analysis indicates that limited RDH54/RDH54 expression underlies the role of MATa/MATα control of Rad51 sensitivity
Given that cell type-specific transcriptional control modulates the sensitivity of both haploid and diploid cells to high RAD51 levels, we searched the Ploidy Regulation of Gene Expression (http://staffa.wi.mit.edu/fink_public/ploidy/) database to identify genes in the HR pathway that are expressed at significantly lower levels in MATa/MATα diploids compared to MATα haploids. Starting with the NEJ1 gene, which is severely reduced in expression in MATa/MATα cells compared to MATα cells (Valencia et al., 2001), we performed a “nearest-neighbor analysis” of 75 genes with a similar pattern of expression. Of these, only five genes are known to be involved in HR or DNA damage repair: RDH54, SRS2, SHU1, MPH1 and PSY3. RDH54 was the top scoring HR gene and ranked eighth below NEJ1; the gene is reported to be expressed at 4–10 fold lower level in diploids compared to haploids. (de Godoy et al., 2008). These observations were of particular interest based on two previous findings. First, it has been shown that slow growth or cell cycle arrest in rdh54 or rdh54 rad54 double mutants can be alleviated by elimination of RAD51 (Raschle et al., 2004; Signon et al., 2001). Second, Dmc1 has been shown to accumulate random, non-recombinogenic complexes on meiotic chromatin in the absence of RDH54 (Holzen et al., 2006). These results led us to hypothesize that the relatively high level of Rdh54 in haploid cells acts to prevent accumulation of Rad51 on chromatin; when Rad51 accumulates due to reduced RDH54 levels in diploids, growth is reduced. These predictions are tested below.
Rdh54 limits growth rate in MATa/MATα cells
To demonstrate that a limiting amount of Rdh54 is primary cause for the sensitivity of MATa/MATα haploid cells to RAD51 overexpression, we introduced a high copy plasmid carrying a functional copy of RDH54, into MATα and MATa/MATα haploid cells, along with the high copy RAD51 plasmid. High copy RDH54 has no phenotype on its own; however, in cells overexpressing RAD51, high copy RDH54 increases the growth rate of MATa/MATα haploids to the rate of MATα haploids. This finding indicates that the growth defect in haploid cells expressing both mating types is due to a limiting amount of Rdh54 (Figure 1B). High copy RDH54 only partially rescues the growth defect conferred by high copy RAD51, suggesting that Rad51 may also affect an Rdh54-independent process, or that the optimum ratio of Rdh54 activity is not achieved by the construct employed.
An rdh54 mutation makes haploid cells sensitive to overexpression of RAD51
To further test the hypothesis that the low level of RDH54 expression is responsible for the sensitivity of cells expressing both mating types to high levels of RAD51, we examined the effect of deleting the RDH54 gene in haploids and diploids. We found that both rdh54 haploids and diploids are profoundly sensitive to high levels of RAD51 expression (Figure 1C; Figure S2). These data indicate that RDH54 is critical for allowing cells to tolerate high levels of Rad51. The results also suggest that the level of RDH54 in normal diploid cells, though lower than in haploid cells, is sufficient to provide significant protection against RAD51 overexpression. Because the RAD54 gene shows partial redundancy with RDH54 in mitotic and meiotic cells (Klein, 1997; Shinohara et al., 1997), we also examined Rad51 sensitivity in a rad54 mutant and an rdh54 rad54 double mutant. The rad54 single mutant has a significant defect in growth on galactose medium compared to the empty vector control. This defect was not nearly as pronounced as that of the rdh54 single mutant. The rdh54 rad54 double mutant defect was at least as severe than the rdh54 single mutant defect. Elevated levels of Rad54 could not substitute for Rdh54 in protecting cells against high levels of Rad51 (data not shown). These results indicate that Rad54 has a more limited role than Rdh54 in protecting cells against the deleterious effects of RAD51 overexpression.
Rdh54 uses its ATPase activity to counteract the growth defect conferred by the ability of Rad51 to bind DNA with high affinity
As Rdh54 promotes the dissociation of Rad51 from DNA and chromatin in vitro, and dissociation of Dmc1 in vivo, we hypothesized that growth inhibition by RAD51 overexpression depends on its ability to stably bind DNA. To test this, we examined the effect of overexpressing rad51K191A, which has reduced DNA binding affinity (Li et al., 2007). We found that rad51K191A did not inhibit growth when expressed under the control of the same galactose inducible promoter used to express the wild type gene (Figure 1D). Western blot analysis was used to confirm that the steady state levels achieved for both the wild type and the mutant protein were equivalent, supporting the hypothesis the deleterious consequences of RAD51 overexpression requires stable DNA binding (Figure S1). Analysis of Rad51 immunostaining provided additional support for the biochemical observation that the Rad51-K191A protein binds chromatin less efficiently than the wild type Rad51 protein (data not shown).
Another prediction of the hypothesis that chromatin-bound Rad51 is responsible for growth inhibition is that the protective effect of Rdh54 should require its DNA-dependent ATPase and translocase activity, as Rdh54 displacement of Rad51 from DNA is dependent on ATP hydrolysis. To test this, we employed the ATPase defective rdh54K352R mutant. We tested the ability of rdh54K352R to promote tolerance to RAD51 overexpression in haploids and found it to be as defective as the rdh54 null mutant (Figure 1E). This finding, in the context of other results described here, suggests that Rdh54 translocation on dsDNA is required for cells to tolerate RAD51 overexpression.
rdh54 mutants overexpressing RAD51 exhibit a severe defect in the growth assay on galactose medium, making it difficult to distinguish between cells that are unable to grow, versus cells that have died as a result of high copy RAD51. To distinguish between these possibilities, rdh54 mutants were returned to growth on glucose medium following induction of RAD51 expression in galactose medium. While rdh54 cells have poor plating efficiency, less than 1%, on glucose medium when maintaining selection for the RAD51 overexpression plasmid, rdh54 mutants have normal plating efficiency, 95–100%, when returned to glucose medium that is not maintaining selection for the RAD51 overexpression plasmid. These results indicate that rdh54 mutant cells did not die following RAD51 overexpression, but were prevented from further growth on selective medium because they no longer contained the RAD51 overexpression plasmid. This was confirmed by Southern blot analysis, which showed on a population basis that rdh54 cells contained only 60% the amount of plasmid as wild type (RDH54+) cells, after 4–5 generations of growth under RAD51-inducing conditions (data not shown).
To confirm that cells can recover from RAD51 overexpression, we examined a strain carrying a chromosomal GAL-RAD51 fusion. Cells were transferred to galactose for six hours and then plated on medium containing either glucose or galactose. In this period of time, the plating efficiency for the GAL-RAD51 fusion-containing strain was not reduced compared to a control strain, confirming that cells are capable of efficient recovery from the effects of RAD51 overexpression.
Rad51 forms multiple subnuclear immunostaining foci when overexpressed in rdh54 haploids and RDH54+ diploids
Previous studies of Rdh54 function in meiosis showed that the protein acts to prevent DSB-independent accumulation of Dmc1 protein on DNA. Given our finding that Rdh54 is required for mitotic cells to continue to grow in the presence of high levels of Rad51, we used immunostaining of spread mitotic nuclei to determine if formation of chromatin-bound Rad51 foci correlated with growth inhibition.
Surface spread nuclei were prepared and immunostained against Rad51 and RPA, the ssDNA binding protein. RPA binds ssDNA preferentially and is therefore a good marker for the tracts of ssDNA associated with replication forks, resected DSBs, and subsequent DSB repair intermediates. Previous studies showed that DNA damage caused by ionizing radiation or by programmed meiotic DSBs result in formation of Rad51 foci that show a high percentage of colocalization with RPA foci (Gasior et al., 1998). In contrast, DSB-independent Dmc1 focus accumulation was shown previously to occur without associated assembly of RPA (Holzen et al., 2006). These findings led us to explore the possibility that Rad51 forms foci independent of ssDNA lesions.
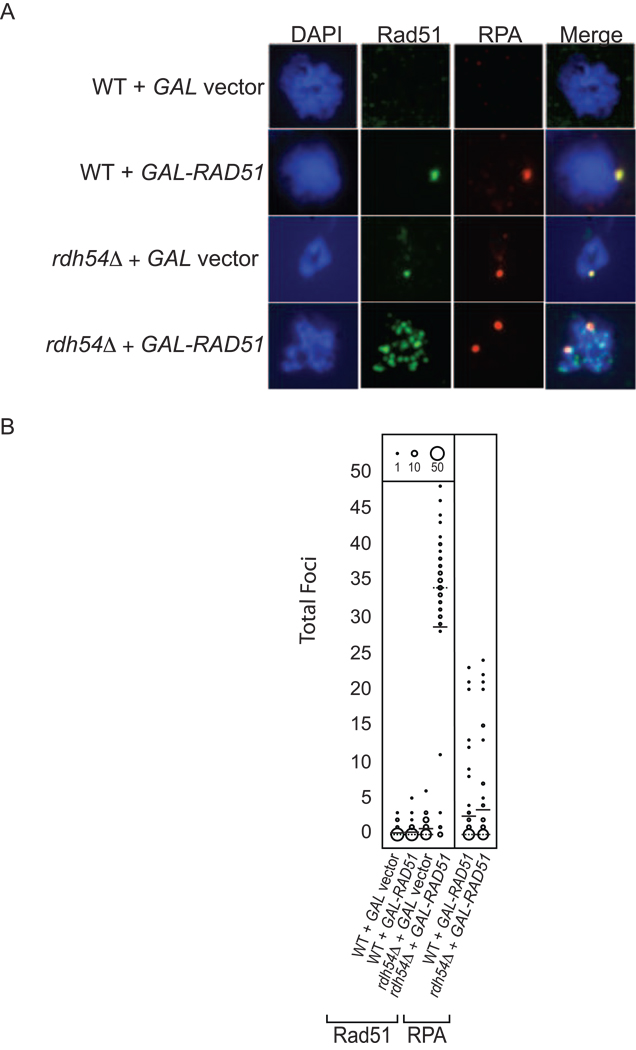
Immunofluorescence microscopy was performed for wild type or rdh54 haploids (both MATa; average focus counts were similar in haploids of either mating type) following overexpression of RAD51 using the GAL-RAD51 plasmid. Representative images of stained spread nuclei are shown in Figure 2A. Rad51 and RPA foci were counted for fifty random nuclei from each strain (Figure 2B). 12% and 18% of wild type nuclei with GAL empty vector or GAL-RAD51, respectively, contained at least one Rad51 focus (Figure 2B, first and second lanes). The average number of foci was 0.22 foci per nucleus for wild type with GAL empty vector (median focus count = 0) and 0.32 foci per nucleus for wild type with GAL-RAD51 (median focus count = 0). The rdh54 GAL empty vector strain also displayed a low level of Rad51 foci (median focus count = 0); however, the number of foci in rdh54 GAL empty vector was significantly higher than that in wild type GAL empty vector (Student’s t-test, p=0.0042; Figure 2B); 60% of rdh54 empty vector nuclei contained at least one Rad51 focus, with an average of 0.8 foci per nucleus. This suggests that under conditions of normal RAD51 expression, Rad51 forms foci that are blocked by the function of Rdh54.
Figure 2.
RPA-independent Rad51 foci form in rdh54 strains overexpressing RAD51. A. Representative images from MATa wild type (WT) or MATa rdh54 strains transformed with GAL empty vector or GAL-RAD51 overexpression plasmid. Left to Right: DAPI DNA stain (blue), Rad51 (green), RPA (red), and Merge. B. Circle plot representation of the total number of Rad51 foci (left hand side of plot) or RPA foci (right hand side of plot) from wild type (WT) and rdh54 strains with GAL empty vector or GAL-RAD51. The center of each circle corresponds to the Rad51 or RPA focus count indicated on the Y-axis; the diameter of each circle corresponds to the number of nuclei that displayed the corresponding focus count. The mean level of foci is given as a solid line, the median is given as a dashed line. Fifty randomly selected nuclei scored per strain.
The rdh54 GAL-RAD51 strain shows a strikingly different staining pattern from the control strains. 88% of nuclei from rdh54 GAL-RAD51 cells contain at least one Rad51 focus, with an average of 32.5 foci per nucleus (median focus count = 34). The staining structures in rdh54 GAL-RAD51 include fine puncta and larger, irregular staining structures, atypical of normal focus formation. This result suggests that Rad51 accumulates on chromatin in the absence of Rdh54 activity and supports the hypothesis that the accumulation of Rad51 is responsible for the growth defect associated with RAD51 overexpression.
In contrast to the increase in Rad51 foci, the number of RPA foci was not altered by RAD51 overexpression in wild type or rdh54 mutants (Figure 2B, RPA lanes). S-phase yeast cells contain up to 25 RPA foci, which are presumed to mark tracts of ssDNA at replication forks. However, only 6–7% of Rad51 foci colocalize with RPA in rdh54 GAL-RAD51 strains. This is compared with a colocalization frequency of 94% RPA-Rad51 among Rad51 foci scored in RDH54+ GAL-RAD51 (Figure 2A, compare merged images from the second and fourth rows). Importantly, the total number of RPA foci per cell is essentially the same in rdh54 GAL-RAD51 and RDH54+ GAL-RAD51, indicating spontaneous Rad51 foci caused by rdh54 deficiency are rarely associated with RPA. This finding is in contrast to Rad51 foci induced by DNA damage, of which 75% colocalize with RPA (Gasior et al., 1998; see below). Cell cycle arrest and release experiments provided additional evidence the Rad51 foci induced by Rad51 overexpression are not associated with ssDNA tracts formed during S-phase (Figure S3).
Having found evidence that Rad51 accumulates on chromatin in rdh54 haploids, we wanted to determine if the modest growth defect in wild type diploids (RDH54+/RDH54+) with RAD51 overexpression was also associated with accumulation of Rad51 on DNA. Rad51 and RPA focus formation were monitored in a wild type diploid strain overexpressing RAD51 (Figure S4). 64% of wild type GAL-RAD51 diploids contained at least one Rad51 focus, with 36% of these nuclei containing 10 or more Rad51 foci. The average number of Rad51 foci in focus-positive wild type GAL-RAD51 diploids is 16.4 ±1.3 (median focus count = 14), and the majority of these foci were not colocalized with RPA.
The Rad51 staining pattern exhibited in wild type GAL-RAD51 diploids is similar to that seen in rdh54 haploid strains with GAL-RAD51. Such foci are indicative of Rad51 binding to undamaged chromatin. The observation of RPA-independent Rad51 foci is consistent with the genetic data showing RAD51 overexpression being inhibitory in normal diploid cells (MATa/MATα, RDH54+/RDH54+) due to the fact that MATa/MATα control maintains a low level of RDH54 expression. The genetic results, taken together with immunostaining results, provide evidence that the growth inhibition effect of RAD51 overexpression is a consequence of accumulation of Rad51 on chromatin.
Redundant activities of three Swi2/Snf2 translocases prevent accumulation of Rad51 on chromatin under normal cellular conditions
The experiments above suggest that Rdh54 can dissociate Rad51 from chromatin when the recombinase is expressed at abnormally high levels. Since we saw only modest Rad51 accumulation in controls that lacked RAD51 overexpression, we hypothesized that one or more Rdh54-related translocases contributes a Rad51-dissociating activity that efficiently substitutes for Rdh54 when Rad51 is present at normal levels, but not when RAD51 is overexpressed.
Two Swi2/Snf2-like translocases, Rad54 and Uls1, were judged to be lead candidates for sharing Rad51 dissociating activity with Rdh54. Rad54 was examined because it is known to have redundant activity with Rdh54 and to be able to dissociate Rad51 from dsDNA. Uls1 was examined because uls1 was found to have a diploid synthetic growth defect when combined with an rdh54 rad54 double mutant (H. Klein, unpublished observations). Furthermore, Uls1 can partially substitute for the repair function of Rad54 (H. Klein, unpublished observations). Uls1 was also found to have dsDNA translocase activity in vitro (P.Sung, personal communication).
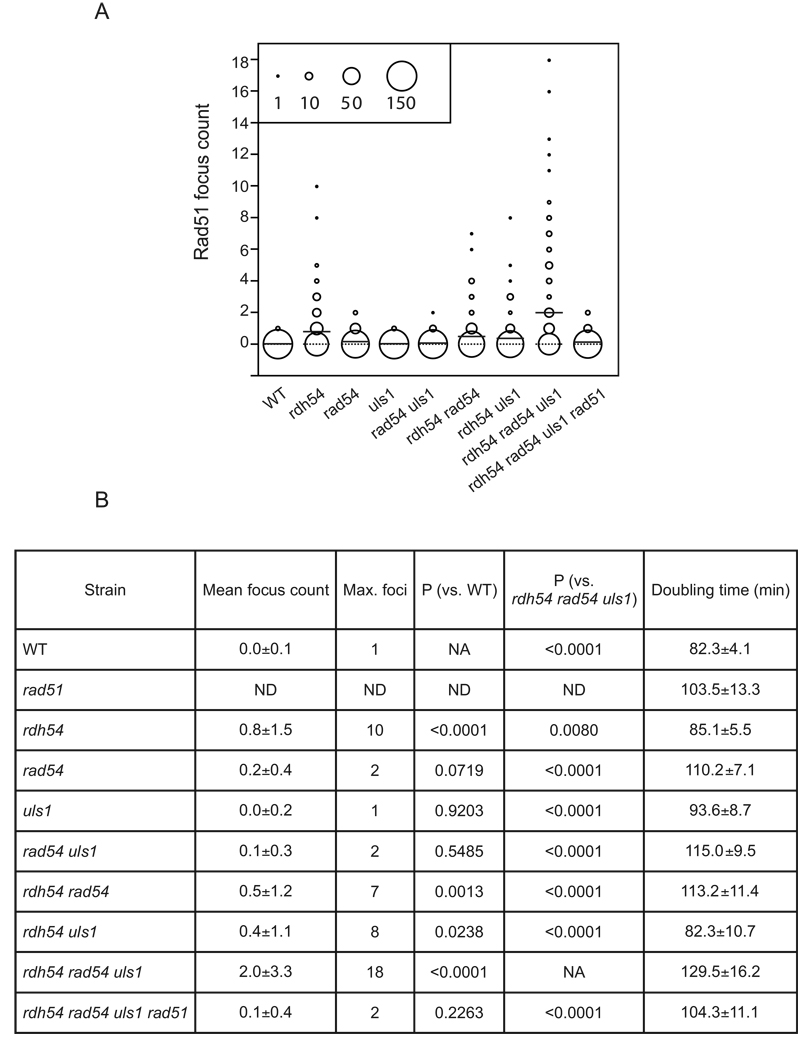
Nuclei from mitotic haploid cells carrying all possible combinations of mutations in RDH54, RAD54, and ULS1 were spread and immunostained for Rad51. As in the empty vector controls from the previously described experiment, the rdh54 single mutant displayed a significantly higher level of spontaneous Rad51 foci relative to the wild type strain (Figure 3A). In contrast to the rdh54 mutant, we did not detect any increase in spontaneous Rad51 foci in either a rad54 or uls1 single mutant or in a rad54 uls1 double mutant, indicating that neither Rad54 nor Uls1 is required for preventing Rad51 focus accumulation if Rdh54 is present (Figure 3A). Furthermore, we saw no enhancement of the spontaneous Rad51 focus accumulation phenotype conferred by an rdh54 mutation when it was combined with either a rad54 or a uls1 mutation; the rdh54 rad54 and rdh54 uls1 double mutants showed the same modest Rad51 focus accumulation defect as the rdh54 single mutant.
Figure 3.
Effects of Rad51 accumulation in rad54 rdh54 uls1 strains. A. Rad51 focus accumulation without overexpression in triple translocase mutant cells. The center of each circle corresponds to the Rad51 focus count indicated on the Y-axis with the diameter of each circle corresponding to the number of nuclei that displayed the corresponding focus count. Note that the data from the quadruple mutant on the right side of the plot provides an indication of the level of background staining in this experiment. There is no significant increase in focus count in rad54, uls1, or rad54 uls1 cells relative to wild type (WT). The rdh54, rdh54 rad54, and rdh54 uls1 strains show the same intermediate level of focus accumulation and the rdh54 rad54 uls1 triple shows the highest level of focus accumulation. 150 nuclei were scored for each sample. B. Number of Rad51 foci and strain doubling times for wild type (WT) and translocase mutants. Strains were a mix of MATa and MATα haploids; focus count results were independent of mating type.
Interestingly, a triple mutant (rdh54 rad54 uls1) lacking all three translocases showed a significantly higher level of Rad51 foci than either the rdh54 uls1 or the rdh54 rad54 double mutant. Spread nuclei from the triple mutant had an average of 2 ±3.3 Rad51 foci per nucleus, with as many as 18 foci observed (Figure3A). These results indicate that DNA translocase activity blocks spontaneous Rad51 focus accumulation during normal mitotic growth.
Analysis of growth rates shows that the rdh54 rad54 uls1 triple mutant grows more slowly than any of the double mutant combinations (Figure 3B). Moreover, a point mutation in any of the three genes that eliminates ATPase activity and DNA translocation has the same phenotype as a deletion mutation, showing that the growth defect results from defective translocase function. The slow growth phenotype in the triple translocase mutant can be rescued by elimination of Rad51; the rdh54 rad54 uls1 rad51 quadruple mutant has a similar doubling time to that of a RAD51 single mutant. This finding is consistent with the view that accumulation of Rad51 foci above a critical threshold inhibits progression of the cell cycle.
Accumulation of Rad51 on chromatin is associated with chromosome instability
The experiments with RAD51 overexpression suggested that loss of Rad51 regulation in the rdh54 mutant was associated with a defect in plasmid maintenance. Therefore we tested the possibility that Rdh54 and its relatives control chromosome stability via effects on Rad51. These experiments employed the rad54 rdh54 uls1 triple mutant without RAD51 overexpression. The rad54 rdh54 uls1 triple mutant is close to inviable as a diploid (H. Klein, unpublished observations), making it impossible to determine chromosome loss rates in the diploid triple translocase mutants. Therefore, we determined the loss rate of a supernumerary chromosome fragment carrying the ADE2 gene in haploid triple translocase mutant strains. While this fragment is relatively stable, it is lost more frequently than a normal chromosome, making it possible to measure loss rates by visual screening for red/white half-sectored colonies (Figure S5). In this system, the frequency of loss is equal to the rate of loss, as loss events are determined for one generation.
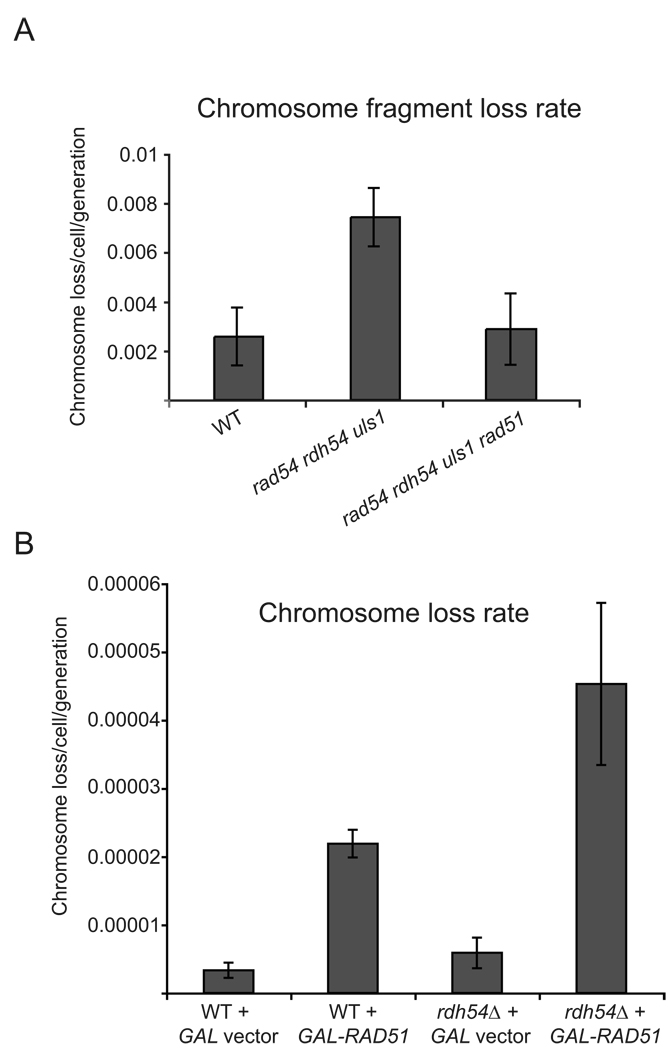
Chromosome fragment loss rate was significantly increased in the triple rad54 rdh54 uls1 haploid mutant over wild type haploids, and this increase was dependent on RAD51 (Figure 4A). RAD51 is responsible for a substantial fraction of this instability phenotype, because the loss rate is significantly reduced in a rad54 rdh54 uls1 rad51 quadruple mutant (p<0.015). The quadruple mutant also showed a faster doubling time compared to the mutant rad54 rdh54 uls1 triple mutant (Figure 3B). It is also important to note that the total fraction of chromosome fragment loss events including those that occurred prior to plating of the cells in addition to the loss event that occurred during the first division of plating (red colonies and red/white half-sectors) was higher in the rad54 rdh54 uls1 triple mutant compared to either wild type or the rad54 rdh54 uls1 rad51 quadruple mutant (p<0.0001; Figure 4A). These results suggest that Rad51 accumulation on chromatin has negative consequences for chromosome stability and overall cell growth.
Figure 4.
Genome instability in strains that accumulate Rad51 foci. A. Chromosome fragment loss rates. Chromosome fragment loss rates were determined by counting red/white half-sectored colonies in wild type (WT), rad54 rdh54 uls1 and rad54 rdh54 uls1 rad51 strains. Differences in chromosome loss rates between wild type (WT) and rad54 rdh54 uls1, and between rad54 rdh54 uls1 and rad54 rdh54 uls1 rad51 were significant by t tests, p<0.015. B. Chromosome loss rates in diploid cells overexpressing Rad51. Chromosome loss rates were determined in diploid cells of the indicated genotypes following Rad51 induction. The rates of chromosome loss between wild type and rdh54 with the GAL empty vector were not different by a t test, p>0.15, but the rates of chromosome loss between wild type and rdh54 with the GAL-RAD51 plasmid were different by a t test, p<0.03.
Finally, we measured the rate of loss of chromosome V (Klein, 2001) in diploid wild type and rdh54 strains (Figure 4B). The chromosome loss rate was about two fold greater in rdh54 cells as compared to wild type (p<0.03). This finding indicates that Rdh54 has a modest, but significant, chromosome stabilizing activity.
rad54 mutants accumulate more Rad51 foci than rdh54 mutants following irradiation
Previous mutant studies have shown that Rad54 plays a more important role in cellular survival of DNA damage than Rdh54 (Klein, 1997). Both translocases have been found to be important for Rad51 removal from DSB-dependent foci in meiosis (Shinohara et al., 1997; Ward et al., 2010), and when taking into consideration the biochemical activity of Rad54, this suggests that Rad54-mediated Rad51 removal could be associated with the role of this translocase in promoting cellular repair of mitotic DSBs. This possibility predicts that Rad54 is more important for preventing accumulation of DNA damage induced Rad51 complexes than Rdh54. To test, we measured Rad51 focus accumulation after irradiation. The effects of rad54 and rdh54 mutations were compared in a uls1 mutant strain background to avoid the possibility that functional substitution of Uls1 for Rad54 or Rdh54 would obscure a phenotypic difference.
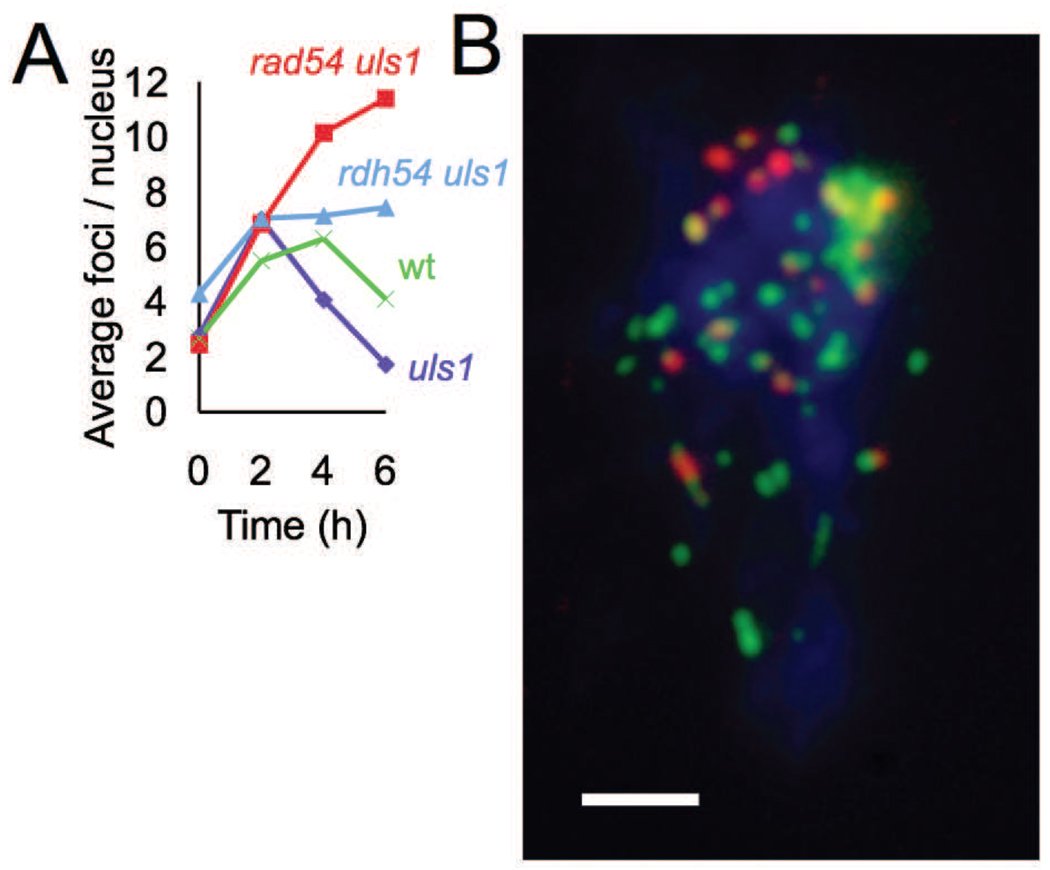
Wild type and mutant cells were gamma-irradiated with 50 krad and incubated for six hours. Spread nuclei were prepared from culture aliquots at 2, 4, and 6 hours after irradiation, and double immunostained for RPA and Rad51 (Figure 5A). In wild type and uls1 single mutant cells, overall Rad51 focus levels were induced 2–3 fold above levels in undamaged cells to a peak level of 6–7 Rad51 foci per nucleus cell on average. After peaking, Rad51 focus levels returned to pre-irradiation levels at 6 hours. In contrast to wild type and the uls1 single mutant, the uls1 rad54 double mutant accumulated foci during the entire 6-hour post irradiation period reaching a level 2.7-fold higher than wild type at 6 hours. The uls1 rdh54 double mutant also accumulated more Rad51 foci than the control strains, although focus accumulation slowed at 2 hours and reached a level only 1.8-fold higher than wild type at 6 hours. These observations indicate that both Rad54 and Rdh54 play roles in removal of Rad51, with the role of Rad54 dominating.
Figure 5.
Analysis of Rad51 foci following gamma irradiation. A. Cultures were irradiated with 50 krad, and cells taken at the times indicated were examined for Rad51 and RPA nuclear immunostaining foci. Data points are mean focus counts from 50 random nuclei. The strains examined are indicated in the figure. B. A uls1 rad54 nucleus 6 hours after irradiation. Staining is shown as follows: Rad51 (Red), RPA (Green), Colocalized Rad51-RPA (Yellow), DAPI-stained DNA (blue). Note several Rad51-RPA colocalizing foci, as well as Rad51 foci that are immediately adjacent to RPA foci. The large structure staining with both Rad51 and RPA in the upper right corner of the DAPI staining area was seen in a significant fraction of uls1 rad54 and uls1 rdh54 nuclei at 4 and 6 hours. These structures were not included in the focus counts. Bar = 2µM.
RPA colocalization with Rad51 foci following gamma irradiation was about 50% (Gasior et al., 2001; Figure 5B, Figure S6, Table S2). An additional 20–25% lay immediately adjacent to an RPA focus, as expected for Rad51 filaments associated with ongoing recombinational repair (Figure 5A, Figure S6, Table S2). Thus the total fraction of RPA associated Rad51 foci was about 70–75%. The 50% of foci showing overlap are likely to include adjacent filaments that are not lying in the focal plane and/or those at closer distances than can be resolved by this method (<250nM). This high level of RPA association is in sharp contrast to the nearly complete absence of RPA association seen for Rad51 foci caused by translocase deficiency in the absence of induced damage (Figure 2, Figure S3).
Discussion
Synthetic dosage lethality identifies Rdh54 as critical in preventing Rad51 complex accumulation
Synthetic dosage lethality has been used to identify a diverse set of interacting proteins (Kroll et al., 1996). In contrast to synthetic lethal phenotypes from combining loss of function mutations, a synthetic dosage lethality interaction studies factors required to keep the toxic effects of overexpression of a non-mutant wild type protein in check. Synthetic dosage lethality can identify functional interactions between two proteins not revealed by standard synthetic lethal tests (Measday et al., 2005).
Although other studies found little effect of overexpressing Rad51 (Ira et al., 2003; Milne et al., 1995; Paffett et al., 2005), we find that under conditions where overexpression results in accumulation of Rad51 complexes, chromosome stability and plasmid retention are compromised. These findings reflect the existence of mechanisms that down-regulate deleterious Rad51 activity. The slightly negative phenotypes of wild type become lethal when the Rdh54 translocase is absent, showing that Rdh54 counters spontaneous Rad51 complexes. Further highlighting the importance of Rdh54 is the observation that genetic conditions that result in lowered Rdh54 levels, such as expression of both mating types in haploid cells, show synthetic dosage lethality with RAD51 overexpression.
Rad51 forms non-productive complexes that can be disassembled by Rdh54
Most cytologically detectable Rad51foci form at the sites of ssDNA tracts associated with DNA breaks (Bishop, 1994). When RDH54 is absent, or present at limiting concentration, overexpression of RAD51 results in focus formation at chromosomal sites that lack colocalization of the ssDNA-specific protein RPA. We will refer to these structures as non–damage-associated foci.
Rad51 spontaneous non–damage-associated foci can also be detected in rdh54 mutants that are not overexpressing RAD51. Such foci may be fully analogous to the Dmc1 foci observed in rdh54 spo11 diploids during meiosis (Holzen et al., 2006). Rad51, like Dmc1, shows almost no binding preference for ssDNA versus dsDNA in vitro (Hong et al., 2001; Ogawa et al., 1993). The finding that Rad51 does not form cytologically detectable non–damage-associated complexes on chromatin in wild type mitotic cells suggests that one or more cellular mechanisms actively prevents the accumulation of Rad51 on chromatin. Although the absence of Rad51 foci in wild type cells could in principle reflect a block to Rad51 association with chromatin or a process that promotes removal of Rad51, removal is mostly likely at play. First, Rdh54 promotes dissociation of the Rad51 relative Dmc1 in vivo (Holzen et al., 2006). Second, Rdh54 and Rad54 promote dissociation of Rad51 from dsDNA in purified systems (Chi et al., 2006; Solinger et al., 2002).
These previous findings, coupled with the overexpression results presented here, make a strong argument that Rdh54 can remove Rad51 from non–damage-associated complexes in yeast. The mutant phenotype shows that failure to remove Rad51 from non–damage-associated complexes is deleterious. Previous interpretations of HR mutant lethalities that are suppressed by loss of Rad51 protein have focused the formation of lethal HR intermediates. We suggest that in addition to such lethal intermediates, accumulation of Rad51 on chromatin at undamaged sites can also cause chromosome loss and block mitotic growth, through a disruption of replication or segregation.
Three Swi2-Snf2-related translocases have redundant activities in Rad51 focus removal
Despite finding that Rdh54 can remove non–damage-associated Rad51 foci, we observed only modest focus accumulation in rdh54 single mutants at normal levels of Rad51 expression. This can be contrasted with rad54 or uls1 single mutants in which no Rad51 accumulation is detected. These findings suggest that Rdh54 plays the predominant role in Rad51 dissociation in wild type cells and that Rad54 and Uls1 become more important only when Rdh54 is absent. Rad54 and Uls1 can substitute for Rdh54, but it is necessary to remove both of these proteins to see accumulation of non-damage associated Rad51 foci above the levels seen in the absence of Rdh54 alone.
Rad54 is critical for removal of damage-associated Rad51 foci. Rdh54 also participates in removal of damage-associated foci, although more foci are removed when Rdh54 is absent than when Rad54 is absent. These findings, compared to the results obtained without the induction of damage, suggest that Rad54 and Rdh54 have evolved distinct functions in Rad51 removal, with Rdh54 being most proficient in removal of non–damage-associated complexes and with Rad54 being most proficient in removal of damage-associated Rad51 complexes (Solinger et al., 2002). Our findings are also consistent with those of Boulton and colleagues who recently reported partial redundancy of translocase functions for removal of Rad51 from sites of DSBs during meiosis in C. elegans (Ward et. al 2010).
The finding that Rdh54 is required for removal of Rad51 from sites of damage was somewhat surprising as neither an rdh54 mutant or an rdh54 uls1 double mutant shows strong sensitivity to induced damage. The relative similarity of Rad51 focus removal phenotype in rdh54 mutants compared to rad54 mutants can be reconciled in different ways. First, Rad54 may be capable of completely clearing Rad51 foci, but its ability to do so may take longer than the duration of the experiments reported here. Second, there may be a qualitative difference between the foci that accumulate in the absence of Rad54 compared to those that accumulate in the absence of Rdh54. It is possible that the Rad51 structures that persist in the absence of Rdh54 do not preclude completion of recombination because the critical sites for polymerase loading are cleared of Rad51 by Rad54. This possibility is supported by biochemical evidence (Li and Heyer, 2009). Alternatively, removal of Rad51 foci might not be essential for cell survival; some other function of Rad54, such as stabilizing nascent homology-dependent interactions or clearing nucleosomes may be critical (Alexeev et al., 2003; Petukhova et al., 1998; Sugawara et al., 2003; Zhang et al., 2007). In such a case, translocase-mediated Rad51 removal could be important for resumption of growth and chromosome stability following completion of repair. Further studies are required to better define translocase functions in Rad51 removal.
A shared mechanism for recombinase removal from non–damage associated complexes
Current data indicate that non-functioning Rad51 and Dmc1 foci are actively removed from duplex DNA by ATP hydrolysis-driven mechanisms that are closely related (this study; (Holzen et al., 2006)). The use of Rdh54 and its relatives in disassembly of non-damage-associated complexes was proposed to be a consequence of the mechanism of strand exchange reactions, which are driven by stability of the filament bound to the duplex product of the reaction (Holzen et al., 2006).
Swi2/Snf2-like translocases may not be the only proteins involved in preventing the accumulation of non–damage-associated complexes. Recent biochemical studies have shown that fragments of the human BRCA2 (hBRCA2) protein can block binding of human RAD51 (hRAD51) to dsDNA in a purified system (Carreira et al., 2009; Shivji et al., 2009). This finding led to the proposal that full-length hBRCA2 functions to dissociate hRAD51 from dsDNA in vivo. However, there is no evidence that non– damage-associated hRAD51 foci accumulate in hBRCA2 defective cells; the hRAD51 foci observed in BRCA2−/− colocalize with RPA (Hatanaka et al., 2005; Tarsounas et al., 2003). Thus, if full-length hBRCA2 does act to prevent association of hRAD51 with dsDNA, its activity may be redundant with other mechanisms.
Non–damage-associated Rad51 complexes and tumorigenesis
A number of studies have shown that human RAD51 mRNA and protein are expressed at high levels in tumor cells (Klein, 2008). High expression levels are associated with both increases and decreases in DNA repair efficiency and can result in aberrant repair events that alter the genome (Richardson et al., 2004). Thus, improperly regulated, hRAD51-mediated repair events have been hypothesized to contribute to tumorigenesis. The work described here raises the new possibility that formation of non–damage-associated hRAD51 complexes drives tumorigenesis. It is worth noting that high hRAD51 levels have been found to be associated with non–damage-dependent hRAD51 foci in tumor cell nuclei (Raderschall et al., 2002). Thus, it will be interesting to determine the extent to which genome instability in tumorigenesis is caused by limited translocase activity.
Experimental Procedures
Media, growth conditions and genetic methods
Standard yeast growth medium and growth conditions were used. Details can be found in the supplemental data.
Yeast strains
All strains used are listed in Table S1.
Protein expression, extraction and western blotting
Standard methods were used. Details of cell growth can be found in the supplemental data.
Chromosome fragment loss
Chromosome fragment loss was determined as the number of red/white half-sectored colonies divided by the total colony number as described previously (Christman et al., 1988).
Chromosome loss rates
Fluctuation tests were performed as described previously in detail (Klein, 2001).
Nucleoid spreading and immunofluorescence microscopy
Preparation of spread yeast nuclei and immunostaining of the slides was performed as described previously (Bishop, 1994; Gasior et al., 1998).
Supplementary Material
Acknowledgements
We thank Patrick Sung, Lorraine Symington, Wolf Heyer and Akira Shinohara for strains, plasmids and antibodies. This work was supported by NIH grants GM50936 to DKB and GM53738 to HLK
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Kwon Y, Seong C, Epshtein A, Lam I, Sung P, Klein HL. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J Biol Chem. 2006;281:26268–26279. doi: 10.1074/jbc.M602983200. [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Fink GR. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Eggleston AK, Kowalczykowski SC. An overview of homologous pairing and DNA strand exchange proteins. Biochimie. 1991;73:163–176. doi: 10.1016/0300-9084(91)90199-b. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Olivares H, Ear U, Hari DM, Weichselbaum R, Bishop DK. Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc Natl Acad Sci U S A. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka A, Yamazoe M, Sale JE, Takata M, Yamamoto K, Kitao H, Sonoda E, Kikuchi K, Yonetani Y, Takeda S. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol Cell Biol. 2005;25:1124–1134. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzen TM, Shah PP, Olivares HA, Bishop DK. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 2006;20:2593–2604. doi: 10.1101/gad.1447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EL, Shinohara A, Bishop DK. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem. 2001;276:41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics. 2001;157:557–565. doi: 10.1093/genetics/157.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Pellicioli A, Malkova A, Foiani M, Haber JE. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr Biol. 2001;11:1053–1057. doi: 10.1016/s0960-9822(01)00296-2. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD. RAD54 controls access to the invading 3'-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:638–646. doi: 10.1093/nar/gkn980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang XP, Solinger JA, Kiianitsa K, Yu X, Egelman EH, Heyer WD. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V, Baetz K, Guzzo J, Yuen K, Kwok T, Sheikh B, Ding H, Ueta R, Hoac T, Cheng B, et al. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc Natl Acad Sci U S A. 2005;102:13956–13961. doi: 10.1073/pnas.0503504102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Kowalczykowski SC. Biochemical properties of the Escherichia coli recA430 protein. Analysis of a mutation that affects the interaction of the ATP-recA protein complex with single-stranded DNA. J Mol Biol. 1990;211:845–855. doi: 10.1016/0022-2836(90)90078-Z. [DOI] [PubMed] [Google Scholar]
- Milne GT, Ho T, Weaver DT. Modulation of Saccharomyces cerevisiae DNA double-strand break repair by SRS2 and RAD51. Genetics. 1995;139:1189–1199. doi: 10.1093/genetics/139.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Paffett KS, Clikeman JA, Palmer S, Nickoloff JA. Overexpression of Rad51 inhibits double-strand break-induced homologous recombination but does not affect gene conversion tract lengths. DNA Repair (Amst) 2005;4:687–698. doi: 10.1016/j.dnarep.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Sung P, Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 2002;62:219–225. [PubMed] [Google Scholar]
- Raschle M, Van Komen S, Chi P, Ellenberger T, Sung P. Multiple interactions with the Rad51 recombinase govern the homologous recombination function of Rad54. J Biol Chem. 2004;279:51973–51980. doi: 10.1074/jbc.M410101200. [DOI] [PubMed] [Google Scholar]
- Richardson C, Stark JM, Ommundsen M, Jasin M. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene. 2004;23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Schmuckli-Maurer J, Rolfsmeier M, Nguyen H, Heyer WD. Genome instability in rad54 mutants of Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:1013–1023. doi: 10.1093/nar/gkg190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Gasior SL, Bishop DK, Shinohara A. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc Natl Acad Sci U S A. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, Klenerman D, Venkitaraman AR. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc Natl Acad Sci U S A. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signon L, Malkova A, Naylor ML, Klein H, Haber JE. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- Ward JD, Muzzini DM, Petalcorin MI, Martinez-Perez E, Martin JS, Plevani P, Cassata G, Marini F, Boulton SJ. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol Cell. 2010;37:259–272. doi: 10.1016/j.molcel.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Fan HY, Goldman JA, Kingston RE. Homology-driven chromatin remodeling by human RAD54. Nat Struct Mol Biol. 2007;14:397–405. doi: 10.1038/nsmb1223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.