Abstract
We performed a chemical screen to look for novel inhibitors of zebrafish caudal fin regeneration. In a pilot screen, 520 compounds were tested. Two compounds, budesonide and AGN192403, abrogated fin regeneration. One compound in particular, AGN192403, targets the imidazoline receptor, a pathway not previously linked to fin regeneration. In addition to inhibiting regeneration of the adult fin, AGN192403 also blocked regeneration of the larval fin fold. Finally, the inhibitory effect of AGN192403 on fin regeneration persisted after removal of the drug. These studies demonstrate that chemical screening is feasible in adult zebrafish and that it is a reasonable strategy to use for exploring the biology of regeneration.
Introduction
Regeneration is a process by which damaged or lost structures are perfectly or near-perfectly replaced.1 In classic models of regeneration, this process follows a pattern of epithelial migration to cover the wound, formation of a regeneration blastema, and ultimately proliferation and cell differentiation leading to replacement of the lost or damaged structure. The zebrafish (Danio rerio) caudal fin has become a valuable model for studying regeneration. Mammals, including humans, have the potential to regenerate only a limited number of structures, including the liver and digit tips.2 In contrast to mammals, urodele amphibians and teleosts have the ability to regenerate multiple structures. Zebrafish are able to regenerate the fin, larval fin fold, optic nerve, scales, heart, retina, liver, pancreas, and spinal cord.3–7 The caudal fin model of regeneration is particularly attractive because the fin is a relatively simple structure, quick to regenerate, and nonessential to the viability of fishes.8 Zebrafish are also amenable to molecular and genetic manipulations, including transgenic techniques and forward genetic screens.9–11 Indeed, many recent details regarding the genetic pathways regulating regeneration have been uncovered using zebrafish as a model system. Predominant among these details are the roles of fibroblast growth factors and Wnt/β-catenin signaling during regeneration.12–14
Forward genetic approaches have proven valuable in dissecting the mechanisms underpinning regeneration in multiple model systems, including zebrafish. An important feature of the forward genetic approach is its unbiased nature: no a priori assumptions are made regarding the genetic pathways underlying the regenerative process. However, a potential limitation arises when studying postembryonic phenotypes such as regeneration, as many of the critical pathways are likely to be essential during embryonic development and therefore difficult to identify in postembryonic screens. This challenge is partly mitigated by the identification of conditional alleles, and all but one zebrafish regeneration mutant reported to date are conditional (see below).
Chemical screens represent an alternative methodology for identifying genes of interest in an unbiased fashion and such screens have been successfully employed in several species, including zebrafish.15 In theory, chemicals have similar advantages to conditional genetic alleles in that they can be added at specific time points. Collections of compounds are commercially available that are designed to satisfy multiple screening strategies, including diverse arrays of synthetic small molecules (e.g., NCI Diversity Set), compounds with known biological activity (e.g., Lopac 1280; Sigma), or FDA-approved drugs in clinical use (e.g., John Hopkins Clinical Compound Library). Chemical screens have been successfully employed in zebrafish to study cardiac development,16 angiogenesis,17 cell cycle,18 and gonadal development.19 Chemical screens using a moderate number of compounds (hundreds) and targeting adult phenotypes in zebrafish have not been reported, nor have chemical screens targeting regeneration in any species.
In this article, we report a chemical screen for novel inhibitors of zebrafish caudal fin regeneration. Further, we demonstrate that screening a commercially available compound library is feasible even with the relatively large volume of drugs required to screen adult animals. Further, we have identified a novel compound, AGN192403, which irreversibly abrogates regeneration of both the adult fin and the embryonic fin fold. AGN192403 acts via a mechanism hitherto unattributed to regeneration; moreover, irreversible inhibition of fin regeneration has not previously been described.
Materials and Methods
Zebrafish husbandry and general methods
Wild-type fish stocks utilized in this study were of the AB strain. Fish were reared using standard techniques, at a constant temperature of 25°C, fed three times daily, and maintained on a 14 h light:10 h dark photoperiod. Microscopy was performed with a Nikon SMZ1500 stereomicroscope. Photographs were taken with a Nikon DXM1200F digital camera. Images were captured using Nikon Act-1 software and processed using Photoshop CS4 (Adobe). All graphics and statistical analyses were performed using Prism 4.0 (Graph Pad).
Fin regeneration experiments
For the primary fin regeneration screen, adult zebrafish (AB strain) were raised to 6–9 months of age. Fish were anesthetized briefly in Tricaine (Sigma), and approximately 50% of the caudal fin was amputated using a straight razor blade. Fish were recovered in fresh water and then divided into 250 mL disposable specimen cups containing 100 mL of system water. Compounds from the Sigma Lopac 1280 panel were added to the cups (one compound per cup) at a final concentration of 10 μM and caudal fin regeneration was followed for 72 h. Fish were maintained in a 33°C incubator during this time. The compound and water were refreshed once daily and any dead fish were removed from the cups. One percent dimethyl sulfoxide (DMSO; diluent) was used as a negative control. After 72 h, fish were removed from the cups, anesthetized, photographed, and then euthanized.
For the secondary screen with individual compounds, drug stocks were made in DMSO such that the final dilution was 1:100 (1% DMSO). One percent DMSO was used as a negative control. Fin amputations and analyses were performed as described above, except that regeneration experiments were conducted in small tanks containing 10 fish and 500 mL of system water. The compound and water were again refreshed daily.
Polymerase chain reaction amplification
Primer sequences for amplification of the zebrafish nischarin gene were F-ACGGCAATGTGAAAGGTGGACA and R-GGGTCAGATGGGGCTTTGGCA. RNA was purified from 72 h postamputation (hpa) fin regenerates by standard Trizol extraction. RNA was used for first-strand cDNA transcription. Five nanograms of cDNA was used as template for the polymerase chain reactions (PCRs).
Detection of proliferating cells using bromodeoxyuridine
After AGN192403 (25 μM) or control (1% DMSO) treatment, fish were labeled in vivo with bromodeoxyuridine (BrdU) by allowing them to swim in fresh water containing BrdU (50 μg/mL) before harvesting the fin regenerates (n = 5 fish per group). Detection of BrdU incorporation was done on whole mounts.20,21 Fins were harvested as described above, and then fixed overnight in Carnoy's solution (60% EtOH, 30% chloroform, and 10% acetic acid) followed by dehydration in 100% MeOH. Fins were subsequently rehydrated into phosphate-buffered saline containing 0.3% Triton X-100 (PBTx), washed in PBTx, rinsed twice in 2 N HCl in PBTx, and then incubated for 30 min in 2 N HCl/PBTx at room temperature. After two more PBTx washes, fins were blocked for several hours in PBTxB (PBTx containing 0.25% bovine serum albumin), and then incubated over night at 4°C in anti-BrdU (1:50 dilution in PBTxB; Roche). Fins were then washed extensively (4–24 h) in multiple changes of PBTx, blocked for 30 min in PBTxB, and then incubated over night at 4°C in secondary antibody (goat anti-mouse conjugated to either Alexa-488 of Alexa-546, 1:200 dilution in PBTxB; Molecular Probes). After 2–24 h of additional PBTx washes, fins were embedded in 1% agarose and sectioned (15 μm) on a cryostat before mounting. BrdU-labeled cells were observed using a Nikon SMZ1500 stereomicroscope. For the quantitative analyses, 20 consecutive sections from each fin were scored. All BrdU-labeled cells that were located beneath the basement membrane and within 1000 μm of the distal tip of the fin were counted.
In situ hybridization
Templates for msx-b sense and antisense probes were cloned by reverse transcription (RT)-PCR. PCR amplicons were gel-purified (Qiagen) and TA-cloned (pCRII; Invitrogen). Insert-containing plasmids were grown and harvested (Maxi-prep; Qiagen). Plasmids were linearized using BamH1 or EcoRV to serve as template for transcription of sense or antisense probes, respectively. Digoxigenin-labeled RNA probes were transcribed from these templates using SP6 (EcoRV) or T7 (BamH1) RNA polymerase. Whole-mount in situ hybridization was performed as described previously.20 After staining, fins were mounted in 50:50 glycerol:PBT and imaged on a Nikon SMZ1500 stereomicroscope.
Regeneration of the embryonic fin fold
For embryonic fin fold experiments, wild-type zebrafish (AB strain) were crossed using in vitro fertilization. Fertilized embryos were raised in the embryo medium at 28.5°C for 72 h. At 72 h postfertilization (hpf ), larvae were anesthetized by adding several drops of Tricaine to the water and arrayed on Petri dishes coated in 1% agarose. A portion of the caudal fin fold, distal to the most caudal portion of the notochord, was then amputated using a scalpel. Larvae were recovered in the fresh embryo medium and arrayed in 12-well plastic tissue culture plates, one larva per well with 2 mL of the embryo medium. Drug dissolved in DMSO was added to each well in a 1:100 final dilution (1% DMSO). One percent DMSO was used as a negative control. Fin folds were allowed to regenerate at 28.5°C for 72 h. Larvae were then euthanized in Tricaine and photographed for analyses.
Results
A screen for novel inhibitors of caudal fin regeneration
We used the Sigma Lopac 1280 panel to screen for novel inhibitors of zebrafish caudal fin regeneration. Adult fish were anesthetized in batches, and then approximately 50% of the caudal fin was amputated from each fish. After fin amputation, adult fish were arrayed in cups (two fish per cup) and treated either with a compound from the Sigma Lopac 1280 panel of compounds or diluent (DMSO). In all, we screened the first 520 compounds from the Lopac 1280 collection (www.sigmaaldrich.com/chemistry/drug-discovery/validation-libraries/lopac1280-navigator.html). We summarized the results of this screen in Table 1. The primary screen for compounds inhibiting fin regeneration was qualitative. Controls (1% DMSO) were run with every set of compounds tested, and fin regeneration in the compound-treated fish was compared to these DMSO-treated controls (Fig. 1). Overall, fin regeneration was a remarkably robust process. In our primary screen, 2.5% (13/520) of the compounds abrogated fin regeneration to a substantive degree (Table 1; Fig. 1). Importantly, adult zebrafish were also relatively resistant to the toxic effects of these compounds. About 10.8% of the compounds (56/520) proved lethal in both fish tested at the screening concentration of 10 μM. Controls (1% DMSO) were performed with every set of compounds tested. In no cases did 1% DMSO affect fin regeneration in comparison to cups wherein no compound or diluent was added. Further, 1% DMSO was never toxic to the fish. All 13 of the compounds that abrogated fin regeneration in the primary screen were retested at a concentration of 10 μM (n = 10 fish for each retested compound) in a secondary screen. In our secondary screen, 2/13 compounds (budesonide and AGN192403) again showed an inhibitory effect on fin regeneration (Table 1). Thus, overall, 0.4% (2/520) of the compounds tested inhibited regeneration of the zebrafish caudal fin.
Table 1.
Compounds Inhibiting Fin Regeneration in the Primary and Secondary Screens
| Compound | Inhibition in primary screen | Inhibition in secondary screen |
|---|---|---|
| Trans U-50488 methanesulfonate | Yes | No |
| R(−)-Propylnorapomorphine hydrochloride | Yes | No |
| (+)-Butaclamol hydrochloride | Yes | No |
| (±)-AMT hydrochloride | Yes | No |
| Alaproclate hydrochloride | Yes | No |
| Rp-cAMPS triethylamine | Yes | No |
| SKF 97541 hydrochloride | Yes | No |
| AGN192403 hydrochloride | Yes | Yes |
| DL-Buthionine-(S,R)-sulfoximine | Yes | No |
| Betaine aldehyde chloride | Yes | No |
| Benserazide hydrochloride | Yes | No |
| Cefmetazole | Yes | No |
| Budesonide | Yes | Yes |
FIG. 1.
Inhibition of fin regeneration by budesonide and AGN192403. Results from treating fish with 10 μM AGN192403, 10 μM budesonide, or 1% dimethyl sulfoxide (DMSO) in the primary screen. Fin regeneration is inhibited by AGN192403 (A, B) and budesonide (D, E) when compared to DMSO controls (C and F, respectively).
The imidazoline receptor antagonist AGN192403 abrogates caudal fin regeneration in a dose-dependent fashion
As outlined above (Table 1; Fig. 1), in a pilot screen we identified 2/520 (0.4%) compounds that showed substantial inhibition of fin regeneration. We further explored the effects of one of these drugs, AGN192403, in additional experiments. We chose to focus on AGN192403 in these follow-up experiments for several reasons. First, the abrogation of fin regeneration was unequivocal at a concentration of 10 μM. Second, AGN192403 appeared completely nontoxic at concentrations that robustly abrogate fin regeneration. Third, the mechanism through which AGN192403 inhibits regeneration is potentially novel. The purported pharmacologic target of AGN192403 is the type 1 imidazoline receptor (I1)22–26; moreover, imidazoline receptor biology has not previously been linked to tissue regeneration. Finally, additional compound was readily available for further experiments.
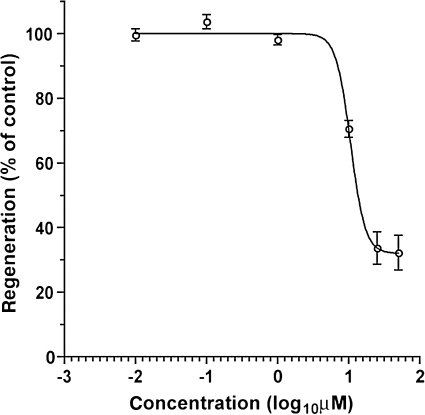
We first examined the dose–response relationship between AGN192043 and fin regeneration. Zebrafish caudal fins were amputated and treated postamputation with AGN192403 at concentrations ranging from 0.001 to 50 μM (n = 10 fish for each group with the experiment performed in duplicate). Fish were maintained at 33°C postamputation and fin regeneration was followed for 72 h. During this time, AGN192403 (or diluent) and water were refreshed daily. In contrast to the primary screen, only a portion (50%) from the ventral half of each caudal fin was amputated, allowing each fish to serve as its own standard for percent regeneration of the fin. At 72 hpa, regeneration was quantified as the percent of the amputated ventral lobe that had regenerated relative to the nonamputated dorsal lobe (length of the longest fin ray in each of the dorsal and ventral lobes was used for consistency of measurements). Data from the AGN192403-treated fish were then normalized against the data from control fish treated with 1% DMSO. AGN192403 significantly inhibited caudal fin regeneration in a dose-dependent manner (p < 0.01 in a one-way ANOVA; data points represent means values ± standard error of the mean) (R2 = 0.9011 for the goodness of fit for a sigmoid dose–response curve) (Fig. 2). No inhibitory effect of AGN192403 on fin regeneration was observed at concentrations less than or equal to 1 μM, although concentrations as low as 0.001 μM were tested (data for 0.001 μM not shown). In addition, the inhibitory effect of AGN192403 on fin ray regeneration was maximal at concentrations equal to or greater than 25 μM. Finally, 50 μM proved to be quite toxic in zebrafish, being lethal in half of the fish treated at this concentration.
FIG. 2.
Dose-dependent inhibition of fin regeneration by AGN192403. Zebrafish caudal fines were amputated and allowed to regenerate for 72 h at 33°C. Fish were treated with AGN192403 at various concentrations for the entire 72 h. AGN192403 significantly inhibited fin regeneration in a dose-dependent manner (p < 0.01, one-way ANOVA).
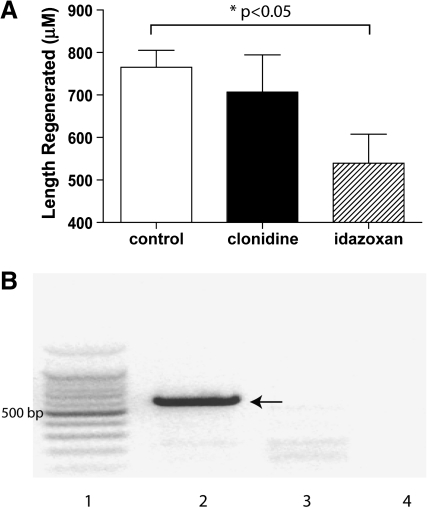
To address the specificity of AGN192403's effect on fin regeneration, we asked whether two additional imidazoline receptor ligands, clonidine and Idazoxan, inhibit fin regeneration. Idazoxan is an α-adrenoceptor antagonist and an imidazoline receptor ligand (I2 > I1).27 Clonidine is an α-adrenoceptor agonist and an imidazoline receptor ligand (I1).28 At a concentration of 50 μM, Idazoxan abrogated fin regeneration by 30% relative to the DMSO control (p < 0.05, n = 10 for each group) (Fig. 3A). There was no significant effect of Idazoxan at 25 μM (not shown). In contrast to Idazoxan, we did not observe an inhibitory effect with clonidine, either at 25 μM (not shown) or at 50 μM (Fig. 3A). As further evidence that imidazoline receptors might play a role in fin regeneration, we asked whether components of the imidazoline receptor complex are expressed in the fin during regeneration. A zebrafish nischarin ortholog (nischarin is a purported protein component of the imidazoline receptor complex) has been identified (BX908397). We designed primers targeting the Zebrafish NISCH gene and show that NISCH transcript is present in the regenerating caudal fin (Fig. 3B).
FIG. 3.
AGN192403 targets the imidazoline receptor. (A) Zebrafish fins were amputated at approximately the level of the first fin ray bifurcation. Fish were treated with 50 μM clonidine, 50 μM Idazoxan, or 1% DMSO (control). Regeneration was allowed to proceed for 72 h postamputation (hpa) at 33°C. At 72 hpa, the length of fin ray regeneration (microns) was measured. For all measurements we used the third dorsal fin ray and measured from the original amputation plane to the distal tip of the regenerating fin. Data represent means ± standard error of the mean (SEM). n = 10 for each group. (B) Qualitative RT-polymerase chain reaction for the zebrafish NISCH gene. cDNA was prepared from 72 hpa fin regenerates and used as template to amplify the zebrafish NISCH gene. A 600–700 bp NISCH band is strongly amplified from regenerating caudal fin (arrow, lane 2). No amplification of NISCH is seen in controls lacking primer (lane 3) or cDNA template (lane 4). Lane 1 contains 100 bp ladder.
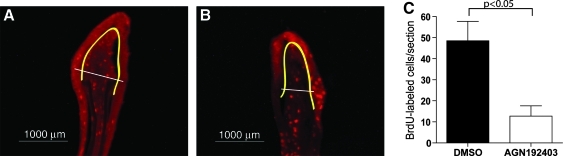
We then asked whether AGN192403 abrogates fin regeneration by inhibiting cell proliferation during blastema formation. Fins were amputated and allowed to regenerate for 72 h (33°C). AGN192403 (or DMSO) was added to the water after blastema formation had begun (12 hpa). Fish were labeled with BrdU 66 hpa and the fins were reamputated for analysis at 72 hpa. We show here (Fig. 4) that AGN192403 decreases the number of BrdU-labeled cells in the regeneration blastema by 71%.
FIG. 4.
AGN192403 blocks cell proliferation. (A, B) Bromodeoxyuridine (BrdU) staining of frozen sections from 72 hpa fin regenerates. Fish were treated with 1% DMSO (A) or 25 μM AGN192403 (B) from 12 to 72 hpa. BrdU-labeled cells were counted if they were deep to the basement membrane (yellow lines) and within 1000 μm of the distal tip of the fin (white lines). (C) AGN192403 significantly decreased the number of BrdU-labeled cells compared to control treated fins (n = 5 fins per group, 20 sections per slide; data represent means ± SEM).
AGN192403 affects blastema formation and outgrowth
We wondered whether the effect of AGN192403 was specific to a particular stage of regeneration, such as wound healing (epithelial migration), blastema formation, or outgrowth. From our pilot screen (Fig. 1) and secondary screen (not shown), fish treated with AGN192403 clearly develop a wound epidermis postamputation, implying that AGN192403 does not inhibit epithelial migration. To ask whether blastema formation is affected by AGN192403 treatment, we performed caudal fin amputations and allowed fins regeneration to proceed at 33°C. Drug (either AGN192403 [25 μM] or 1% DMSO) was added beginning 12 hpa (after epithelial migration has occurred) and fins were reamputated 72 hpa and assayed for the blastemal marker msx-b (n = 10 for each group) by whole-mount in situ hybridization. As expected, msx-b transcript was easily observed at 72 hpa in control fins (Fig. 5A); however, no msx-b transcript was observed in the AGN192403-treated fins (Fig. 5B). To address the outgrowth phase of fin regeneration, we amputated zebrafish caudal fins and followed regeneration for 12 days (at 25°C). These fish were treated with AGN192403 (25 μM, or 1% DMSO) either on days 2–7 or 7–12 postamputation. Amputations were again performed only on the ventral half of the caudal fin and regeneration was quantified by comparison with the nonamputated dorsal lobe as above. These data demonstrate that AGN192403 disrupts the outgrowth phase of fin regeneration (Fig. 5C) (n = 10 for each group and time point; data represent means ± standard error of the mean).
FIG. 5.
AGN192403 inhibits fin regeneration at multiple stages. (A, B) AGN192403 inhibits msx-b expression in the regenerating fin ray blastema. Whole-mount in situ hybridization demonstrates a dramatic reduction in msx-b transcript (arrows) in AGN192403-treated fish (A) compared with controls (B). (C) AGN192403 inhibits the outgrowth stage of fin regeneration. After fin amputation, zebrafish were treated with AGN192403 (or DMSO [open squares]) either from days 2–7 (open triangles) or days 7–12 (open circles) postamputation. Regeneration was inhibited regardless of whether AGN192403 was added during the early (days 2–7) or late (days 5–12) stages of outgrowth.
Inhibition of fin regeneration by AGN192403 is not completely reversible
To date, all but one14 of the genetic mutants that have been identified that inhibit fin regeneration are represented by conditional alleles7,14,29–31; moreover, fin regeneration recovers when the fish are shifted to a permissive temperature. Transgenic techniques have also been successfully employed13,32 to conditionally inhibit fin regeneration by abrogating either fgf or wnt/β-catenin signaling via the inducible heat shock promoter. Again, regeneration resumes normally upon cessation of the heat shock treatments. Thus, it appears that while regeneration can be temporarily disrupted via a number of mechanisms, permanently inhibiting regeneration is substantially more difficult. We wondered whether this might also be true for fins treated with AGN192403.
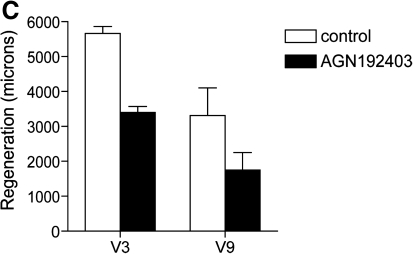
On the basis of the dose–response curve for inhibition of fin regeneration by AGN192403 (Fig. 2), we amputated zebrafish caudal fins and treated the fish postfin amputation with AGN192403 or 1% DMSO at a concentration of 25 μM for 1–7 days postamputation (n = 10 for each group). When fish were treated with AGN192403 for 1–4 days postamputation, fin regeneration recovered normally after washing out the drug (data not shown). Interestingly, when fish were treated with AGN192403 for periods of 5 days or longer postamputation, failed regeneration of the caudal fin rays was sometimes irreversible, even 30 days after drug treatment (i.e., regeneration failed to resume upon removal of the compound). Regeneration was patchy with some fin rays (within a given fin) regenerating normally and others not regenerating at all (Fig. 6A). However, if the same fin was then reamputated proximal to the original amputation plane, subsequent regeneration of the fin (in the absence of drug) occurred normally (Fig. 6B). To quantitate the long-term effects of AGN192403 treatment on the regenerating fin, we amputated caudal fins at the first fin ray bifurcation and treated these fish with either 25 μM AGN192403 of 1% DMSO for 7 days. Fish were then returned to fresh water and fin regeneration was allowed to proceed for an additional 2 weeks (25°C). We then measured the total length of fin ray regeneration (microns) in the ventral lobe of each caudal fin for both the longest (V3) and the shortest (V9) fin ray (Fig. 6C). AGN192403 treatment decreased regeneration in the longest and shortest fin rays by approximately 38% and 45%, respectively, compared with controls (n = 10 fish for each group). After 2 more weeks, minimal additional growth had occurred in the treatment (AGN192403) group (not shown).
FIG. 6.
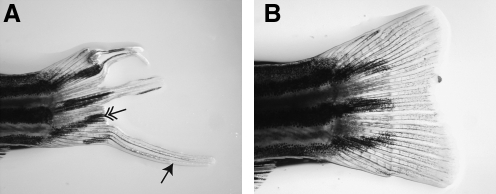
AGN192403 can permanently inhibit regeneration of some fin rays. (A) Fish were treated with AGN192403 for 7 days after fin amputation. Compound was then removed and regeneration was allowed to proceed for 30 days (25°C). Regeneration proceeded normally in some rays (arrow) but was completely absent in others (double arrow). (B) When these same fins were reamputated proximal to the original amputation plane, regeneration proceeded normally. (C) After fin amputation and 7 days of AGN192403 or DMSO treatment, fish were recovered for 2 weeks in fresh water (no drug) and the degree of fin regeneration was quantified for the longest (V3) and shortest (V9) fin rays. AGN192403-treated fish failed to completely restore their fins. These data were significant for both V3 and V9 (p < 0.05) (data represent means ± SEM).
AGN abrogates regeneration of the embryonic fin fold
Historically, regeneration studies in urodeles and fishes have focused on adult tissues. However, regeneration of the larval fin fold has been studied in zebrafish33,34 with the notion that common mechanisms might underpin the general process of tissue regeneration. We wondered whether the effect of AGN192403 on caudal fin regeneration was generalizable to the regeneration of a different, larval structure. Such a finding might suggest that AGN192403 targets a mechanism common to regeneration in multiple different tissues. To address this, we asked whether AGN192403 could abrogate regeneration of the larval zebrafish fin fold.
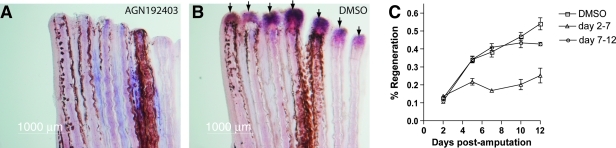
Zebrafish larvae were raised to 72 hpf for fin fold regeneration experiments. We chose 72 hpf because at this stage zebrafish larvae have a well-developed caudal fin fold that is easy to surgically manipulate. At 72 hpf, a small piece of the caudal fin fold was amputated. The plane of amputation was always posterior to the caudal-most tip of the notochord. Larvae were then treated postamputation either with AGN192403 (at concentrations ranging between 3 and 25 μM [in 1% DMSO]) or with 1% DMSO alone. The precision for fin fold amputations is less than for a fin amputations; moreover, in contrast to the fin, it is impossible to discern the plane of amputation once regeneration proceeds. Thus, larvae were photographed immediately postamputation and again after allowing 3 days for the fin fold to regenerate. In addition, larvae were raised individually postamputation, allowing us to unequivocally follow regeneration in individual larvae. After amputation, regeneration of the larval fin fold was allowed to proceed for 3 days at 28.5°C and then the extent of regeneration was qualitatively analyzed. Previous work34 has shown that regeneration of the zebrafish caudal fin fold is essentially complete by 3 days postamputation. This observation was confirmed in our 1% DMSO controls, wherein regeneration of the larval fin fold was complete in 100% of larvae at 72 h postamputation (n = 10 larvae) (Fig. 7). AGN192403 abrogated regeneration of the larval fin fold in a dose-dependent manner (Fig. 7). There was some effect at an AGN192403 concentration of 3 μM as demonstrated by the fact that 50% of the larvae completed fin fold regeneration (n = 10 larvae). At AGN192403 concentrations of 10 and 25 μM, fin fold regeneration occurred in only 20% (n = 10) and 10% (n = 10) of zebrafish larvae, respectively (Fig. 7).
FIG. 7.
AGN192403 inhibits fin fold regeneration. Larval fin folds were amputated (A, B) and then allowed to regenerate for 3 days (28.5°C) (C, D). (A, C) 1% DMSO alone had no effect on fin fold regeneration. (B, D) An example of inhibition of larval fin fold regeneration when larvae are treated with 10 μM AGN192403 following amputation of the fin fold.
Discussion
Regenerative medicine has been identified1 as one of the most significant challenges facing medical science. In contrast to mammals, zebrafish are capable of regenerating multiple tissues, including both adult and larval structures. In this article, we report the use of the zebrafish caudal fin model of regeneration to screen for novel chemical inhibitors of regeneration. This study represents the first chemical screen targeting regeneration ever published in a vertebrate organism.
Overall, 0.4% (2/520) of the compounds that we tested inhibited fin regeneration at a concentration of 10 μM, without being toxic to the fish. There are, of course, caveats to our screening methodology. It is possible that additional compounds would have inhibited regeneration if tested at concentrations higher than 10 μM. Alternatively, some compounds that were lethal at 10 μM might have inhibited regeneration at lower (sub-lethal) concentrations. However, the fact that almost 90% of the compounds tested were not toxic and that a manageable number of compounds inhibited fin regeneration in the primary and secondary screens (2.5% and 0.4%, respectively) suggests that 10 μM was an appropriate concentration to target in the primary screen.
Two compounds, budesonide and AGN192403, inhibited fin regeneration in both the primary and secondary screen. Interestingly, neither compound has been previously reported as an inhibitor of tissue regeneration, although the inhibitory effects of glucocorticoids on wound healing are well described.35 In light of recent data relating signaling from the overlying epidermis to fin regeneration,36 it is perhaps not surprising that budesonide demonstrated inhibitory effects. We chose to focus our secondary experiments on AGN192403 because its effect on regeneration is potentially novel. AGN192403 is a purported imidazoline receptor antagonist22–26 and no published data connect imidazoline receptor signaling to the biology of regeneration. Although the precise role of the imidazoline receptor in regeneration biology remains unresolved, two additional pieces of data support the specificity of the AGN192403 effect. First, we show that a second imidazoline receptor ligand, Idazoxan, also abrogates fin regeneration. Second, the zebrafish nischarin (a component of the imidazoline receptor) ortholog is expressed in the fin during regeneration. Finally, we describe a plausible mechanistic basis for the impairment of fin regeneration, demonstrating that AGN192403 inhibits cell proliferation in the regeneration blastema. Additional information regarding the mechanism by which AGN192403 inhibits regeneration must await further experimentation.
Limited data33,34 suggest that some commonalities might underlie the mechanisms supporting larval fin fold and adult fin ray regeneration. Our data demonstrating that AGN192403 inhibits regeneration of both the adult fin and the larval fin fold lend further support to this hypothesis. In addition, our data further validate the use of the larval zebrafish fin fold as a model of regeneration. This model may prove important in future chemical screens as it is robust and requires far less compound than the adult fin regeneration studies.
Conclusion
We report here the first chemical screen targeting fin regeneration in adult zebrafish. Further, our data support a plausible role for imidazoline receptor signaling in fin regeneration, although further experiments are necessary to conform this. These data extend the important role that zebrafish have played in dissecting developmental and physiological processes via unbiased genetic and chemical screening.
Acknowledgments
The authors would like to thank Casey Allen and Crystal Dao for assistance in performing the primary fin regeneration screen and the fin fold regeneration experiments, respectively. We would also like to thank Bendi Gong for assistance with the NISCH PCR amplification. This work was supported by the NIH (K08 HD-046656).
Disclosure Statement
No competing financial interests exist.
References
- 1.Brockes JP. Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–1923. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- 2.Khan AZ. Mudan SS. Liver regeneration: mechanisms, mysteries and more. ANZ J Surg. 2007;77:9–14. doi: 10.1111/j.1445-2197.2006.03981.x. [DOI] [PubMed] [Google Scholar]
- 3.Becker T. Wullimann MF. Becker CG. Bernhardt RR. Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J Comp Neurol. 1997;377:577–595. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 4.Bereiter-Hahn J. Zylberberg L. Regeneration of teleost fish scale. Comp Biochem Physiol. 1993;105A:625–641. [Google Scholar]
- 5.Bernhardt RR. Tongiorgi E. Anzini P. Schachner M. Increased expression of specific recognition molecules by retinal ganglion cells and by optic pathway glia accompanies the successful regeneration of retinal axons in adult zebrafish. J Comp Neurol. 1996;376:253–264. doi: 10.1002/(SICI)1096-9861(19961209)376:2<253::AID-CNE7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Morgan TH. Regeneration. First. New York: The Macmillan Company; 1901. [Google Scholar]
- 7.Poss KD. Nechiporuk A. Hillam AM. Johnson SL. Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 8.Iovine MK. Johnson SL. Genetic analysis of isometric growth control mechanisms in the zebrafish caudal fin. Genetics. 2000;155:1321–1329. doi: 10.1093/genetics/155.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higashijima S. Okamoto H. Ueno N. Hotta Y. Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 10.Nasevicius A. Ekker SC. Effective targeted gene “knockdown” in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 11.Padhi BK. Joly L. Tellis P. Smith A. Nanjappa P. Chevrette M, et al. Screen for genes differentially expressed during regeneration of the zebrafish caudal fin. Dev Dyn. 2004;231:527–541. doi: 10.1002/dvdy.20153. [DOI] [PubMed] [Google Scholar]
- 12.Poss KD. Shen J. Nechiporuk A. McMahon G. Thisse B. Thisse C. Keating MT. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- 13.Stoick-Cooper CL. Weidinger G. Riehle KJ. Hubbert C. Major MB. Fausto N. Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 14.Whitehead GG. Makino S. Lien C-L. Keating MT. Fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- 15.Kaufman CK. White RM. Zon L. Chemical genetic screening in the zebrafish embryo. Nat Protoc. 2009;4:1422–1432. doi: 10.1038/nprot.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina G. Vogt A. Bakan A. Dai W. Queiroz de Oliveira P. Znosko W, et al. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol. 2009;5:680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalen M. Wallgard E. Asker N. Nasevicius A. Athley E. Billgren E, et al. Combination of reverse and chemical genetic screens reveals angiogenesis inhibitors and targets. Chem Biol. 2009;16:432–441. doi: 10.1016/j.chembiol.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphey RD. Stern HM. Straub CT. Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Des. 2006;68:213–219. doi: 10.1111/j.1747-0285.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- 19.Bauer MP. Goetz FW. Isolation of gonadal mutations in adult zebrafish from a chemical mutagenesis screen. Biol Reprod. 2001;64:548–554. doi: 10.1095/biolreprod64.2.548. [DOI] [PubMed] [Google Scholar]
- 20.Goldsmith MI. Fisher S. Waterman R. Johnson SL. Saltatory control of isometric growth in the zebrafish caudal fin is disrupted in long fin and rapunzel mutants. Dev Biol. 2003;259:303–317. doi: 10.1016/s0012-1606(03)00186-6. [DOI] [PubMed] [Google Scholar]
- 21.Nechiporuk A. Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- 22.Bousquet P. Feldman J. Schwartz J. Central cardiovascular effects of alpha adrenergic drugs: differences between catecholamines and imidazolines. J Pharmacol Exp Ther. 1984;230:232–236. [PubMed] [Google Scholar]
- 23.Doxey JC. Gadie B. Lane AC. Tulloch IF. Evidence for pharmacological similarity between alpha 2-adrenoceptors in the vas deferens and central nervous system of the rat. Br J Pharmacol. 1983;80:155–161. doi: 10.1111/j.1476-5381.1983.tb11061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doxey JC. Lane AC. Roach AG. Virdee NK. Comparison of the alpha-adrenoceptor antagonist profiles of idazoxan (RX 781094), yohimbine, rauwolscine and corynanthine. Naunyn Schmiedebergs Arch Pharmacol. 1984;325:136–144. doi: 10.1007/BF00506193. [DOI] [PubMed] [Google Scholar]
- 25.Michel MC. Ernsberger P. Keeping an eye on the I site: imidazoline-preferring receptors. Trends Pharmacol Sci. 1992;13:369–370. [PubMed] [Google Scholar]
- 26.Reis DJ. Li G. Regunathan S. Endogenous ligands of imidazoline receptors: classic and immunoreactive clonidine-displacing substance and agmatine. Ann NY Acad Sci. 1995;763:295–313. doi: 10.1111/j.1749-6632.1995.tb32416.x. [DOI] [PubMed] [Google Scholar]
- 27.Dabire H. Idazoxan: a novel pharmacological tool for the study of alpha 2-adrenoceptors. J Pharmacol. 1986;17:113–118. [PubMed] [Google Scholar]
- 28.Ernsberger P. Meeley MP. Mann JJ. Reis DJ. Clonidine binds to imidazole binding sites as well as alpha 2-adrenoceptors in the ventrolateral medulla. Eur J Pharmacol. 1987;134:1–13. doi: 10.1016/0014-2999(87)90125-7. [DOI] [PubMed] [Google Scholar]
- 29.Johnson SL. Weston JA. Temperature-sensitive mutations that cause stage-specific defects in zebrafish fin regeneration. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino S. Whitehead GG. Lien C-L. Kim S. Jhawar P. Kono A, et al. Heat-shock protein 60 is required for blastema formation and maintenance during regeneration. Proc Natl Acad Sci USA. 2005;102:14599–14604. doi: 10.1073/pnas.0507408102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nechiporuk A. Poss KD. Johnson SL. Keating MT. Positional cloning of a temperature-sensitive mutant emmental reveals a role for sly1 during cell proliferation in zebrafish fin regeneration. Dev Biol. 2003;258:291–306. doi: 10.1016/s0012-1606(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 32.Lee Y. Grill S. Sanchez A. Murphy-Ryan M. Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- 33.Kawakami A. Fukazawa T. Takeda H. Early fin primordia of zebrafish larvae regenerate by a similar growth control mechanism with adult regeneration. Dev Dyn. 2004;231:693–699. doi: 10.1002/dvdy.20181. [DOI] [PubMed] [Google Scholar]
- 34.Yoshinari N. Ishida T. Kudo A. Kawakami A. Gene expression and functional analysis of zebrafish larval fin fold regeneration. Dev Biol. 2009;325:71–81. doi: 10.1016/j.ydbio.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 35.Anstead GM. Steroids, retinoids, and wound healing. Adv Wound Care. 1998;11:277–285. [PubMed] [Google Scholar]
- 36.Lee Y. Hami D. De Val S. Kagermeier-Schenk B. Wills AA. Black BL, et al. Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev Biol. 2009;331:270–280. doi: 10.1016/j.ydbio.2009.05.545. [DOI] [PMC free article] [PubMed] [Google Scholar]