Abstract
Background
The transport and catabolism of sialic acid, a critical virulence factor for nontypeable Haemophilus influenzae, is regulated by two transcription factors, SiaR and CRP.
Results
Using a mutagenesis approach, glucosamine-6-phosphate (GlcN-6P) was identified as a co-activator for SiaR. Evidence for the cooperative regulation of both the sialic acid catabolic and transport operons suggested that cooperativity between SiaR and CRP is required for regulation. cAMP was unable to influence the expression of the catabolic operon in the absence of SiaR but was able to induce catabolic operon expression when both SiaR and GlcN-6P were present. Alteration of helical phasing supported this observation by uncoupling SiaR and CRP regulation. The insertion of one half-turn of DNA between the SiaR and CRP operators resulted in the loss of SiaR-mediated repression of the transport operon while eliminating cAMP-dependent induction of the catabolic operon when GlcN-6P was present. SiaR and CRP were found to bind to their respective operators simultaneously and GlcN-6P altered the interaction of SiaR with its operator.
Conclusions
These results suggest multiple novel features for the regulation of these two adjacent operons. SiaR functions as both a repressor and an activator and SiaR and CRP interact to regulate both operons from a single set of operators.
Background
Sialic acid (5-N-acetylneuraminic acid, Neu5Ac) is used by nontypeable Haemophilus influenzae (NTHi) to assist in the evasion of the host innate immune response. Sialic acid is used to decorate the cell surface, primarily as the terminal non-reducing sugar on the lipooligosaccharride (LOS) and the biofilm matrix [1,2]. The presence of sialic acid on the cell surface protects the cell from complement-mediated killing, although the precise mechanism of this protection is unknown and may even vary among strains of NTHi [3-5]. Regardless, the acquisition and utilization of sialic acid is a crucial factor in the virulence of the majority of NTHi [3,4,6-8].
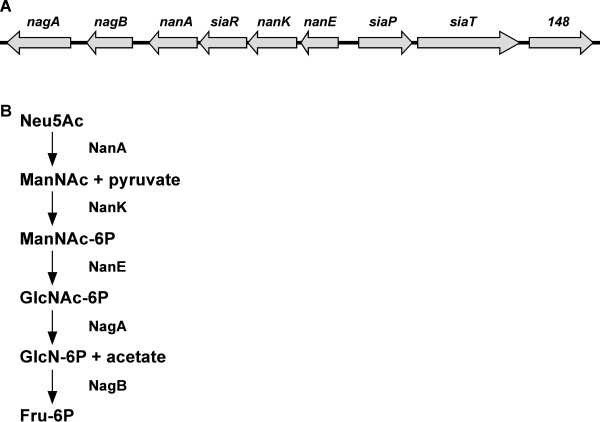
NTHi cannot synthesize sialic acid and therefore must scavenge it from the host. NTHi possess a high-affinity transporter for sialic acid, encoded by siaPT (also referred to as siaPQM) [6,9,10]. The SiaPT transporter is a member of the TRAP transporter family, with SiaP functioning as the solute-binding protein and SiaT functioning as the transmembrane transporter protein. An ortholog of the E. coli sialic acid mutarotase nanM is found downstream of the siaPT operon (HI0148) [11], although nanM does not appear to be co-transcribed with siaPT in H. influenzae strain Rd [12]. The genes required for the catabolism of sialic acid are found in the adjacent, divergently transcribed nan operon (Figure 1A). The genes of the nan operon encode all the enzymes required to convert sialic acid to fructose-6-phosphate (Figure 1B), which can then enter the glycolysis pathway [13]. Prior to the decoration of the cell surface, sialic acid must be activated by SiaB, the CMP-sialic acid synthetase, forming the nucleotide sugar donor used by sialyltransferases [4]. Once transported into the cell, sialic acid is either catabolized by the enzymes of the nan operon or activated by SiaB. Thus, these two pathways compete for the same substrate [13]. The organism must therefore maintain a balance between these two pathways, ensuring that a sufficient amount of sialic acid is available to decorate the cell surface and adequately protect the cell from the host immune response.
Figure 1.
The sialic acid catabolic and transport operons and pathway. A. Schematic diagram of the nan and siaPT operons. The nan operon encodes for the entire catabolic pathway and the transcriptional regulator SiaR. The siaPT operon encodes for the sialic acid transporter and YjhT, a sialic acid mutarotase. The accession numbers for the KW-20 Rd sequence are indicated below each gene. B. The sialic acid catabolic pathway.
Also present in the nan operon is the transcriptional regulator SiaR. SiaR is involved in the regulation of the nan and siaPT operons [14]. SiaR was found to repress the expression of both the siaPT and nan operons, thus regulating both transport and catabolism. Binding of SiaR to the intergenic region between these two operons was demonstrated and the region of DNA protected by SiaR was identified. As expected, it was found that inactivation of siaR lead to a reduction in surface sialylation, demonstrating the need to control the expression of sialic acid catabolism. In addition to SiaR, the cAMP receptor protein (CRP) was identified as a regulator of the siaPT operon, however a role in the regulation of the nan operon was not observed [12,14]. This is in part consistent with the observation that sialic acid is a cAMP-independent sugar [15]. In H. influenzae, CRP has been shown to regulate utilization of galactose, ribose, xylose, and fucose [15], in addition to regulating the development of competence [16].
We now report on the role of intermediates in the Neu5Ac catabolic pathway in SiaR-mediated regulation. Also, the potential interaction between SiaR and CRP was investigated. SiaR was found to utilize glucosamine-6-phosphate (GlcN-6P) as a co-activator in the presence of the CRP-cAMP complex. SiaR and CRP were found to act in a cooperative manner to regulate the expression of the divergent transporter and catabolic operons. Our results reveal a unique mechanism of regulation of two divergent operons regulated by two transcription factors from a single location.
Results
Promoter structure of the nan and siaPT operons
The transcriptional start sites of the nan and siaPT operons were identified using primer extension analysis. Primers that bound in the nanE and siaP open reading frames were used. Two major start sites were identified for the nan operon, 104 (TS-1nan) and 20 (TS-2nan) bp from the start codon of nanE (Figure 2A). The presence of additional minor bands may be the result of addional start sites or RNA degradation or processing. The analysis identified a single transcriptional start site (TS-1siaPT) 107 bp upstream of the start codon of siaP (Figure 2B). This organization leaves 140 bp in between TS-1nan and TS-1siaPT. The putative CRP binding site is located at -59 to -80 relative to TS-1siaPT and at -59 to -80 relative to TS-1nan (Figure 2C). This organization suggests that the siaPT promoter falls into the class I group of CRP-dependent promoters [17]. A consensus -10 sequence was identified for TS-1nan and was found to partially overlap the SiaR binding site, consistent with the role SiaR plays in repression of the nan operon. The relative location of TS-1nan to the SiaR operator, in addition to the identification of a consensus -10 box, suggests that this start site would be primarily involved in SiaR-mediated regulation, however, the relative contribution of the two nan promoters will need to be examined in more detail.
Figure 2.
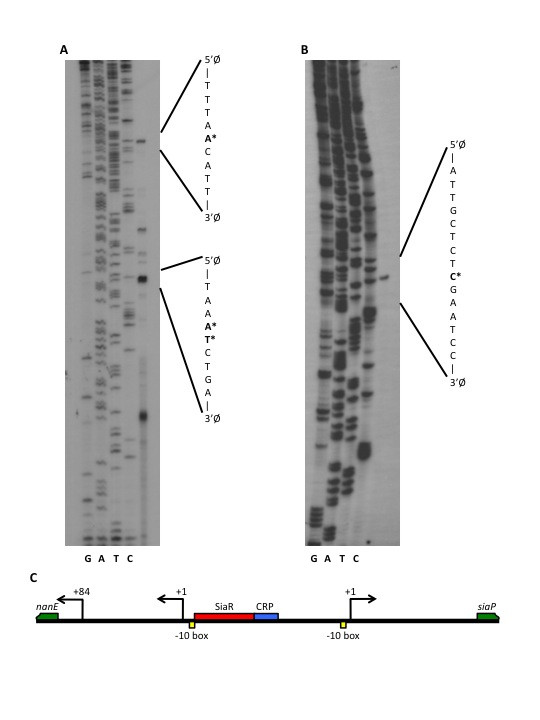
Primer extension analysis of the nan and siaPT operons. Complement of sequence surrounding the transcriptional start sites included with the sites indicated in bold and with asterisks. A. Primer extension analysis identified at least two major transcriptional start sites for the nan operon. Two bands were present for TS-2nan as indicated. B. Primer extension identified one start site for the siaPT operon. C. Schematic diagram of the nan and siaPT promoters. Binding sites for SiaR (red box) and CRP (blue box) are indicated as well as putative -10 boxes for TS-1nan and TS-1siaPT (yellow boxes).
Glucosamine-6-phosphate is a co-activator for SiaR
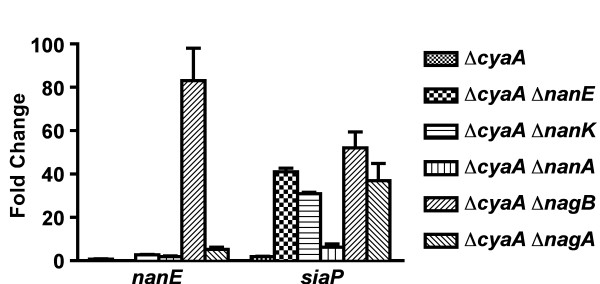
Previous studies found limited activation of SiaR-regulated operons by sialic acid [14]. The potential for intermediates in the sialic acid catabolic pathway to influence regulation by SiaR was explored. H. influenzae is unable to transport any of the intermediate sugars or phosphosugars of the sialic acid catabolic pathway [13,18], therefore a mutagenesis strategy was necessary. Each gene encoding an enzyme in the catabolic pathway was deleted in an adenylate cyclase (cyaA) mutant strain, resulting in a series of double mutants. The ΔcyaA mutant strain was used to allow for CRP to be activated only by the addition of cAMP in subsequent experiments. In each mutant, sialic acid can be catabolized, but the sugar or phosphosugar immediately upstream of the inactivated enzyme should accumulate (Figure 1B).
The mutants were grown to early exponential phase and then either sialic acid, cAMP, or both were added. Expression levels of nanE and siaP, the first genes of the catabolic and transport operons, respectively, were compared using real time quantitative RT-PCR (qRT-PCR). RNA from a culture that received neither sialic acid nor cAMP served as a reference for each experiment. When both sialic acid and cAMP were added to cultures, expression of nanE was only moderately affected in strains 2019ΔcyaA, 2019ΔcyaA ΔnanK, 2019ΔcyaA ΔnanA, and 2019ΔcyaA ΔnagA (0.7- to 5-fold change). The most striking change in nanE expression occurred in 2019ΔcyaA ΔnagB, with expression elevated 83-fold (Fig, 3). This mutant would be unable to convert GlcN-6P to fructose-6P, thus accumulating GlcN-6P. These results suggest that GlcN-6P is a major co-activator in SiaR-mediated regulation.
Figure 3.
Impact of metabolic impairment on the expression of nanE and siaP. 2019ΔcyaA and derived double mutants were grown in sRPMI. Sialic acid and cAMP were added 30 min prior to RNA extraction. Expression of nanE and siaP were measured by qRT-PCR. Results are presented as fold change relative to a culture that received neither sialic acid nor cAMP.
The regulation of siaP appears to be more complex. Expression of siaP was elevated 30- to 52-fold in strains 2019ΔcyaA ΔnanE, 2019ΔcyaA ΔnanK, 2019ΔcyaA ΔnagB, and 2019ΔcyaA ΔnagA (Figure 3). In contrast, increases of only 2- and 6-fold were observed in 2019ΔcyaA and 2019ΔcyaA ΔnanA, respectively (Figure 3). While SiaR can repress siaP expression [14], transcription of the transporter operon is more directly influenced by CRP. Despite this, siaP expression was not as responsive to cAMP in 2019ΔcyaA and 2019ΔcyaA ΔnanA. These results indicate that in these strains, SiaR is able to exert some control over siaP expression, however the mechanism in which this is accomplished is unclear.
SiaR and CRP interact to regulate the adjacent nan and siaPT operons
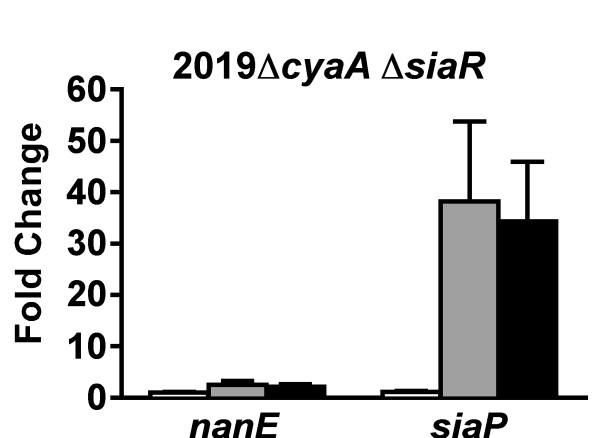
Previous work demonstrated that in a siaR mutant, CRP and cAMP are unable to influence nan operon expression [14]. Since the current studies were performed using different mutant constructs, the experiments were repeated with the double deletion mutant to confirm the previously observed phenotype. The 2019ΔcyaA ΔsiaR mutant was examined by qRT-PCR (Figure 4) and regardless of whether sialic acid or cAMP was added, expression of nanE did not change relative to the control. In the absence of SiaR, cAMP activated the expression of the siaPT operon, while the nan operon was unaffected.
Figure 4.
Expression of nanE and siaP in 2019ΔcyaA ΔsiaR. Cultures grown with sialic acid (open bars), cAMP (gray bars), and both sialic acid and cAMP (black bars) were compared to a reference culture that received neither.
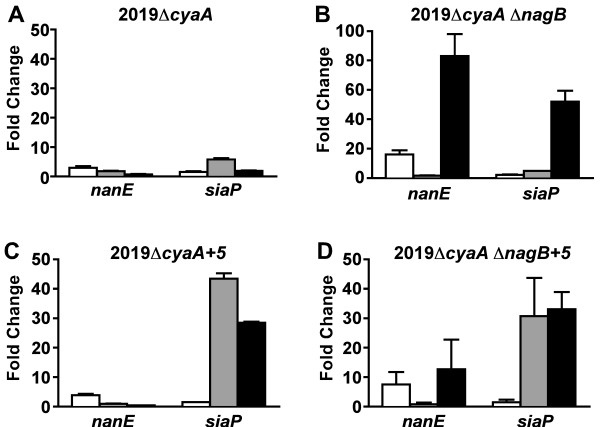
Examination of the results obtained from 2019ΔcyaA ΔnagB revealed a large change in the expression of nanE that was cAMP-dependent (Figure 5B). The addition of sialic acid alone (which would be converted to GlcN-6P) led to a 16-fold induction of nanE while the addition of cAMP alone had no effect. The addition of both sialic acid and cAMP resulted in an 83-fold induction of nanE, indicating that the combination of GlcN-6P and cAMP significantly increase the induction of the nan operon. These results provide evidence of cAMP-dependent activation of both the nan and siaPT operons. Since cAMP does not induce nanE expression in a siaR mutant, this suggests that cAMP-dependent activation of nanE requires SiaR. SiaR and CRP may physically interact to activate nan operon expression.
Figure 5.
Expression of nanE and siaP is altered by helical phasing. Expression of nanE and siaP in 2019ΔcyaA (A), 2019ΔcyaA ΔnagB (B), 2019ΔcyaA+5 bp (C), and 2019ΔcyaA ΔnagB+5 bp (D). Cultures grown with sialic acid (open bars), cAMP (gray bars), and both sialic acid and cAMP (black bars) were compared to a reference culture that received neither.
To demonstrate that SiaR and CRP interact to regulate the nan and siaPT operons, alteration of helical phasing was used. Alteration of helical phasing is accomplished by the insertion of one half turn to the helix between the SiaR and CRP operators. Briefly, 5 bp was inserted between the SiaR and CRP binding sites in strains 2019ΔcyaA and 2019ΔcyaA ΔnagB, resulting in strains 2019ΔcyaA+5 and 2019ΔcyaA ΔnagB+5, respectively. These strains were examined by qRT-PCR and the results were compared with those obtained from the parent strains.
In the 2019ΔcyaA ΔnagB background, altered helical phasing resulted in a steep reduction in nanE expression (from 83-fold in 2019ΔcyaA ΔnagB to 13-fold in 2019ΔcyaA ΔnagB+5) in the presence of both sialic acid and cAMP (Figures 5B and 5D). The induction level of nanE in the presence of sialic acid and cAMP was similar to the expression observed when sialic acid alone was added. The 5 bp insertion eliminated the cAMP-dependent activation of nanE that was observed in the 2019ΔcyaA ΔnagB strain.
In both the 2019ΔcyaA and 2019ΔcyaA ΔnagB backgrounds, altered helical phasing also resulted in the induction of siaP when cAMP was added (Figures 5A and 5C). In the 2019ΔcyaA+5 strain, the 5 bp insertion led to a 43-fold increase in siaP expression in the presence of cAMP (from 6-fold in 2019ΔcyaA) and a 29-fold increase (from 2-fold in 2019ΔcyaA) when both cAMP and sialic acid were present.
Taken together, these results indicate that altering the helical phasing succeeded in uncoupling SiaR- and CRP-mediated regulation of the nan and siaPT operons. It resulted in nanE expression becoming unresponsive to cAMP, much like it is in the 2019ΔcyaA ΔsiaR mutant. Altered helical phasing also prevented SiaR from exerting a negative influence on the expression of siaP. We conclude that the insertion eliminated the ability of SiaR and CRP to interact to regulate both the nan and siaPT operons.
SiaR and CRP bind to their respective operators simultaneously
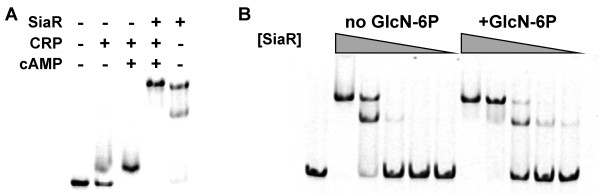
Binding of SiaR to an operator in the intergenic region between nanE and siaP was demonstrated previously [14]. The putative operator of CRP was identified in silico and was found to overlap the region protected by SiaR in a DNase I protection assay by three base pairs. The ability of both proteins to bind to their operators was examined using the electrophoretic mobility shift assay (EMSA). Both proteins were able to bind to a probe comprising the region between the two operons and CRP binding was dependent on the addition of cAMP (Figure 6A). When both proteins were included in the binding reaction, the DNA probe was shifted slightly higher than the SiaR-bound probe. This indicates that both proteins bind to their operators simultaneously, further supporting the hypothesis that the two regulators interact to regulate the adjacent nan and siaPT operons.
Figure 6.
Electrophoretic mobility shift assay. A. Binding of both SiaR and CRP to the nan-siaPT intergenic region. Both SiaR and CRP bind to the probe individually and CRP binding is dependent on the presence of cAMP. Both proteins bind the probe simultaneously as indicated by the higher shift of the probe when both proteins are added. B. GlcN-6P enhances binding of SiaR. Two-fold serial dilutions of SiaR were added to binding reactions in the absence and presence of 100 μM GlcN-6P. More probe was shifted when GlcN-6P was present.
GlcN-6P alters binding of SiaR to its operator
Many transcriptional regulators exhibit altered binding affinity for their operator sequences when a co-regulator is bound. To determine the effect of GlcN-6P on SiaR binding, EMSA was used. Serial dilutions of SiaR were incubated with DNA probes in the absence and presence of GlcN-6P (Figure 6B). In the presence of GlcN-6P, SiaR bound the probe and GlcN-6P slightly increased the binding affinity. While the presence of GlcN-6P did not result in a major change in the binding affinity of SiaR, the change in the shift does suggest that GlcN-6P is interacting with SiaR and impacting its ability to bind to its operator. Other phosphosugars of the sialic acid catabolic pathway (sialic acid, ManNAc, and GlcNAc-6P) nor GlcN-1P altered SiaR-binding (unpublished data) [14]. Taken together with the expression data, this demonstrates that GlcN-6P interacts with SiaR and has an effect on its DNA-binding properties. SiaR is not displaced from the DNA, but instead functions as an activator with GlcN-6P as a co-activator.
As in our previous studies [14], the binding of SiaR to the EMSA probe resulted in the appearance of two shifted bands (Figure 6). This was even more apparent when lower concentrations of SiaR were present in the binding reaction. The double shift is possibly caused by the binding of multiple SiaR proteins to the probe. This is a likely explanation, considering that the region protected by SiaR is large (53 bp) [14]. Further work will be necessary to determine the exact cause for the double shift.
GlcN-6P accumulates in a nagB mutant
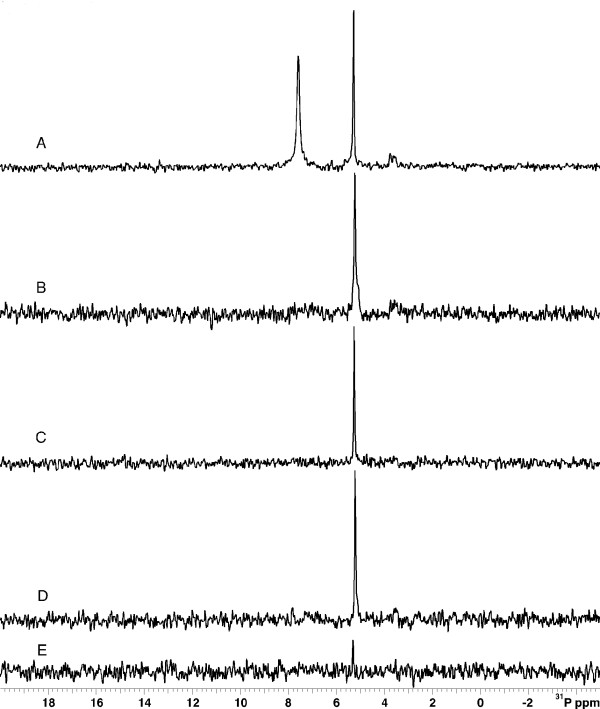
To confirm that Neu5Ac was transported and catabolized in the 2019ΔcyaA ΔnagB mutant strain, 31P NMR spectroscopy of intact cells was used. Cultures of wild-type 2019 and 2019ΔcyaA ΔnagB were grown to early exponential phase and cAMP and/or Neu5Ac were added and the 31P spectrum was obtained (Figure 7). A peak was detected near 5 ppm when cAMP was added to either strain. When Neu5Ac was added, a peak was detected near 7 ppm in the 2019ΔcyaA ΔnagB mutant that was absent in the wild-type strain. This peak was also absent in either strain when Neu5Ac was omitted. This indicated the accumulation of a significant amount of a phosphorylated compound in the mutant strain when exogenous Neu5Ac was present. Since the Neu5Ac catabolic pathway is blocked at NagB in the mutant strain, Neu5Ac would be converted to GlcN-6P, but not Fru-6P. Taken together with the interaction of GlcN-6P with purified SiaR, this indicates that GlcN-6P is accumulating in the 2019ΔcyaA ΔnagB mutant and is responsible for the activation of the nan operon.
Figure 7.
Detection of intracellular GlcN-6P by 31P NMR spectroscopy. 31P NMR spectra were obtained following the growth of cells in the presence of exogenous cAMP and/or Neu5Ac. A. 2019ΔcyaA ΔnagB with Neu5Ac and cAMP. B. 2019 wild-type with Neu5Ac and cAMP. C. 2019ΔcyaA ΔnagB with cAMP. D. 2019 wild-type with cAMP. E. 2019 wild-type without supplement.
Discussion
The importance of sialic acid in the protection of NTHi from the host immune response requires that most of the sialic acid transported into the cell is activated by SiaB and utilized for the decoration of the LOS and biofilm matrix. Therefore, it is important that the catabolism of sialic acid be tightly regulated. We previously identified SiaR as a repressor for these two operons, in addition to the role of CRP in activating the expression of the transporter [14]. In this study, we present data that expands on our previous work, providing key details about the unique regulation of these adjacent operons. The two operons required for the transport and catabolism of sialic acid were found to be simultaneously regulated by SiaR and CRP in a novel mechanism for cooperative regulation. SiaR functions as both a repressor and activator, utilizes GlcN-6P as a co-activator, and interacts with CRP to regulate two adjacent and divergently transcribed promoters.
Since H. influenzae cannot transport the intermediates of the sialic acid catabolic pathway [13,18], mutants in each gene of the pathway were used to examine the role of the sugar and phosphosugar intermediates in the expression of the SiaR-regulated operons. Increased expression of the nan operon in the 2019ΔcyaA ΔnagB double mutant suggested that GlcN-6P functions as a co-activator. This is unusual because catabolic pathways are typically regulated by the presence of the substrate. SiaR likely uses GlcN-6P as a co-activator because sialic acid is utilized rapidly after transport by H. influenzae, either by activation with SiaB or catabolism beginning with NanA. Thus, sialic acid never accumulates to levels that would allow for sufficient expression of the transporter. In contrast, using GlcN-6P allows for moderate activation of siaPT to provide for transport of sialic acid. Since GlcN-6P can also be synthesized by the cell, expression of the transporter is not reliant on the presence of high levels of sialic acid, while increased sialic acid and catabolism will elevate levels of GlcN-6P and increase expression of the nan and siaPT operons. Even though GlcN-6P is not an endpoint in the catabolic pathway, transient levels of the phosphosugar likely allow for sufficient expression of the two operons.
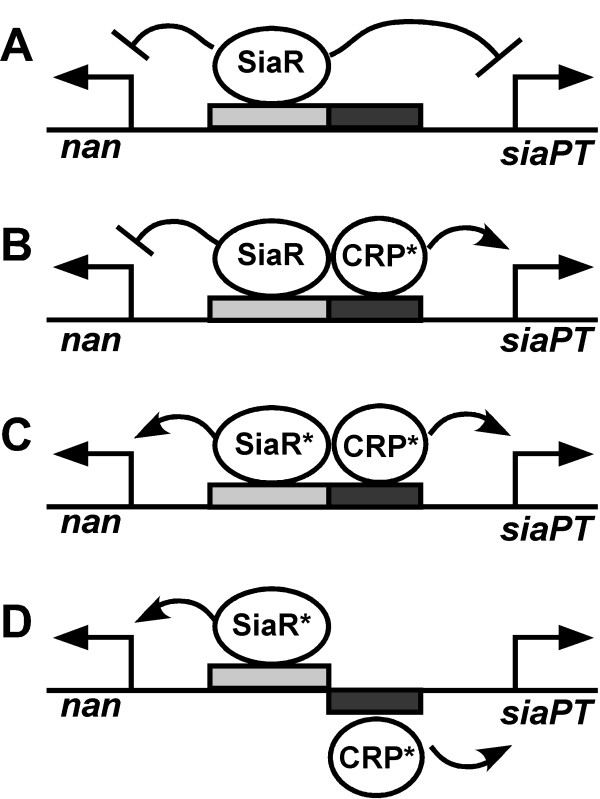
In addition to identifying GlcN-6P as a co-activator, we found that SiaR and CRP interact to regulate both the nan and siaPT operons. Both regulators were able to bind to their operators simultaneously, demonstrating that binding of one protein does not prevent the binding of the other. cAMP-dependent activation of nanE requires SiaR. Furthermore, regulation of the two operons was uncoupled by the insertion of one half-turn of DNA between the SiaR and CRP operators. This insertion resulted in the loss of SiaR influence on siaPT expression and the loss of nan induction by cAMP. Based on this data and the proximity of the two operators, it can be concluded that SiaR and CRP interact to impact the expression of the two operons. This interaction may be the result of direct contacts between the two regulators or cooperative effects on DNA topography, however we cannot make any conclusions on the mechanism at this time. Our results have led to the description of a novel regulatory mechanism for two regulators of adjacent operons. This model, in the context of the experiments carried out in this study, is displayed in Figure 8. SiaR by itself functions as a repressor of both the nan and siaPT operons (Figure 8A). When cAMP levels are elevated, the CRP-cAMP complex can bind to its operator and partially activate expression of the transport operon, but not the catabolic operon (Figure 8B). When both GlcN-6P and CRP-cAMP are present, an activating complex is formed with SiaR that induces expression of the two adjacent operons (Figure 8C). When the helical phase of the two operators is altered, SiaR can only regulate the nan operon while CRP can only regulate the siaPT operon (Figure 8D). Interaction between SiaR and CRP is necessary for regulation.
Figure 8.
Model of SiaR and CRP regulation of the nan and siaPT operons. A. In the absence of sialic acid and cAMP, SiaR is bound to its operator and expression of the nan and siaPT operons is repressed. B. When cAMP is present, CRP binds to its operator and is able to activate the siaPT operon, but not the nan operon. C. When both GlcN-6P and cAMP are present, SiaR and CRP are active and interact to form a complex that activates both the nan and siaPT operons. D. In helical phasing experiments, insertion of one half-turn in between the SiaR and CRP operators prevents the regulators from interacting and thus maximal activation of the nan operon is not achieved.
The interaction of CRP with another transcriptional regulator is not an unusual phenomenon, however the regulation of the adjacent nan and siaPT operons by CRP and SiaR appears to operate via a novel regulatory mechanism. What makes this regulatory region unique is that it appears that the two operons are regulated by one set of operators. Other examples of divergent operons regulated by CRP and additional regulators operate by distinctly different mechanisms. The most common mechanism is the formation of a repression loop. An example of this is in the glp regulon of E. coli [19]. As with the siaPT operon of NTHi, only one of the divergent glp operons is induced by CRP [19]. The difference between these two systems is that the repressor GlpR binds to four operators in the intergenic region and forms a repression loop [19]. The two divergent operons of the L-rhamnose catabolic regulon of E. coli utilize yet another mechanism. In addition to having multiple CRP binding sites, the two rha operons are regulated by separate transcriptional regulators, RhaR and RhaS [20]. RhaR and CRP interact to regulate the rhaSR operon while RhaS and CRP interact to regulate the rhaBAD operon [20,21].
SiaR shares functional similarity to NagC, the regulator of N-acetylglucosamine catabolism in E. coli. Like SiaR, NagC regulates the expression of nagA and nagB, as well as a number of additional genes. Also, SiaR and NagC both regulate divergently transcribed operons and CRP is involved in regulation [22,23]. There are a number of striking differences as well. GlcNAc-6P is the inducer of the NagC regulon. Just as inactivation of nagB causes induction of SiaR-regulated genes, the inactivation of nagA, and the subsequent accumulation of GlcNAc-6P, induces NagC-related genes [22]. NagC is displaced from its binding site in the presence of GlcNAc-6P [22] while SiaR appears to always be bound to its operator. In E. coli, the alteration of phasing between NagC operator sequences results in derepression of both divergently transcribed operons. This is due to the inability of NagC to form a repression loop that is required for NagC-mediated repression [24]. This differs significantly with what we observed in SiaR regulation. In our studies, the alteration of phasing did not result in derepression, but instead uncoupled SiaR- and CRP-mediated regulation of the nanE and siaP genes. The differences between SiaR and NagC suggest that, while some functional similarity exists between the two regulators, they both employ different mechanisms.
Given the nature of regulation by SiaR and CRP, the nan and siaPT operons will never be maximally expressed when H. influenzae is in its natural environment. This is due to a number of factors, including the low abundance of sialic acid in the host and the rapid utilization of intracellular sialic acid. Instead, regulation acts to subtly modulate expression of the operons, keeping expression under constant control so that catabolism does not outpace utilization and the expression of the transporter is appropriate for the availability of the ligand. These requirements are also in balance with the need to prevent the accumulation of inhibitory amounts of sialic acid, however, this need is likely minimal considering the factors of sialic acid availablity and utilization discussed above.
The role of CRP in the regulation of sialic acid transport and catabolism suggests that sialic acid is utilized as an emergency carbon source in the host. H. influenzae can use sialic acid as a sole carbon source as efficiently as glucose [10]. Sialic acid catabolism is not required for virulence as a nanA mutant exhibits increased fitness in multiple infection models [13]. However, the fact that catabolism is present and conserved among H. influenzae strains suggests that it provides some advantage to the organism. The previous study examining virulence of a nanA mutant was performed using an encapsulated, invasive type B strain rather than a non-typeable strain and did not test all possible environments within the host [13]. Additionally, intranasal mixed-challenge experiments did not reveal an advantage for either the wild-type or nanA mutant strain [13]. Therefore, it is possible that sialic acid catabolism is advantageous in certain conditions or has increased importance for non-typeable strains. The need for cell surface sialylation has been well established in multiple infection models for NTHi, but it is unclear at this time if and when catabolism is required.
Conclusions
GlcN-6P, an intermediate in the catabolism of sialic acid, was found to function as a co-activator of SiaR in the regulation of the catabolic and transport operons for sialic acid in NTHi. SiaR functions as both a repressor and an activator, depending on conditions, and is required for CRP-dependent activation of the catabolic operon. Direct interactions between SiaR and CRP are likely involved in regulation.
Methods
Bacterial strains, media and growth
The strains used in this study are listed in Table 1. E. coli was grown at 37°C in Luria-Bertani (LB) medium with or without agar (2%) and supplemented with antibiotics as needed. NTHi strain 2019 [25] and derivatives thereof were used in this study. H. influenzae was grown at 37°C in the presence of 5% CO2 on brain heart infusion agar (Difco Laboratories, Detroit, MI) supplemented with 10 μg/ml hemin and 10 μg/ml β-NAD (sBHI). Kanamycin-resistant H. influenzae were selected on sBHI agar containing 15 μg/ml ribostamycin in the absence of additional CO2. Spectinomycin was added to sBHI at a concentration of 25 μg/ml. RPMI 1640 media (Sigma-Aldrich, Saint Louis, MO) was used as a sialic acid-free chemically defined media. Supplemented RPMI (sRPMI) was prepared with protoporphyrin IX (1 μg/ml), hypoxanthine (0.1 mg/ml), uracil (0.1 mg/ml), β-NAD (10 μg/ml), and sodium pyruvate (0.8 mM). Neu5Ac (100 μM) and cAMP (1 mM) were added as indicated.
Table 1.
Strains and plasmids
| Strain or plasmid | Genotype, relevant phenotype or selection marker | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Invitrogen | |
| E. coli BL21 Star | Invitrogen | |
| NTHi 2019 | Clinical respiratory isolate | [25] |
| JWJ091 | NTHi 2019ΔcyaA mutant | This study |
| JWJ093 | NTHi 2019ΔcyaA ΔsiaR mutant, kanamycin resistant | This study |
| JWJ112 | NTHi 2019ΔcyaA ΔnanA mutant | This study |
| JWJ114 | NTHi 2019ΔcyaA ΔnagA mutant | This study |
| JWJ116 | NTHi 2019ΔcyaA ΔnagB mutant | This study |
| JWJ118 | NTHi 2019ΔcyaA ΔnanK mutant | This study |
| JWJ120 | NTHi 2019ΔcyaA ΔnanE mutant | This study |
| JWJ159 | NTHi 2019ΔcyaA mutant with 5 bp insertion between SiaR and Crp operators | This study |
| JWJ160 | NTHi 2019ΔcyaA ΔnagB mutant with 5 bp insertion between SiaR and Crp operators | This study |
| Plasmids | ||
| pGEM-T Easy | PCR-cloning vector | Promega |
| pGEM-T | PCR-cloning vector | Promega |
| pCR2.1 | PCR-cloning vector | Invitrogen |
| pCR2.1_443 | pCR2.1 with PCR fragment spanning | This study |
| pUC19 | General cloning vector | New England Biolabs |
| pET-24(+) | Expression vector | Novagen |
| pUC19-142del | nanA deletion construct | This study |
| pUC19-142sacB | nanA::tetR-sacB/kanR | This study |
| pJJ150 | siaR knockout vector | [14] |
| pJJ185 | C-terminal his-tagged SiaR expression vector | [14] |
| pJJ260 | tetR-sacB/kanR cassette | |
| pJJ276 | C-terminal his-tagged CRP expression vector | This study |
| pJJ279 | cyaA deletion construct | This study |
| pJJ290 | cyaA::tetR-sacB/kanR | This study |
| pJJ308 | nagA deletion construct | This study |
| pJJ309 | nagB deletion construct | This study |
| pJJ310 | nanK deletion construct | This study |
| pJJ311 | nanE deletion construct | This study |
| pJJ313 | nagA::tetR-sacB/kanR | This study |
| pJJ314 | nagB::tetR-sacB/kanR | This study |
| pJJ315 | nanK::tetR-sacB/kanR | This study |
| pJJ316 | nanE::tetR-sacB/kanR | This study |
| pJJ321 | pCR2.1_443 with 5 bp deletion | This study |
| pJJ331 | pJJ321 with tetR-sacB/kanR | This study |
Construction of mutants
Genetic manipulations were performed using established techniques. Restriction enzymes, Antarctic Phosphatase, and T4 polymerase were obtained from New England Biolabs (Beverly, MA) and were used following established protocols. The Expand High Fidelity PCR System (Roche Applied Science, Indianapolis, IN) was used for PCR reactions. Oligonucleotide primers were designed and ordered from Integrated DNA Technologies (Coralville, IA) and are listed in Table 2. Plasmids were transformed and maintained in MAX Efficiency DH5α Chemically Competent cells (Invitrogen). Competent H. influenzae cells were prepared using the M-IV method and transformed as described previously [26]. cAMP (1 mM final concentration) was added to cyaA mutant strains 30 minutes prior to the addition of DNA to aid in the development of competence.
Table 2.
Oligonucleotide primers
| Oligonucleotide primer | Sequence1 | Use |
|---|---|---|
| 140F3 | gaa ttc CTG CTT CTT CAT TAA GTT CTC GC | nagA upstream |
| 140R5 | ccc ggg CAT ATT CTG TTC CTA ATA TCA ACA TCA GTT | nagA upstream |
| 140F6 | ccc ggg TAA TAG TAA ACA CTT AAA TAG TTA ATT GAT TTA AAA ATC | nagA downstream |
| 140R6 | gca tgc TCA AAA ACA GCA ACA CGG TGC | nagA downstream |
| 141F1 | gaa ttc CAT CAT CGC TGA AAC AGG C | nagB upstream |
| 141R3 | ccc ggg CAT ATT AGC CTT CCT TTA TTA TTG ACC G | nagB upstream |
| 141F3 | ccc ggg TAG AGA TCT ATT CTT CAT CTT TAT GTA GGG | nagB downstream |
| 141R4 | gca tgc GGT TTC AAC GCT AGT TTG GTC G | nagB downstream |
| 144F1 | gaa ttc CCG TCC TTT TGT GAA TGT CC | nanK upstream |
| 144F3 | ccc ggg CAT AAC TTA TCC TTA TAG TGT AAA GTC TTT TCT CAC | nanK upstream |
| 144F2 | ccc ggg AAG CCT GAA GGA ACA ATT TAT GGC TAA | nanK downstream |
| 144R2 | gca tgc GCC GTT TCA GCA GAA TAA CCA G | nanK downstream |
| 145F5 | gaa ttc CGC TCC TGT GTG AAC TTA TG | nanE upstream |
| 145R5 | ccc ggg CAT ATA ACA CCC CTC ATT TAA ATC TGA AT | nanE upstream |
| 145F6 | ccc ggg TAG TCG TAA GAC GTG TGA GAA AAG ACT T | nanE downstream |
| 145R6 | gca tgc CGA ACG CAA AAT CGT ATC GGC | nanE downstream |
| 604F1 | gag ctc CAT TTT GCT GAC GAG GAA CTG | cyaA upstream |
| 604R1 | ccc ggg CAT TAC ATA AAC ACC TAA AAT TGG TGG | cyaA upstream |
| 604F2 | ccc ggg TAA TAT TTT CCT GTG GTT GAT AGG TTA CC | cyaA downstream |
| 604R2 | aag ctt AAA GCA ATG GAG TGG ACC ACA ATT | cyaA downstream |
| 145R8 | CCG CAG CAA TTT TTG TCC | PCR SOEing |
| 145M2 | TTT ATG AAA AAA CAC TTC AAA AAT | PCR SOEing |
| 145M3 | ATT TTT GAA GTG TTT TTT CAT AAA TTA AAT GTG ATC AAC TTC TC | PCR SOEing |
| 146R2 | CCA TTA CGG CAC ACT AAA GAG G | PCR SOEing |
| 957F2 | aag ctt AAT AAA ACG GAA TTT TTG AAA CAG G | CRP expression |
| 957R2 | ctc gag TCT TGC GCC ATA TAC AAC GAT TGT | CRP expression |
| P146F1 | ACA CCC CTC ATT TAA ATC TGA ATA AAT CAC | EMSA |
| P146R4 | CCC CCA AAA TAG GAT TCG | EMSA |
| 145R7 | CGA CAG GTT GGC AAG AAG AAA TAA GAC C | Primer extension |
| 145R1 | ATC AGC GGC AAG AAC AGC AG | Primer extension |
1Restriction sites added to aid in subcloning are indicated by lowercase letters. Bases added for 5 bp insertion are italicized.
All mutants used in this study were constructed as non-polar deletions using a counter-selectable cassette. The cassette used was a variation of sacB cassettes that have been described previously [27,28]. In this cassette, which is described in more detail in work to be submitted elsewhere, sacB is under the control of the tetracycline promoter and Tet repressor. The cassette also contains genes for the repressor, tetR, and nptII, a kanamycin resistance marker. This allows for inducible expression of sacB in the presence of the tetracycline analog chlortetracycline. Constructs for mutagenesis were prepared for each gene using the design detailed as follows. Regions flanking the target gene were amplified by PCR using primers that had restriction sites added to the 5'-end. These primers were designed to contain the start codon for the upstream fragment and stop codon for the downstream fragment. These products were cloned into pGEM-T (Promega, Madison, WI) and sequentially subcloned into pUC19 using the primer-encoded restriction sites. The resulting plasmid contained the flanking regions ligated to form an open reading frame consisting of a start codon, SmaI site, and stop codon. This plasmid would serve as the deletion construct. SmaI was then used to open the plasmid and the sacB-KanR cassette was inserted. The resulting plasmid was transformed into the desired NTHi strain, selecting for resistance to ribostamycin. A RibR, SucS isolate was then transformed with the deletion construct and transformants were selected on LB agar supplemented with 5% sucrose, chlortetracycline (1 μg/ml), hemin, and NAD. Deletions were confirmed by PCR. Confirmed mutants were then able to be transformed with the sacB-KanR cassette to delete additional genes.
PCR SOEing and mutagenesis
PCR splicing by overlap extension (PCR SOEing) was used to insert 5 bp between SiaR and CRP operators. Primers were designed to insert 5 bp between the operators of SiaR and CRP while conserving the 3 bp that are shared between the two (Table 2). The junction primers contained a 24 bp overlap to allow for splicing. Fragments were amplified by PCR with primer pairs 145R8/145M2 and 145M3/146R2 and products were purified using the QiaQuick PCR Clean Up Kit (Qiagen). PCR products were quantified with NanoDrop and mixed to yield a final concentration of 5 ng/μl of each and this mixture was used as the template in the SOEing reaction with primers 145R8 and 146R2. The product from the splicing reaction was cleaned up and used for transformation.
Transformation of NTHi strains was performed as detailed above. JWJ091 and JWJ116 were transformed with the plasmid pJJ331, a construct that spans from within the nan operon and into the siaPT operon and has the sacB-KanR cassette inserted near the insertion target. pJJ331 had an unintentional mutation in the CRP binding site that allowed for the plasmid to be maintained in E. coli. KanR, SucS transformants were then transformed with the PCR SOEing product and selected for growth on sucrose. Transformants were then screened by PCR and sequenced to confirm the presence of the 5 bp insertion and the absence of additional mutations. The resultant strains, JWJ159 (2019cyaA+5 bp) and JWJ160 (2019cyaAnagB+5 bp) were used for subsequent analysis.
RNA extraction and transcriptional analysis
RNA was extracted using the hot acid phenol method as described previously [29]. DNA was removed from extracted RNA by digestion with DNase I (New England Biolabs) and cleaned up with the RNeasy Mini Kit (Qiagen, Valencia, CA). RNA quality was assessed with an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA) and the concentration was determined using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). For real time RT-PCR analysis, primer/probe sets were obtained using the Custom TaqMan Gene Expression Service (Applied Biosystems, Foster City, CA). Primer/probe sets were designed using the sequence of HI0145 and HI0146 from H. influenzae 2019. A primer/probe set for the 16S rRNA of H. influenzae was designed and used as a control. The TaqMan RNA-To-CT 1-Step Kit (Applied Biosystems) was used following the manufacturer's protocol. Reactions were set up in triplicate using 20 ng of RNA. Reactions were carried out using the StepOnePlus Real Time PCR System (Applied Biosystems) with StepOne analysis software. Results were calculated using the comparative CT method to determine the relative expression ratio between RNA samples. The primer and probe set for HI16S rRNA was used as the endogenous reference to normalize the results. Two independent sets of RNA samples were used for each experiment and the mean fold change is reported. Data are expressed as mean +/- SD.
Protein expression and purification
SiaR was expressed and purified as described previously [14], with modified buffers to enhance stability of the purified protein and an additional purification step. Cells were resuspended in the SiaR lysis and equilibration buffer (10 mM Tris, pH 8.0, 300 mM NaCl, 0.1% CHAPS) prior to lysis by French press. After protein binding, the resin was washed with the SiaR wash buffer (10 mM Tris, pH 8.0, 1,150 mM NaCl, 10% glycerol, 0.1% CHAPS, 5 mM imidazole) and protein was eluted with the SiaR elution buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% CHAPS, 500 mM imidazole). The purified protein was concentrated using an Amicon Ultra centrifugation filter (Millipore, Billerica, MA) with a 10 kDa molecular weight cutoff. The protein sample was then desalted into the SiaR storage buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% CHAPS) using FPLC through a 10 ml (2-5 ml) HiTrap Desalting Column (GE Healthcare, Piscataway, NJ). Protein concentration was determined using the NanoDrop ND-1000 Spectrophotometer and an extinction coefficient of 7,575 M-1 cm-1. Aliquots of the purified protein were frozen in liquid nitrogen and stored at -80°C until needed.
CRP was expressed and purified in a similar manner. Primers were used to amplify crp with the restriction sites HindIII and XhoI on the 5' and 3' ends, respectively (Table 2). The 41 bases immediately upstream of crp were included to ensure that the native bacterial translation signals were present. The downstream primer included the last codon of the crp open reading frame, excluding the stop codon, to allow for the fusion of a multiple-histidine tag. The PCR product was cloned into pGEM-T and subsequently subcloned into pET-24(+) (Novagen, Madison, WI) using the HindIII and XhoI sites. The resulting plasmid, pJJ276, was expected to express CRP with a carboxy-terminal His•Tag.
Protein expression was induced using the Overnight Express Autoinduction System 1 (Novagen) grown at 37°C overnight. Expressed protein was purified using the BD TALON Metal Affinity Resin (BD Biosciences, Palo Alto, CA). Purification was performed in native conditions following the manufacturer's protocol and using the suggested TALON buffers. Eluted fractions were examined by SDS-PAGE and fractions containing CRP were pooled. Protein was concentrated using an Amicon Ultra centrifugation filter and desalted as described above. The protein concentration was determined using the NanoDrop ND-1000 Spectrophotometer and an extinction coefficient of 21,555 M-1 cm-1. Purified protein was stored at 4°C.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was used to study the binding of SiaR and CRP to potential promoter sequences as done previously [14]. The probe for EMSA was amplified by PCR using primer pairs P146F1 and P146R4 (Table 2), resulting in a probe that spans the region from the nanE start codon to +18 of the siaPT transcript. Binding reactions were prepared using the EMSA Kit (Molecular Probes, Eugene, OR) following the manufacturer's directions with some modifications. Binding reactions consisted of the binding buffer (150 mM KCl, 0.1 mM DTT, 0.1 mM EDTA, 10 mM Tris, pH 7.4), the DNA probe (15 nM), and 1 μM SiaR and/or CRP. Control reactions without protein were set up for each probe. Reactions were incubated at room temperature for 20 minutes. After incubation, 6× EMSA gel-loading solution was added and reactions were loaded onto a 6% DNA Retardation Gel (Invitrogen) with prechilled 0.5× TBE buffer and run at 200 V for 60 minutes. After electrophoresis, the gel was stained with SYBR Green EMSA gel stain and bands were visualized by UV transillumination. Images were captured using a Kodak EDAS 120 camera with an EDAS 590 mm filter (Eastman Kodak Company, Rochester, NY). cAMP was added to reactions when indicated to a final concentration of 100 μM.
Primer extension analysis
Primer extension analysis was used to identify the transcriptional start sites for both nan and siaPT operons. Primers 145R7 (nan) and 146R1 (siaPT) were labeled with 32P using T4 polynucleotide kinase (New England Biolabs) and γ-[32P]-ATP (GE Healthcare). Illustra Microspin G-25 columns (GE Healthcare) were used to remove unincorporated 32P. The primer extension reaction was performed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen) following the supplied protocol. After first-strand synthesis RNA was degraded by incubation with RNase A (New England Biolabs) at 37°C for 15 min. Nucleic acids were precipitated by the addition of 300 μl of chilled ethanol, incubation in a dry ice bath for 15 min, and centrifugation at 4°C. Dried samples were dissolved in loading buffer (98% deionized formamide, 10 mM EDTA, 0.025% xylene cyanol FF, 0.025% bromophenol blue) prior to loading on sequencing gel. Sequencing reactions were set up for each labeled primer using the SequiTherm EXCEL II DNA Sequencing Kit (Epicentre Technologies, Madison, WI). A PCR fragment amplified with the primers 145R7 and 146R1 was used as a template. Sequencing and primer extension reactions were loaded onto an 8% sequencing gel. After electrophoresis, the gel was dried and exposed to film at -80°C.
NMR spectroscopy
Strains 2019 wild-type and the 2019ΔcyaA ΔnagB mutant were grown in 100 ml cultures of sRPMI without Neu5Ac to early exponential phase. Neu5Ac, cAMP, or both were added and cultures were incubated for 20 min. Cells were pelleted and resuspended in 0.5 ml of MOPS buffer (40 mM MOPS, pH 7.3, with 50 μl D2O). Phosphorus NMR spectra were acquired at 162 MHz on a 400 MHz Varian Inova spectrometer in a 5 mm probe. Spectra were obtained upon excitation with at 45° pulse and digitization of 0.8 s followed by a delay of 1.7 s for recovery between scans. Spectra 20 kHz wide were collected and processed with gaussian line-broadening of 0.1 s prior to Fourier transformation. Samples were maintained at 15°C, 2048 transients were averaged in an experiment lasting 1.5 hours. For each sample, two such spectra were collected one after the other. These were not significantly different, indicating that relatively minor changes take place on the time scale of data collection. However a third spectrum collected some 13 hours later indicated significant change in some cases. Chemical shifts were referenced relative to external 85% phosphoric acid at 0 ppm.
Authors' contributions
JWJ constructed all mutant strains, designed and carried out the transcriptional analysis, performed the EMSA analysis, and wrote the primary draft of the manuscript. HS and AFM performed the NMR studies and assisted in data analysis. MAA assisted in the conception of the study and contributed to data analysis and manuscript editing. All authors read and approved the final manuscript.
Contributor Information
Jason W Johnston, Email: jason.johnston@uky.edu.
Haider Shamsulddin, Email: dochaider83@yahoo.com.
Anne-Frances Miller, Email: afm@uky.edu.
Michael A Apicella, Email: michael-apicella@uiowa.edu.
Acknowledgements
This work was supported by funding from NIAID Grants AI024616 and AI30040 and NIH grant GM085302.
References
- Greiner LL, Watanabe H, Phillips NJ, Shao J, Morgan A, Zaleski A, Gibson BW, Apicella MA. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect Immun. 2004;72(7):4249–4260. doi: 10.1128/IAI.72.7.4249-4260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrell RE, McLaughlin R, Aba Kwaik Y, Lesse A, Yamasaki R, Gibson B, Spinola SM, Apicella MA. Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect Immun. 1992;60(4):1322–1328. doi: 10.1128/iai.60.4.1322-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira MA, Ram S, Goldstein R, Hood DW, Moxon ER, Pelton SI. Role of complement in defense of the middle ear revealed by restoring the virulence of nontypeable Haemophilus influenzae siaB mutants. Infect Immun. 2007;75(1):325–333. doi: 10.1128/IAI.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood DW, Makepeace K, Deadman ME, Rest RF, Thibault P, Martin A, Richards JC, Moxon ER. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol Microbiol. 1999;33(4):679–692. doi: 10.1046/j.1365-2958.1999.01509.x. [DOI] [PubMed] [Google Scholar]
- Williams BJ, Morlin G, Valentine N, Smith AL. Serum resistance in an invasive, nontypeable Haemophilus influenzae strain. Infect Immun. 2001;69(2):695–705. doi: 10.1128/IAI.69.2.695-705.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S, Zaleski A, Johnston JW, Gibson BW, Apicella MA. Novel sialic acid transporter of Haemophilus influenzae. Infect Immun. 2005;73(9):5291–5300. doi: 10.1128/IAI.73.9.5291-5300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet V, Hood DW, Li J, Brisson JR, Randle GA, Martin A, Li Z, Goldstein R, Schweda EK, Pelton SI. et al. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc Natl Acad Sci USA. 2003;100(15):8898–8903. doi: 10.1073/pnas.1432026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun. 2005;73(6):3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JW, Coussens NP, Allen S, Houtman JC, Turner KH, Zaleski A, Ramaswamy S, Gibson BW, Apicella MA. Characterization of the N-acetyl-5-neuraminic acid-binding site of the extracytoplasmic solute receptor (SiaP) of nontypeable Haemophilus influenzae strain 2019. J Biol Chem. 2008;283(2):855–865. doi: 10.1074/jbc.M706603200. [DOI] [PubMed] [Google Scholar]
- Severi E, Randle G, Kivlin P, Whitfield K, Young R, Moxon R, Kelly D, Hood D, Thomas GH. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol Microbiol. 2005;58(4):1173–1185. doi: 10.1111/j.1365-2958.2005.04901.x. [DOI] [PubMed] [Google Scholar]
- Severi E, Muller A, Potts JR, Leech A, Williamson D, Wilson KS, Thomas GH. Sialic acid mutarotation is catalyzed by the Escherichia coli beta-propeller protein YjhT. J Biol Chem. 2008;283(8):4841–4849. doi: 10.1074/jbc.M707822200. [DOI] [PubMed] [Google Scholar]
- Jenkins GA, Figueira M, Kumar GA, Sweetman WA, Makepeace K, Pelton SI, Moxon R, Hood DW. Sialic acid mediated transcriptional modulation of a highly conserved sialometabolism gene cluster in Haemophilus influenzae and its effect on virulence. BMC Microbiol. 2010;10:48. doi: 10.1186/1471-2180-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E, Lichtensteiger C, Steenbergen S. Sialic acid metabolism's dual function in Haemophilus influenzae. Mol Microbiol. 2000;36(5):1113–1123. doi: 10.1046/j.1365-2958.2000.01925.x. [DOI] [PubMed] [Google Scholar]
- Johnston JW, Zaleski A, Allen S, Mootz JM, Armbruster D, Gibson BW, Apicella MA, Munson RS Jr. Regulation of sialic acid transport and catabolism in Haemophilus influenzae. Mol Microbiol. 2007;66(1):26–39. doi: 10.1111/j.1365-2958.2007.05890.x. [DOI] [PubMed] [Google Scholar]
- Macfadyen LP, Redfield RJ. Life in mucus: sugar metabolism in Haemophilus influenzae. Res Microbiol. 1996;147(6-7):541–551. doi: 10.1016/0923-2508(96)84010-1. [DOI] [PubMed] [Google Scholar]
- Redfield RJ, Cameron AD, Qian Q, Hinds J, Ali TR, Kroll JS, Langford PR. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J Mol Biol. 2005;347(4):735–747. doi: 10.1016/j.jmb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293(2):199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- MacFadyen LP, Dorocicz IR, Reizer J, Saier MH Jr, Redfield RJ. Regulation of competence development and sugar utilization in Haemophilus influenzae Rd by a phosphoenolpyruvate:fructose phosphotransferase system. Mol Microbiol. 1996;21(5):941–952. doi: 10.1046/j.1365-2958.1996.441420.x. [DOI] [PubMed] [Google Scholar]
- Larson TJ, Cantwell JS, van Loo-Bhattacharya AT. Interaction at a distance between multiple operators controls the adjacent, divergently transcribed glpTQ-glpACB operons of Escherichia coli K-12. J Biol Chem. 1992;267(9):6114–6121. [PubMed] [Google Scholar]
- Wickstrum JR, Santangelo TJ, Egan SM. Cyclic AMP receptor protein and RhaR synergistically activate transcription from the L-rhamnose-responsive rhaSR promoter in Escherichia coli. J Bacteriol. 2005;187(19):6708–6718. doi: 10.1128/JB.187.19.6708-6718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan SM, Schleif RF. A regulatory cascade in the induction of rhaBAD. J Mol Biol. 1993;234(1):87–98. doi: 10.1006/jmbi.1993.1565. [DOI] [PubMed] [Google Scholar]
- Plumbridge JA. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol Microbiol. 1991;5(8):2053–2062. doi: 10.1111/j.1365-2958.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge JA. Induction of the nag regulon of Escherichia coli by N-acetylglucosamine and glucosamine: role of the cyclic AMP-catabolite activator protein complex in expression of the regulon. J Bacteriol. 1990;172(5):2728–2735. doi: 10.1128/jb.172.5.2728-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J, Kolb A. DNA loop formation between Nag repressor molecules bound to its two operator sites is necessary for repression of the nag regulon of Escherichia coli in vivo. Mol Microbiol. 1993;10(5):973–981. doi: 10.1111/j.1365-2958.1993.tb00969.x. [DOI] [PubMed] [Google Scholar]
- Campagnari AA, Gupta MR, Dudas KC, Murphy TF, Apicella MA. Antigenic diversity of lipooligosaccharides of nontypable Haemophilus influenzae. Infect Immun. 1987;55(4):882–887. doi: 10.1128/iai.55.4.882-887.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriott RM, Meyer EM, Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970;101(2):517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Pericone CD, Lysenko E, Goldfine H, Weiser JN. Multiple mechanisms for choline transport and utilization in Haemophilus influenzae. Mol Microbiol. 2003;50(2):537–548. doi: 10.1046/j.1365-2958.2003.03703.x. [DOI] [PubMed] [Google Scholar]
- Copass M, Grandi G, Rappuoli R. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect Immun. 1997;65(5):1949–1952. doi: 10.1128/iai.65.5.1949-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J Bacteriol. 2000;182(21):6192–6202. doi: 10.1128/JB.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]