Abstract
Saliniketals A and B are unusual polyketides from the marine actinomycete Salinispora arenicola that inhibit ornithine decarboxylase induction. The structural similarities between the saliniketals and the ansa chain of the potent rifamycin antibiotics, which co-occur in the fermentation broth, suggest a common origin between the two compound classes. Using PCR-directed mutagenesis, chemical complementation studies, and stable isotope feeding experiments, we showed that the saliniketals are byproducts of the rifamycin biosynthetic pathway diverging at the stage of 34a-deoxyrifamycin W. Our results suggest that a single enzyme, the cytochrome P450 monooxygenase encoded by sare1259, catalyzes multiple oxidative rearrangement reactions on 34a-deoxyrifamyin W to yield both the saliniketal and rifamycin structural classes.
Introduction
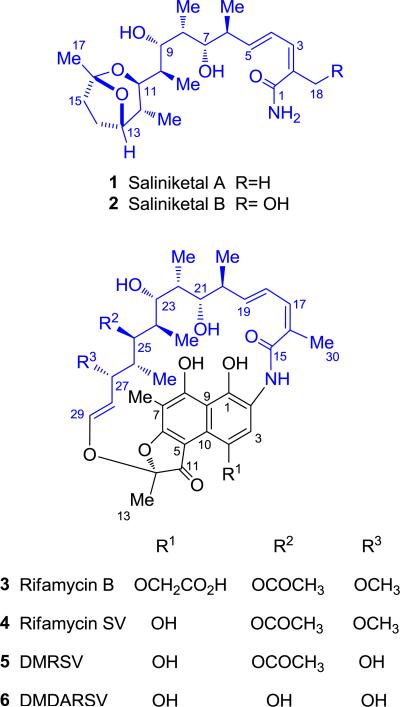
Saliniketals A (1) and B (2), originally isolated by Fenical and co-workers in 2007, are novel bicyclic polyketides produced by the obligate marine actinomycete Salinispora arenicola (Figure 1).1 These inhibitors of ornithine decarboxylase induction,1 an effective treatment strategy for cancer chemoprevention,2 harbor unusual structural features that have inspired three total syntheses to date.3-5 The saliniketals contain an unprecedented 1,4-dimethyl-2,8-dioxabicyclo[3.2.1]octane ring system with a polyketide side-chain at C11 that terminates in a primary amide. The global structure of 1 and 2 is also notable because of the striking similarity, including the exact stereochemistry, to the ansa chain of the potent rifamycin antibiotics (3–6), which co-occur in the fermentation broth (Figure 1).1 While the rifamycin polyketides are assembled from the aromatic starter unit 3-amino-5-hydroxybenzoic acid (AHBA) with two acetate and eight propionate extender units,6 the presence of the primary amide on the saliniketals suggested that they are not biosynthetic shunt products nor degradation products of rifamycin because cleavage of the C–N bond of the AHBA-derived aromatic starter unit was unlikely.1 Instead, the saliniketals were proposed to originate from a three carbon starter unit that is extended by a disparate polyketide synthase (PKS) with two acetate and five propionate units to form the ketide with the exact stereochemistry as in 3–6.1 However, prior to this study, this hypothesis had not been explored experimentally.
Figure 1.
Structures of the saliniketals (1–2) and rifamycins (3–6) from S. arenicola CNS-205. The numbering of 1–6 is consistent with prior publications.1,42
The rifamycins were first identified by Sensi and co-workers in 1959 from a terrestrial actinomycete that would eventually be classified as Amycolatopsis mediterranei.6,7 In 2006, Kim et al. first reported the production of rifamycins B (3) and SV (4) from a strain of Salinispora isolated from an Australian marine sponge.8 Later, Jensen and co-workers showed that the Salinispora exhibit species-specific secondary metabolite production trends in which saliniketal and unspecified rifamycins along with the staurosporines were observed in all examined strains of S. arenicola, thus comprising the S. arenicola core chemotype.9 Bioinformatic analysis of the Palauan S. arenicola CNS-205 genome and polymerase chain reaction (PCR) based screening experiments confirmed genes predicted to be involved in the biosynthesis of rifamycins.8,10 The high sequence identity between genes in the S. arenicola rifamycin cluster (SA-rif) and genes from the well characterized A. mediterranei S699 rifamycin biosynthetic gene cluster (AM-rif) suggests that the rif locus has undergone horizontal gene transfer.10
In the present study, we explored the biosynthetic relationship between the rifamycins and saliniketals by a multidisciplinary approach involving bioinformatic analysis, in vivo mutagenesis, chemical complementation, and stable isotope incorporation studies. In doing so, we probed the following questions: Does the structural similarity between the saliniketals and rifamycins originate from a common biosynthetic machinery? If so, are the saliniketals shunt products of the rifamycin polyketide assembly or molecules selectively generated from a rifamycin-type biosynthetic precursor by a distinct enzyme in the pathway? Herein we report that the saliniketals are unexpected products of the central rif pathway intermediate 34adeoxyrifamycin W (7, see Figure 4) in which the cytochrome P450 monooxygenase (CYP) Sare1259 is responsible for dual oxidative rearrangement reactions that lead to the formation of the mature rifamycins 3–6 and the truncated saliniketals 1–2.
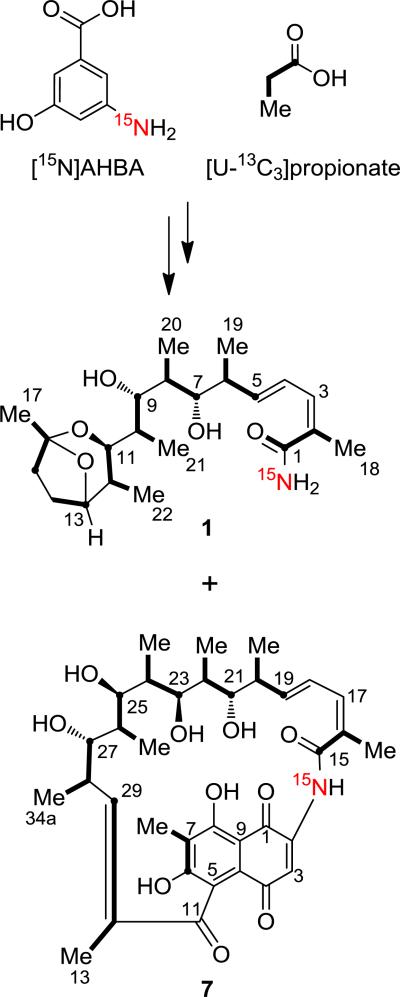
Figure 4.
Stable isotope labeling pattern observed for 1 and proposed for 7 (see reference 43) from the incorporation of [15N]AHBA and [U-13C3]propionate.
Results
Chemical analysis of S. arenicola CNS-205 and A. mediterranei S699
Organic extracts of S. arenicola CNS-205 and A. mediterranei S699 were analyzed by liquid chromatography/mass spectrometry (LC/MS) to directly compare their rif chemistry with known saliniketal and rifamycin chemical standards. In addition to 1 and 2, S. arenicola CNS-205 produces a mixture of rifamycin SV (4), 27-O-demethyl-25-O-desacetylrifamycin SV (5) and 27-O-demethylrifamycin SV (6) when cultured in A1 production media (Figure 1). Amycolatopsis mediterranei S699, on the other hand, primarily produces rifamycin B (3).11 After a closer inspection of extracts from the fermentation broth of A. mediterranei S699 grown in YMG liquid media, we learned that 1 and 2 are not unique to S. arenicola but are also minor components of the original rifamycin producer (Figure S1, Supporting Information). The co-production of both compound classes by these distantly related actinomycetes suggested that the saliniketals and rifamycins may arise from a common biosynthetic pathway.
Bioinformatic analysis of the S. arenicola CNS-205 genome
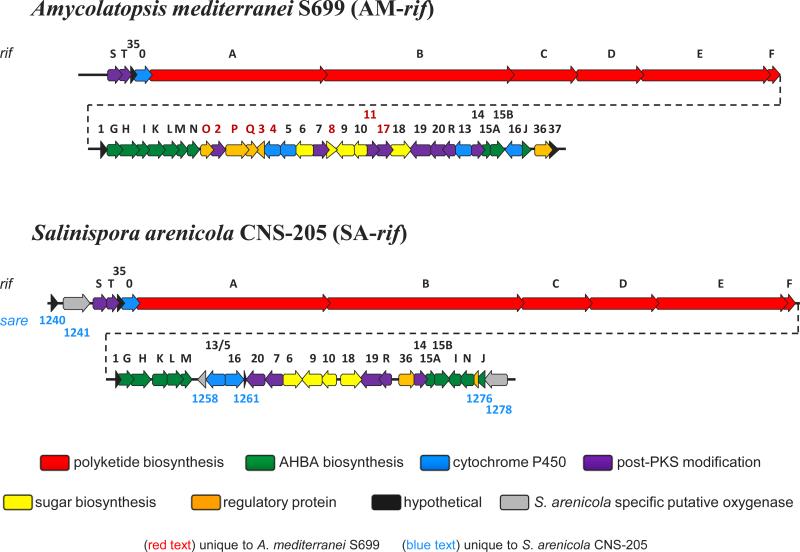
In early 2007, the completion of the S. arenicola CNS-205 genome sequencing project yielded a 5,786,361 bp genome (CP000850) with at least 30 distinct secondary metabolite gene clusters.10 Initially excluding the rifamycin biosynthetic gene cluster (SA-rif), none of the remaining twelve polyketide synthase (PKS)-associated pathways had a domain architecture consistent with the predicted assembly of the saliniketals. We therefore were left with no other viable option but the 92 kb SA-rif cluster for saliniketal biosynthesis. The SA-rif pathway is comprised of 39 open reading frames (ORFs), 33 of which are homologous to genes in the AM-rif cluster with identities >50% (Table 1). The SA-rif cluster is organized similarly to the AM-rif cluster with a distinct 10 module PKS operon (rifA–E), an AHBA biosynthetic sub-cluster, and a 24 kb tailoring and regulation region (Figure 2). All genes determined essential for the biosynthesis of rifamycin B in A. mediterranei S699 share a homolog in the SA-rif cluster.6,12-17 There are, however, nine genes in the tailoring region of the AM-rif cluster that do not have homologs in the SA-rif cluster (rifO, rif-orf2, rifP, rifQ,rif-orf3, rif-orf4, rif-orf8, rif-orf11, rif-orf17, and rif-orf37) and conversely, the SA-rif cluster contains six genes without counterparts in AM-rif (sare1240, sare1241, sare1258, sare1261, sare1276, and sare1278). The unique S. arenicola genes are annotated as a multicopper oxidase (sare1241), a Rieske-domain protein (sare1258), a FAD-dependent oxidoreductase (sare1278), two hypothetical proteins (sare1240 and sare1276), and a ferrodoxin-domain containing protein (sare1261). The regions upstream and downstream of the SA-rif locus also differ from the AM-rif cluster in that none of the transport, membrane protein, ribosomal protein, or RNA polymerase genes are shared. Homologs of many of these genes are, however, located elsewhere in the S. arenicola genome (data not shown). Interestingly, SA and AM rif share genes related to aminodeoxysugar biosynthesis (sare1263–7 corresponding to rif-orf7, rif-orf6, rif-orf9, rif-orf10, and rif-orf18, respectively) while no glycosylated rifamycins have been reported from either strain. The co-production of the closely related glycosylated compound, tolypomycin Y, with rifamycins B (3) and O by Amycolatopsis tolypomycina 18 suggests that these genes may be silent in SA-rif, as proposed for the aminodeoxysugar locus in AM-rif.19
Table 1.
Summary of SA-rif cluster and chemotypes of genetically mutated strains.
| SA-rif gene | annotation | AM-rif homolog | % identity | chemotype of mutant |
|---|---|---|---|---|
| Sare_1240 | hypothetical protein | N/A | N/A | |
| Sare_1241 | multicopper oxidase type 2 | N/A | N/A | |
| Sare_1242 * | oxidoreductase domain protein | rifS | 76 | No production of 1–8 |
| Sare_1243 * | oxidoreductase domain protein | rifT | 57 | No production of 1–8 |
| Sare_1244 | hypothetical protein | orf35 | 59 | |
| Sare_1245 * | cytochrome P450 | orf0 | 82 | No production of 1–8 |
| Sare_1246 | AMP-dependent synthetase and ligase | rifA | 72 | |
| Sare_1247 | beta-ketoacyl synthase | rifB | 73 | |
| Sare_1248 | beta-ketoacyl synthase | rifC | 71 | |
| Sare_1249 | beta-ketoacyl synthase | rifD | 69 | |
| Sare_1250 | acyltransferase | rifE | 72 | |
| Sare_1251 | N-acetyltransferase | rifF | 68 | |
| Sare_1252 | hypothetical protein | orf1 | 50 | |
| Sare_1253 | 3-dehydroquinate synthase | rifG | 79 | |
| Sare_1254 | 3-deoxy-7-phosphoheptulonate synthase | rifH | 66 | |
| Sare_1255 * | AHBA synthase | rifK | 86 | No production of 1–8 |
| Sare_1256 | oxidoreductase domain protein | rifL | 79 | |
| Sare_1257 | putative phosphatase | rifM | 83 | |
| Sare_1258 | rieske [2Fe-2S] domain protein | N/A | N/A | |
| Sare_1259 * | cytochrome P450 | orf5 | 71 | 7 |
| Sare_1260 * | cytochrome P450 | orf16 | 73 | WT |
| Sare_1261 | protein of unknown function DUF1271 | N/A | N/A | |
| Sare_1262 * | acetyltransferase, PapA5 | orf20 | 58 | 1, 2, & 6 |
| Sare_1263 * | protein of unknown function DUF1205 | orf7 | 66 | WT |
| Sare_1264 * | DegT/DnrJ/EryC1/StrS aminotransferase | orf6 | 86 | WT |
| Sare_1265 * | aminotransferase class-III | orf9 | 79 | WT |
| Sare_1266 * | oxidoreductase domain protein | orf10 | 70 | WT |
| Sare_1267 * | NDP-hexose 2,3-dehydratase | orf18 | 79 | WT |
| Sare_1268 | monooxygenase FAD-binding | orf19 | 74 | |
| Sare_1269 | type II thioesterase | rifR | 70 | |
| Sare_1270 | regulatory protein LuxR | orf36 | 64 | |
| Sare_1271 | methyltransferase type 11 | orf14 | 65 | |
| Sare_1272 * | transketolase domain protein | orf15A | 80 | No production of 1–8 |
| Sare_1273 * | transketolase central region | orf15B | 70 | No production of 1–8 |
| Sare_1274 * | shikimate dehydrogenase | rifI | 55 | WT |
| Sare_1275 | ROK family protein | rifN | 53 | |
| Sare_1276 | DoxX family protein | N/A | N/A | |
| Sare_1277 | 3-dehydroquinate dehydratase | rifJ | 77 | |
| Sare_1278 | FAD-dependent oxidoreductase | N/A | N/A |
indicates genes inactivated in this study, Abbreviations: (WT) wild-type chemistry observed (compounds 1–2 and 4–6).
Figure 2.
Comparison of the rifamycin biosynthetic gene clusters from A. mediterranei S699 (AF040571) and S. arenicola CNS-205 (CP000850).
Inactivation of the AHBA synthase (rifK) and the transketolase (rif-orf15)
The bioinformatic analysis of the SA and AM rif clusters did not support the previously proposed mechanism for saliniketal formation involving a distinct three-carbon primer unit intercepting the rifamycin synthase in lieu of the aromatic precursor AHBA. Since the saliniketals are not unique to S. arenicola and are also produced by A. mediterranei, we explored the biosynthetic scenario that the saliniketals are directly related to the rifamycins as either shunt or degradative products. In order to initially probe their putative biosynthetic relationship, we employed a PCR-directed gene replacement methodology to disrupt the AHBA synthase encoding gene rifK (sare1255) with an apramycin resistance cassette (acc(3)IV) as described previously for cyclomarin biosynthesis in S. arenicola.20 As predicted, inactivation of rifK in S. arenicola completely abolished the production of all rifamycins. In addition, the saliniketals were also not produced in the rifK mutant (rifK::aprR), providing unequivocal evidence that these natural products share a common biosynthetic origin from AHBA (Figure 3A–B). This conclusion was further substantiated through the successful chemical complementation of rifK::aprR with synthetically prepared AHBA that restored both saliniketal (1–2) and rifamycin (3–6) production to wild-type levels (Figure 3C). Similar results were also observed when we inactivated, with a single replacement, the SA-rif transketolase A (sare1272) and B (sare1273) subunits corresponding to rif-orf15A and rif-orf15B from AM-rif, respectively (Figure S2, Supporting Information). Based on in vitro reconstitution of genes involved in AHBA biosynthesis with the E. coli transketolase (tktA),21 Orf15 is predicted to catalyze the conversion of 3-amino-3-deoxy-D-fructose-6-phosphate to imino-erythrose-4-phosphate during the biosynthesis of AHBA.21-23 To the best of our knowledge, these are the first experimental data to confirm the function of Orf15 in vivo.
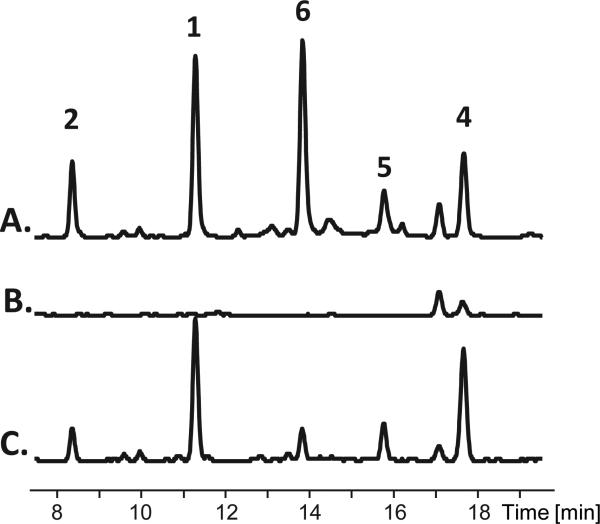
Figure 3.
HPLC-MS analysis of the inactivation the S. arenicola AHBA synthase (rifK, sare1255) and chemical complementation of resulting mutant (rifK::aprR). Extracted ion chromatogram (EIC) for masses corresponding to the m/z of the sodium adducts of the saliniketals (1–2) and rifamycins (4–6) from Salinispora arenicola (A) wild-type, (B) rifK::aprR, and (C) rifK::aprR mutant chemically complemented with synthetically prepared AHBA (25 mM).
To explore whether the nitrogen atom of saliniketal's primary amide originates from AHBA or from an alternate source, we synthesized and then administered [15N]AHBA to production cultures of the rifamycin deficient mutant rifK::aprR. We observed by MS similar isotopic enrichments of the saliniketal and rifamycin biosynthetic products as in the synthetic [15N]AHBA precursor of 25 atom% 15N, thereby confirming that the primary amide nitrogen of the saliniketals is derived from the “unlikely”1 C–N bond cleavage of the AHBA unit (Figure 4). These observations also suggested that 1 and 2 are not shunt products formed during polyketide extension but rather originate from a mature rifamycin macrocyclic product since the priming AHBA nitrogen atom ultimately resides at the terminating carbonyl C1 group of saliniketal.
Identification of genes directly involved in saliniketal production
Once we established that 1–6 share a single post-PKS assembly biosynthetic precursor, we methodically inactivated genes in the SA-rif cluster with homologous representatives in AM-rif to determine which are involved with the production of 1 and 2 and to elucidate the point at which the rifamycin and saliniketal pathways diverge. We inactivated four genes with predicted functions based on characterization or inactivation of their corresponding AM-rif homologs (sare1245, sare1259, sare1260, andsare1262 corresponding to rif-orf0, rif-orf13/rif-orf5, rif-orf16, and rif-orf20, respectively).
We first explored rif-orf20, which encodes an acetyltransferase that catalyzes the acetylation of the C25 hydroxyl of 6 to form 5.16 Based on an early mechanistic proposal for the formation of 1, we envisioned the assimilation of the 25-O-acetyl unit of 4 or 5 into the bicyclic ring of 1 at C16/C17. Deletion of sare1262 yielded a mutant that accumulated the desacetylated 6 without any effect on the production of 1 and 2, suggesting that the pathways must diverge prior to the formation of 6. This result was corroborated through an isotope experiment with [U-13C]propionate that enriched 1 with six intact propionate units and one incomplete propionate unit at C13/C14 (Figure 4) corresponding to the formal loss of C34a of 34a-deoxyrifamycin W (7). Importantly, C16/C17 of 1 originate from a propionate unit versus acetate as we originally contemplated.
We next turned our attention to the other three genes with characterized AM-rif homologs (sare1245, sare1259, and sare1260) that code for CYPs. Previous inactivation of the CYP rif-orf16 in A. mediterranei S699 produced a mixture of 3 and 4,13 and upon further analysis, we showed that this mutation had no effect on the production of 1 and 2. Similarly, inactivation of the S. arenicola homolog, sare1260 (73% identity), did not alter the production of 1, 2, and 4–6.
The CYP encoded by sare1245 shares 82% identity with rif-orf0, and unlike the other rif CYPs, they are similarly positioned directly upstream of the rifamycin PKS gene rifA rather than in the downstream region that primarily harbors the tailoring and regulatory genes (Figure 3). Lee et al. suggest that the AM rif-orf0 CYP is responsible for the introduction of the C34a hydroxyl group in rifamycin W (8, Figure 5a) based on their observation that inactivation of rif-orf0 in A. mediterranei resulted in a mutant that no longer produced rifamycin B yet accumulated proansamycin X.24 Our attempt to inactivate the homologous sare1245 resulted in a mutant that no longer produced either the rifamycins or saliniketals. Subsequent chemical complementation of the sare1245 mutant (sare1245::aprR) with AHBA failed to restore saliniketal/rifamycin production. However, complementation of sare1245::aprR with 7 rescued the production of the saliniketals and rifamycins, suggesting that the gene product of sare1245 is not involved in the post-PKS modification of rifamycin nor the biosynthesis of 1 and 2 (Figure S3, Supporting Information). Identical results were observed for the inactivation of the rifS (sare1242) and rifT (sare1243) homologs, which are located immediately upstream of sare1245 on the same putative transcriptional operon. This suggested that inactivation of rif-orf0, rifS, and rifT produced a polar effect on the transcription of the rifamycin polyketide genes, rifA–F, directly downstream of sare1245, thus resulting in the observed chemotype for all respective mutant strains.
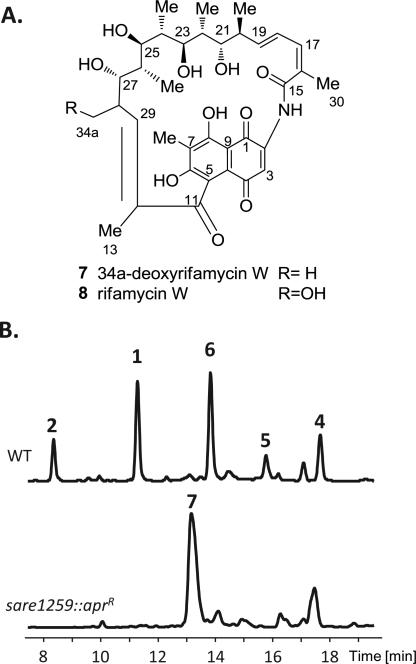
Figure 5.
Inactivation of the cytochrome P450, sare1259. (A) Structure of rifamycin biosynthetic intermediates 34a-deoxyrifamycin W (7) and rifamycin W (8). (B) EIC for masses corresponding to the m/z of the sodium adducts of the saliniketals (1–2) and rifamycins (4–8) produced by S. arenicola wild-type and sare1259::aprR strains, showing the loss of saliniketal (1–2) production and accumulation of 7 in the mutant.
The third and final SA-rif CYP sare1259 is homologous to two AM-rif CYPs, namely orf13 and orf5, at 79% and 71% identity, respectively (Table 1). Xu et al. showed, using in vivo mutagenesis, that the gene product of rif-orf13 in A. mediterranei S699 is not involved in rifamycin B (3) production, while inactivation of rif-orf5 yielded a mutant that accumulated the biosynthetic intermediate rifamycin W (8, Figure 5a) in lieu of 3.13 The authors concluded that the CYP encoded by rif-orf5 catalyzes the multistep oxidative conversion of 8 to 6.13 Further analysis of the AM-rif-orf5 mutant in this study, however, revealed that the mutant also produced 1, 2, 4, and 6 as minor components (Figure S4, Supporting Information). With this knowledge in hand, we genetically inactivated sare1259 in S. arenicola. The resulting mutant (sare1259::aprR) not only lost the biosynthetic ability to produce rifamycins 4–6, as expected, but significantly the saliniketals as well (Figure 5b). This suggested that the CYP encoded by sare1259 is a key enzyme in the biosynthesis of 1 and 2 in S. arenicola and confirms our hypothesis that the saliniketals are products of an enzymatic cleavage of a rifamycin macrocyclic intermediate versus a shunt product of the polyketide elongation. Interestingly, sare1259::aprR accumulated an unexpected product that was confirmed by NMR analysis to be the rifamycin biosynthetic intermediate 34a-deoxyrifamycin W (7) instead of the predicted product 8. The presence of two homologs to sare1259 in the AM-rif pathway may account for the accumulation of 8 in the AM-rif-orf5 mutant, where the CYP encoded by rif-orf13 may hydroxylate C34a of 7, and perhaps even perform the conversion of 7 to 1, 2, and 6.
The mechanism proposed for the function of the CYP encoded by rif-orf5 involves the oxidative cleavage of the C12/C29 double bond followed by rearrangement of the ansa chain to form 6 from 8.13 If Sare1259 also performs the same oxidative cleavage of the C29/C12 olefin to yield 1, then C16/C17 of 1 would have to come from an alternate 2-carbon source such as the acetate unit in 5 that we previously ruled out. Analysis of the [U-13C3]propionate-labeled 1, however, unequivocally showed that C15/C16/C17 originates from an intact propionate molecule, thereby suggesting that this 3-carbon unit is most likely derived from C12/C13/C29 in 7 (Figure 4). Consequently, these data suggest that Sare1259 has dual function to oxidatively cleave 7 at the C12/C29 olefin to yield the rifamycins and at the C11/C12 bond to give the saliniketals. The biosynthesis of 2 may similarly proceed from a 30-hydroxyrifamycin W precursor, which was previously observed in a recombinant strain of A. mediterranei,25 to account for its C18 hydroxyl group.
In order to determine if any gene products other than the CYP encoded by sare1259 are also necessary for the biosynthesis of 1, an additional eight genes (sare1242, sare1243, sare1263–sare1267, and sare1274), for which no function has been either confirmed or evaluated in A. mediterranei (Table 1), were inactivated in S. arenicola. While inactivation of sare1242 (rifS) and sare1243 (rifT) created putative polar effects on the downstream PKS resulting in the total loss of rifamycin/saliniketal biosynthesis, inactivation of the six remaining genes in either S. arenicola CNS-205 (this study) or A. mediterranei S699 (see ref. 13) had no effect on the biosynthesis of 1–8 (Figure S2, Supporting Information).
Chemical complementation of the rifK mutant with rifamycin macrocyclic intermediates
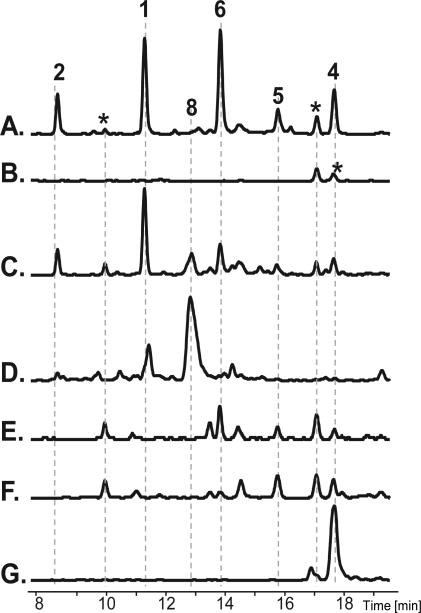
In order to confirm 7 as an immediate precursor to the saliniketals, we complemented cultures of rifK::aprR with successive biosynthetic intermediates from the rifamycin pathway 7→8→6→5→4. Complementation with both 7 and 8 were able to restore production of the saliniketals, while intermediates 4–6 did not rescue their production (Figure 6). While the complementation of rifK::aprR with 8 was able to restore saliniketal production, the bioconversion of 8 to 1 and 2 does not appear to be as efficient as the conversion of 7 to 1 and 2 (Figure 6C and D). These findings support our mutagenesis results indicating that the saliniketals are synthesized during the bioconversion of 7 to 6 by Sare1259 with 8 as a potential intermediate. The intermediates that were not able to rescue saliniketal production are presumed to be taken up by the cells based on the observation that the early biosynthetic intermediates can undergo bioconversion into later stage rifamycins (Figure 6C–E).
Figure 6.
HPLC-MS analysis of the chemical complementation of rifK::aprR with rifamycin biosynthetic intermediates. EIC for all masses corresponding to the m/z of the sodium adducts of the saliniketals (1–2) and rifamycins (3–8) for (A) wild-type S. arenicola CNS-205, (B) rifK::aprR negative control (C) rifK::aprR complemented with 7, (D) rifK::aprR complemented with 8, (E) rifK::aprR complemented with 6, (F) rifK::aprR complemented with 5, and (G) rifK::aprR complemented with 4. (* indicates unrelated compounds that share an m/z ratio with one of the compounds 1–8)
Discussion
The great structural diversity of naturally occurring rifamycins is largely due to the extensive post-PKS enzymatic tailoring carried out by enzymes encoded by pathway specific genes. After more than 50 years of study, the rifamycin pathway continues to reveal new and unusual biosynthetic potential. The saliniketals are another example of how enzymatic tailoring of natural products by CYPs can produce novel chemical structures through complex chemical reactions.
CYPs are known to catalyze a multitude of diverse oxidative modifications (e.g. hydroxylation, epoxidation, oxidative bond cleavage, or dehydrogenation) on a wide range of molecular substrates, often activating or detoxifying the chemical species.26-28 The CYP encoded by sare1259 is most closely related to rif-orf13 and rif-orf5 from the AM-rif cluster as well as other CYPs belonging to the CYP107 family. This superfamily includes a large number of bacterial CYPs, many of which are associated with xenobiotic degradation and secondary metabolite tailoring.29-32
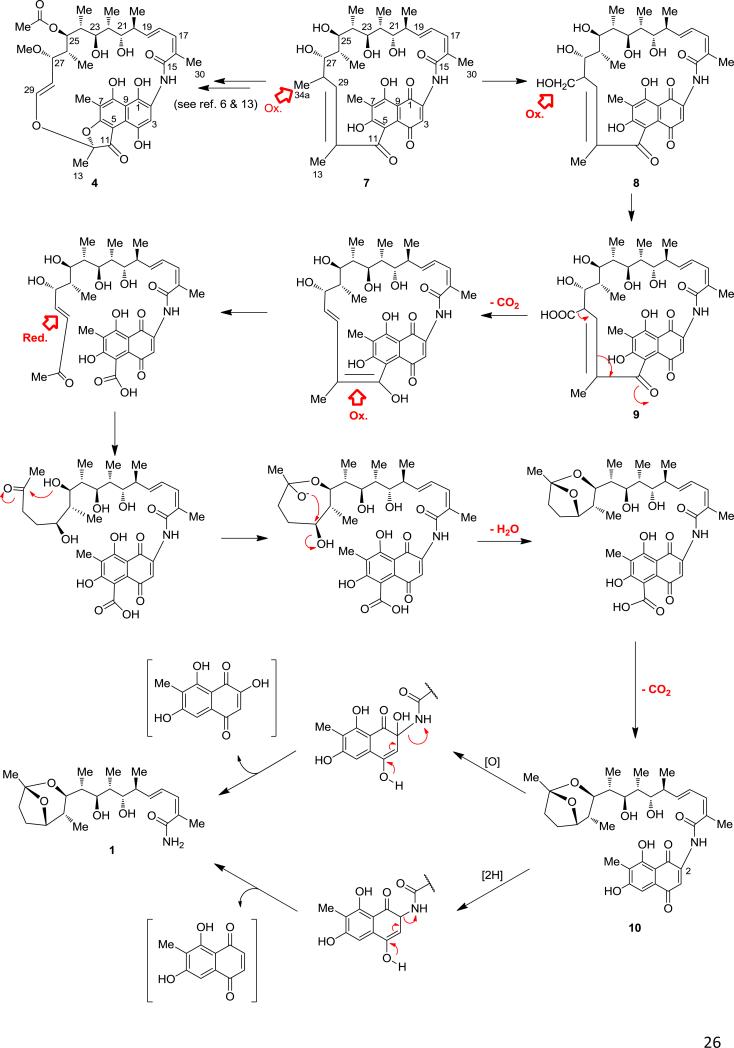
Our data suggest that Sare1259 catalyzes multiple oxidations of 7 at variable positions for its conversion to either the saliniketals or rifamycins. During the biosynthesis of 3, C34a of 7 is proposed to be sequentially oxidized twice and then lost via a decarboxylation of the resulting carboxyl.13,33 We hypothesize that the timing of this decarboxylation reaction determines whether the final product of the SA-rif pathway yields the rifamycins (3–6) or the saliniketals (1–2). The current mechanism proposed for the function of the CYP encoded by rif-orf5 in A. mediterranei involves the oxidative cleavage of the C12/C29 double bond of 7 followed by formation of the ketal of 6 and decarboxylation of the C34a carboxyl group.13 In contrast, for the production of 1 and 2, we propose that the decarboxylation reaction occurs prior to oxidative bond cleavage, shifting the C12/C29 olefin to the C11/C12 position where it is then cleaved in a similar fashion by Sare1259 (Figure 7). Reduction of the resulting C28/C29 olefin and rearrangement of the ansa chain would form the 5-carboxy intermediate of the salinisporamycin A (10), a recently described compound isolated from S. arenicola YM23-082.34 Our proposed mechanism is further supported by the [U-13C]propionate tracer experiments and by the isolation of (5-carboxy)salinisporamycin A from fermentations of S. arenicola (J. B. MacMillan, personal communication). The isolation of 10 by Matsuda and co-workers also suggests the sequence of transformation from 7 to 1 proceeds with the formation of the bicyclic ketal prior to the construction of the primary amide. Mechanisms for this latter reaction may involve an oxidation of the quinone at C2 or reduction of the quinone to the hydroquinone followed by a retro-Michael cleavage of the C–N bond to yield 1 and the respective naphthoquinone (Figure 7). Attempts to identify and isolate the resulting naphthoquinone moiety have thus far been unsuccessful. Additionally, we did not identify a rif pathway gene associated with the putative conversion of (5-carboxy)salinisporamycin A to 1, which may suggest that if this reaction is indeed catalyzed by a dedicated enzyme, that it may be encoded outside the rif cluster.
Figure 7.
Proposed mechanism of saliniketal biosynthesis facilitated by CYP Sare1259 (intermediates with published structures are numbered).1,25,33,34
In conclusion, we took a multidisciplinary approach involving a combination of genomics, in vivo mutagenesis, chemical complementation, stable isotope incorporation studies, and chemical analyses to interrogate the biosynthetic origin of the saliniketals. Such a multifaceted approach was required to effectively explore the biosynthesis of the saliniketals that ultimately led to the identification of the Sare1259 CYP as the key biosynthetic gene product that governs the diversification of the rif pathway at the stage of 34a-deoxyrifamycin W (7) leading to the truncated saliniketals and mature rifamycins (Figure 7). We are presently probing the multifunctional Sare1259 to explore its preferred substrates, its reaction profile, and its molecular mechanism that likely will lead to new P450 catalyzed chemistry.
Experimental Section
General Methods
Low-resolution LC/MS was carried out on a Hewlett-Packard series 1100 LC/MS system in positive ion mode with a linear gradient of 10–90% MeCN at a flow rate of 0.7 ml/min over 24 min on a RP C18 column (Phenomenex Luna, 4.6 mm × 100 mm, 5 μm). 1H, heteronuclear multiple bond correlation (gHMBC) and heteronuclear single quantum coherence (gHSQC) NMR spectral data were obtained on a Bruker DRX600 spectrophotometer equipped with a 1.7 mm cryoprobe. 13C-NMR spectral data were obtained on a Varian VX-500 instrument equipped with an XSens™ cold probe. Rifamycin SV (4) sodium salt was purchased from Sigma Aldrich (St. Louis, MO). Saliniketal A (1) was graciously provided by William H. Fenical.1 Rifamycin W (7), 27-O-demethyl-24-O-desacetylrifamycin SV (6, DMDARSV), and 27-O-demethylrifamycin SV (5, DMRSV) were isolated and confirmed based on literature precedence.13,16,17
Bacterial strains, culture conditions, and extraction of natural products
Salinispora arenicola strain CNS-205 and Amycolatopsis mediterranei strains S699, MT45025H, and MT1601KH were obtained as described previously.13,20 All S. arenicola seed cultures were grown in A1 liquid media (10 g starch, 5 g yeast extract, and 2 g peptone per liter of seawater) and production cultures were grown in A1BFeC liquid media (10 g starch, 5 g yeast extract, 2 g peptone, 100 mg KBr, 40 mg Fe2(SO4)3·4H2O, 1 g CaCO3 per 1 L seawater) at 28°C and 225 rpm.20 Amycolatopsis mediterranei S699 seed and production cultures were both grown in YMG liquid media35 at 28°C and 225 rpm, unless otherwise noted. Extraction of saliniketal and rifamycin compounds was achieved by acidifying the culture broth to pH 2–3 with 1 N HCl and extracting the cultures 3X with equal volumes of EtOAc. The organic fraction was dried under vacuum, resuspended in MeOH, and analyzed by LC/MS. Escherichia coli strains EPI300, BW25113/pKD2036 and S17-137 used for mutagenesis experiments were grown in Luria-Bertani (LB) media with appropriate antibiotics.
Genetic Manipulations
Inactivation of rifamycin and saliniketal biosynthetic genes was performed using REDIRECT© PCR targeting technology as described previously for S. arenicola.20,38 E. coli EPI300 (Epicentre) carrying fosmids BPPW5227, BPAF1230, and BPAF1361 were provided by the Joint Genome Institute, Walnut Creek, CA. Each of the genes targeted for inactivation was replaced with an apramycin resistance (acc(3)IV) cassette by double crossover homologous recombination on the fosmid containing the gene of interest. The mutant fosmid was then transformed into E. coli S17-1 and transferred to S. arenicola CNS-205 via conjugation. Exconjugates were confirmed by PCR analysis and restriction digest.
Isolation of 34a-Deoxyrifamycin W (7) from the sare1259::aprR Mutant
The sare1259::aprR mutant was cultured in 8 × 1 L Fernbach flasks of A1BFeC media for 7 days at 28°C while shaking at 225 rpm. XAD-7 resin (30 g) was added to each flask on day 7 and continued to shake for 5 hours before collecting the resin by filtration. The resin was then extracted with 3 × 1 L of acetone and concentrated under vacuum. The residue was separated by silica gel flash chromatography under isocratic conditions with MeOH/CHCl3 (1:9). Silica fractions were further purified by HPLC on a RP C18 column (Phenomenex Luna, 10 mm × 250 mm, 5 μm, MeCN/H2O 56:44, v/v).
34a-Deoxyrifamycin W (7)
HPLC ESI-MS: [M+Na]+ 662.3; 1H NMR (600 MHz, DMSO-d6) δ 12.40 (s, 8-OH), 10.54 (br s, 6-OH), 9.59 (s, NH), 7.47 (s, H-3), 6.32 (dd, J = 15.5, 11.2 Hz, H-18), 6.16 (d, J = 10.8 Hz, H-17), 5.99 (dd, J = 15.6, 6.9 Hz, H-19), 4.76 (d, J = 6.5 Hz, 23-OH), 4.35 (s, 21-OH), 4.31 (d, J = 5.8 Hz, 27-OH), 3.84 (d, J = 5.6 Hz, H-27), 3.80 – 3.73 (m, H-25), 3.68 (d, J = 6.5 Hz, 25-OH), 3.35 – 3.28 (m, H-23), 2.40 (dd, J = 15.1, 7.3 Hz, H-28), 2.20 (dd, J = 14.6, 7.2 Hz, H-20), 2.13 (s, H-14), 2.00 (s, H-30), 1.89 (s, H-13), 1.65 (br d, J = 6.8 Hz, H-22), 1.59 – 1.51 (m, H-24), 1.25 – 1.15 (m, H-26), 0.95 (d, J = 7.0 Hz, H-34a), 0.91 (d, J = 6.9 Hz, H-32), 0.81 (d, J = 6.8 Hz, H-31), 0.55 (d, J = 6.7 Hz, H-33), 0.18 (d, J = 6.9 Hz, H-34).
Chemical Complementation of S. arenicola Mutants
Chemical complementation studies were carried out in 50 ml cultures of the S. arenicola rifK::aprR mutant in A1 production media. Compounds 4–8 (2–6 mg) were individually added in triplicate to 1 day old production cultures of rifK::aprR. The cultures were then allowed to grow for 4 more days before being extracted as described above. Bioconversion of the rifamycin intermediates was monitored by HPLC-MS. Retention times, m/z ratios, and UV profiles of the biotransformation products were compared to authentic standards.
Synthesis of 15N-AHBA
AHBA was synthesized by reacting 3,5-dihydroxybenzoic acid (2.0 g, 13 mmol) with NH4Cl (1.7 g, 32 mmol) and aqueous NH4OH (14.8N, 6 mL, 89 mmol) in a sealed high-pressure reaction tube at 180°C for 40 hours.39,40 After cooling, the volatile components of the reaction mixture were removed in vacuo and the remaining solid redissolved in 100 mL 6N HCl. The resulting solution was heated under reflux for 16 hours, filtered, extracted with EtOAc (3 × 40 mL) to remove unreacted starting material, and concentrated to 25 mL. The desired product crystallized from this solution yielding grey crystals, which were collected and recrystallized from 6N HCl to furnish pure AHBA hydrochloride as white crystals in 68% overall yield (1.67 g, 8.8 mmol). The [15N]AHBA was prepared following the same synthetic route but substituting NH4Cl with 15NH4Cl. The statistically expected 15N-enrichment of 25% in the product was verified by mass spectrometric analysis. 1H NMR (500 MHz, CD3OD) δ 7.04 (m, 1H), 7.48 (m, 1H), 7.51 (m, 1H). HPLC ESI-MS: AHBA [M+H]+ 154.1; [15N]AHBA [M+H]+ 154.1 (75%), 155.2 (25%).
Stable Isotope Labeling Experiments of 1
[U-13C3]Propionate incorporation experiments were carried out in 2 × 1 L cultures of S. arenicola CNS-205 grown in A1BFeC liquid media at 225 rpm and 30°C for 7 days. 50 mg/L of sodium [U-13C3]propionate (Cambridge Isotopes Laboratories, Incorporated) was aseptically added to each liter of culture after 48 hrs. The cultures were extracted with EtOAc, and 1 was isolated as described previously1 and analyzed by 13C-NMR (Figure S5 and S6, Supporting Information).
[15N]AHBA incorporation studies were carried out in 50 mL cultures of rifK::aprR in A1BFeC at 225 rpm and 30°C for 5 days. 3 mg of [15N]AHBA was added to each culture after 24 hrs and extracted as described above on day five. Crude extracts were analyzed by HPLC-MS. Incorporation of 15N into AHBA and 1 were calculated from m/z.41 HPLC ESI-MS: saliniketal A (1) [M+Na]+ 418.3; [15N]saliniketal A (1) [M+Na]+ 418.2 (75%), [M+Na]+ 419.2 (25%).
Supplementary Material
Acknowledgment
We thank William H. Fenical and Paul R. Jensen for kindly providing authentic standards of saliniketal A (1) and S. arenicola CNS-205, respectively. We also thank Heinz G. Floss for his comments and assistance in the preparation of this manuscript. This work was generously supported by a grant from the NIH (GM085770 to B.S.M.) and a postdoctoral fellowship from the German Academic Exchange Service (DAAD) to T.A.M.G.
Footnotes
Supporting Information Available: HPLC-MS spectra for A. mediterranei S699 and rif-orf5 mutant strain, HPLC-MS spectra for SA-rif mutants generated in this study, HPLC-MS spectra for sare1245::aprR chemical complementation study, 13C NMR spectra for 13C-labeled 1, 1D and 2D NMR spectra (1H, gHSQC, gHMBC) for 7, table of primers use in this study, and complete Ref. 30. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Williams P, Asolkar R, Kondratyuk T, Pezzuto J, Jensen P, Fenical W. J. Nat. Prod. 2007;70:83. doi: 10.1021/np0604580. [DOI] [PubMed] [Google Scholar]
- 2.Pegg A, Shantz L, Coleman C. J. Cell. Biochem. 1995:59. doi: 10.1002/jcb.240590817. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, De Brabander JK. J. Am. Chem. Soc. 2009;131:12562. doi: 10.1021/ja9061757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paterson I, Razzak M, Anderson EA. Org. Lett. 2008;10:3295. doi: 10.1021/ol801148d. [DOI] [PubMed] [Google Scholar]
- 5.Yadav JS, Hossain SS, Madhu M, Mohapatra DK. J. Org. Chem. 2009;74:8822. doi: 10.1021/jo901913h. [DOI] [PubMed] [Google Scholar]
- 6.Floss HG, Yu TW. Chem. Rev. 2005;105:621. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 7.Sensi P, Margalith P, Timbal M. Farmaco Ed. Sci. 1959;14:146. [PubMed] [Google Scholar]
- 8.Kim TK, Hewavitharana AK, Shaw PN, Fuerst JA. Appl. Environ. Microbiol. 2006;72:2118. doi: 10.1128/AEM.72.3.2118-2125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Appl. Environ. Microbiol. 2007;73:1146. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Moore BS, Jensen PR. ISME J. 2009;3:1193. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim CG, Kirschning A, Bergon P, Zhou P, Su E, Sauerbrei B, Ning S, Ahn Y, Breuer M, Leistner E, Floss HG. J. Am. Chem. Soc. 1996;118:7486. [Google Scholar]
- 12.Floss H, Yu T. Curr. Opin. Chem. Biol. 1999;3:592. doi: 10.1016/s1367-5931(99)00014-9. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Wan E, Kim CJ, Floss HG, Mahmud T. Microbiology. 2005;151:2515. doi: 10.1099/mic.0.28138-0. [DOI] [PubMed] [Google Scholar]
- 14.Yu T, Muller R, Muller M, Zhang X, Draeger G, Kim C, Leistner E, Floss H. J. Biol. Chem. 2001;276:12546. doi: 10.1074/jbc.M009667200. [DOI] [PubMed] [Google Scholar]
- 15.Yu TW, Shen Y, Doi-Katayama Y, Tang L, Park C, Moore BS, Richard Hutchinson C, Floss HG. Proc. Natl. Acad. Sci., U. S. A. 1999;96:9051. doi: 10.1073/pnas.96.16.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y, Wu X, Mahmud T. ChemBioChem. 2005;6:834. doi: 10.1002/cbic.200400387. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Mahmud T, Floss HG. Arch. Biochem. Biophys. 2003;411:277. doi: 10.1016/s0003-9861(03)00004-3. [DOI] [PubMed] [Google Scholar]
- 18.Kishi T, Yamana H, Muroi M, Harada S, Asai M. J Antibiot (Tokyo) 1972;25:11. doi: 10.7164/antibiotics.25.11. [DOI] [PubMed] [Google Scholar]
- 19.August PR, Tang L, Yoon YJ, Ning S, Müller R, Yu T-W, Taylor M, Hoffmann D, Kim C-G, Zhang X, Hutchinson CR, Floss HG. Chem. Biol. 1998;5:69. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 20.Schultz AW, Oh DC, Carney JR, Williamson RT, Udwary DW, Jensen PR, Gould SJ, Fenical W, Moore BS. J. Am. Chem. Soc. 2008;130:4507. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 21.Guo J, Frost JW. J. Am. Chem. Soc. 2002;124:528. doi: 10.1021/ja016963v. [DOI] [PubMed] [Google Scholar]
- 22.Arakawa K, Muller R, Mahmud T, Yu TW, Floss HG. J. Am. Chem. Soc. 2002;124:10644. doi: 10.1021/ja0206339. [DOI] [PubMed] [Google Scholar]
- 23.Guo J, Frost JW. Org. Lett. 2004;6:1585. doi: 10.1021/ol049666e. [DOI] [PubMed] [Google Scholar]
- 24.Lee SK, Choi CY, Ahn JS, Cho JY, Park CS, Yoon YJ. J. Microbiol. Biotechnol. 2004;14:356. [Google Scholar]
- 25.Schupp T, Traxler P, Auden JA. J. Antibiot. (Tokyo) 1981;34:965. doi: 10.7164/antibiotics.34.965. [DOI] [PubMed] [Google Scholar]
- 26.O'Keefe D, Harder P. Mol. Microbiol. 1991;5:2099. doi: 10.1111/j.1365-2958.1991.tb02139.x. [DOI] [PubMed] [Google Scholar]
- 27.Meunier B, de Visser SP, Shaik S. Chem. Rev. 2004;104:3947. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 28.Guengerich FP. Chem. Res. Toxicol. 2008;21:70. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 29.Weber JM, Leung JO, Swanson SJ, Idler KB, McAlpine JB. Science. 1991;252:114. doi: 10.1126/science.2011746. [DOI] [PubMed] [Google Scholar]
- 30.Trefzer A, et al. Appl. Environ. Microbiol. 2007;73:4317. doi: 10.1128/AEM.02676-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujii Y, Kabumoto H, Nishimura K, Fujii T, Yanai S, Takeda K, Tamura N, Arisawa A, Tamura T. Biochem. Biophys. Res. Commun. 2009;385:170. doi: 10.1016/j.bbrc.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 32.Prior JE, Shokati T, Christians U, Gill RT. Appl. Microbiol. Biotechnol. 2010;85:625. doi: 10.1007/s00253-009-2135-0. [DOI] [PubMed] [Google Scholar]
- 33.Traxler P, Schupp T, Fuhrer H, Richter WJ. J. Antibiot. (Tokyo) 1981;34:971. doi: 10.7164/antibiotics.34.971. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda S, Adachi K, Matsuo Y, Nukina M, Shizuri Y. J. Antibiot. (Tokyo) 2009;62:519. doi: 10.1038/ja.2009.75. [DOI] [PubMed] [Google Scholar]
- 35.Kim CG, Yu TW, Fryhle CB, Handa S, Floss HG. J. Biol. Chem. 1998;273:6030. doi: 10.1074/jbc.273.11.6030. [DOI] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. Proc. Natl. Acad. Sci., U. S. A. 2000;97:6640. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon R, Priefer U, Puhler A. Bio-Technol. 1983;1:784. [Google Scholar]
- 38.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. Proc. Natl. Acad. Sci., U. S. A. 2003;100:1541. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker AM, Rickards RW, Brown RFC. Tetrahedron. 1983;39:4189. [Google Scholar]
- 40.Wang Z, Silverman RB. Bioorg. Med. Chem. 2006;14:2242. doi: 10.1016/j.bmc.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Biemann K. Mass spectrometry: organic chemical applications. McGraw-Hill; New York: 1962. [Google Scholar]
- 42.Oppolzer W, Prelog V, Sensi P. Experientia. 1964;20:336. doi: 10.1007/BF02171084. [DOI] [PubMed] [Google Scholar]
- 43.White RJ, Martinelli E, Lancini G. Proc. Natl. Acad. Sci., U. S. A. 1974;71:3260. doi: 10.1073/pnas.71.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.