Abstract
The activation of pro-inflammatory gene programs by nuclear factor-κB (NF-κB) is primarily regulated through cytoplasmic sequestration of NF-κB by the inhibitor of κB (IκB) family of proteins1. IκBβ, a major IκB isoform, can sequester NF-κB in the cytoplasm2, although its biological role remains unclear. While cells lacking IκBβ have been reported3,4, in vivo studies have been limited and suggested redundancy between IκBα and IκBβ5. Like IκBα, IκBβ is also inducibly degraded, however upon stimulation by LPS, IκBβ is degraded slowly and resynthesized as a hypophosphorylated form that can be detected in the nucleus6–11. The crystal structure of IκBβ bound to p65 suggested this complex might bind DNA12. In vitro, hypophosphorylated IκBβ can bind DNA with p65 and cRel, and the DNA-bound NF-κB:IκBβ complexes are resistant to IκBα, suggesting hypophosphorylated, nuclear IκBβ may prolong the expression of certain genes9–11. We now report that in vivo IκBβ serves to both inhibit and facilitate the inflammatory response. IκBβ degradation releases NF-κB dimers which upregulate pro-inflammatory target genes such as tumor necrosis factor-α (TNFα). Surprisingly absence of IκBβ results in a dramatic reduction of TNFα in response to lipopolysaccharide (LPS) even though activation of NF-κB is normal. The inhibition of TNFα mRNA expression correlates with the absence of nuclear, hypophosphorylated-IκBβ bound to p65:c-Rel heterodimers at a specific κB site on the TNFα promoter. Therefore IκBβ acts through p65:c-Rel dimers to maintain prolonged expression of TNFα. As a result, IκBβ−/− mice are resistant to LPS-induced septic shock and collagen-induced arthritis. Blocking IκBβ might be a promising new strategy for selectively inhibiting the chronic phase of TNFα production during the inflammatory response.
To better understand the biological function of IκBβ we decided to study mice lacking the IκBβ gene. Homologous recombination was used to delete the majority of the IκBβ coding sequences (30–308 aa) including elements essential for binding to NF-κB (Supplementary Fig. 2)6,12,13. Absence of IκBβ was confirmed by immunoblotting of mouse embryonic fibroblasts (MEFs; Supplementary Fig. 2). Although IκBβ is expressed broadly including in hematopoietic organs (Supplementary Fig. 3a), the IκBβ knockout mice breed and develop normally without any obvious phenotypic defects.
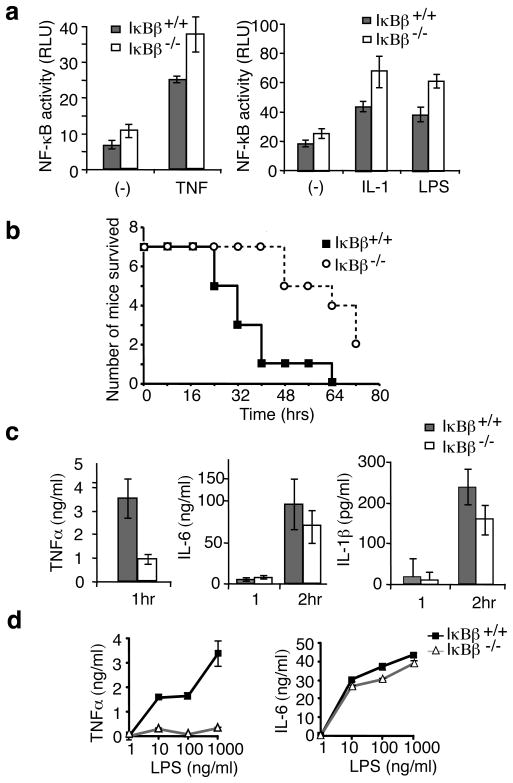
NF-κB and IκB proteins function in an integrated network and hence reduced expression of one component may cause compensatory changes in levels of other proteins 14,15. However, expression levels of IκBα, IκBε, p65, RelB, c-Rel, p105 and p100 were unaffected in IκBβ−/− mice (Supplementary Fig. 3b). Increased NF-κB activity has been observed in other IκB knockouts16–18, and increased basal NF-κB reporter activity was observed in IκBβ−/− MEFs (Fig. 1a). Electrophoretic mobility shift assays (EMSA) demonstrated increased basal NF-κB activity in IκBβ−/− cells (60%) (Supplementary Fig. 3c). Conversely, overexpression of IκBβ inhibits NF-κB activation (Supplementary Fig. 3d). Thus IκBβ inhibits NF-κB and degradation or loss of IκBβ contributes to NF-κB activity. NF-κB reporter assays reveal that absolute NF-κB activity in response to LPS, IL-1β or TNFα is slightly higher in the IκBβ−/− than wild type (WT) cells (Fig. 1a). However, the kinetics of NF-κB activation by EMSA, and the pattern of IκB degradation by immunoblotting, in cells stimulated with LPS, IL-1β or TNFα were not demonstrably different in IκBβ−/− cells (Supplementary Fig. 4). Thus, loss of IκBβ results in a modest elevation in basal NF-κB activity, while inducible NF-κB activation is relatively unaffected.
Figure 1. Mice lacking IκBβ are resistant to LPS-induced endotoxin shock.
a, WT and IκBβ−/− MEF cells transfected with pBIIx-luc reporter and Renilla luciferase vectors were treated with TNFα, IL-1β or LPS for 4 hours and analyzed for luciferase activity. Results are expressed as relative luciferase unit (RLU) normalized by Renilla luciferase activity; error bars indicate ±s.d (n=3). b, Age and sex matched mice received intra-peritoneal injection of LPS and survival rates were scored every 8 hours for 3 days(n=7). c, Serum TNFα, IL-6 and IL-1β 1 hour and/or 2 hour after IP injection of LPS was examined by ELISA; error bars indicate ±s.d (n=5). d, TEPMs from littermate mice were treated for 20 hours with LPS as indicated, and TNFα and IL-6 in the media was determined by ELISA; error bars indicate ±s.d (n=3).
NF-κB regulates the expression of many genes, in particular those involved in inflammation and immune responses19. To determine whether IκBβ has a role in the inflammatory response, IκBβ−/− and IκBβ−/+ mice were challenged with LPS. Surprisingly, IκBβ−/− mice were significantly resistant to the induction of shock (Fig. 1b). We therefore examined the serum levels of the key acute phase cytokines TNFα, IL-1β and IL-620 following LPS injection. In wild type mice TNFα production peaked 1 hour after LPS injection, while IL-6 and IL-1β production peaked around 2 hours, in agreement with previous studies21. Although serum IL-6 and IL-1β were reduced (~25%) in the IκBβ−/− mice, the reduction of TNFα levels (>70%) was more striking (Fig. 1C). As the peak of serum TNFα precedes that of IL-1β and IL-6, it is likely that the reduction of IL-1β and IL-6 is secondary. As monocytes and macrophages are major sources for systemic TNFα, we analyzed LPS induced cytokines in thioglycollate-elicited peritoneal macrophages (TEPM). While equivalent macrophage populations were obtained from the mice (Supplementary Fig. 5a), TNFα, but not IL-6, production was drastically reduced in IκBβ−/− TEPM (Fig. 1d).
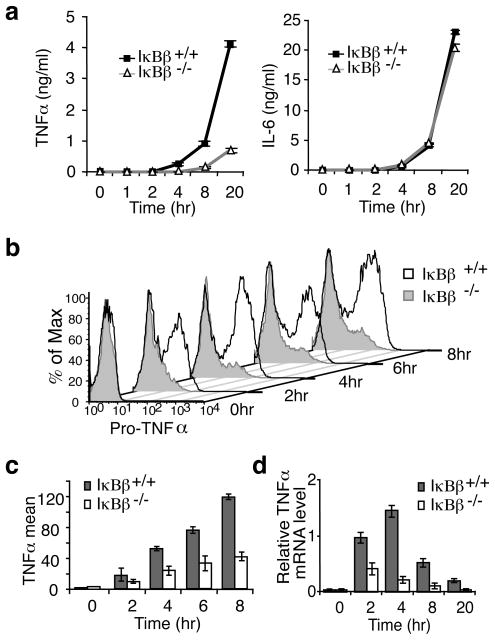
To understand how IκBβ affects TNFα synthesis we examined each step of TNFα production. Secreted TNFα was detectable by ELISA after 2 hours of LPS stimulation and by 4 hours was significantly impaired in IκBβ−/− TEPM (Fig. 2a). IL-6 production was equivalent (Fig. 2a). We examined the level of pro-TNFα by intracellular FACS and found there was very little pro-TNFα detected in the IκBβ−/− TEPMs even after 8 hours of LPS stimulation (Fig. 2b). The average amount of pro-TNFα produced was 2–3 fold higher in WT compared to IκBβ−/− TEPM (Fig. 2c). Consistent with this difference in protein levels, steady-state TNFα was decreased 2–6 fold in the IκBβ−/− TEPM compared to WT cells (Fig. 2d). Although TNFα mRNA is known to be regulated22,23, there was no difference in TNFα mRNA stability between WT and IκBβ−/− TEPM (Supplementary Fig. 5b). Therefore, IκBβ promotes TNFα transcription.
Figure 2. Deficient TNFα transcription in IκBβ−/− macrophages.
a, TEPMs from littermate WT and IκBβ−/− mice were treated with LPS and secreted TNFα and IL-6 were determined by ELISA; error bars indicate ±s.d. (n=3). b, TEMPs from littermate mice were treated as in (a) in the presence of Brefeldin A, and intracellular pro-TNFα was examined with flow cytometry. c, Intracellular pro-TNFα production was examined as in B with macrophages isolated from 3 pairs of littermate mice; error bars indicate ±s.d. d, TEMPs were stimulated with LPS as in A and relative TNFα mRNA level was determined by qRT-PCR; error bars indicate ±s.d (n=3).
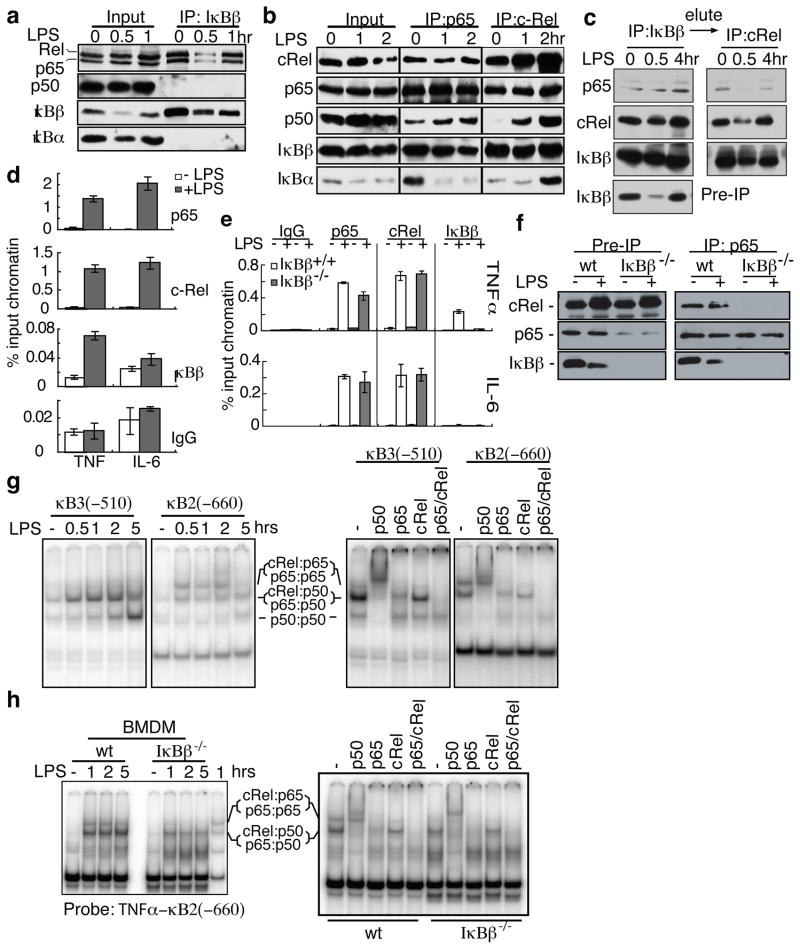
To understand how IκBβ affects TNFα transcription, we investigated which NF-κB subunits were associated with IκBβ in macrophages. It is known that IκBβ associates with p65:p50 and c-Rel:p50 complexes24 through direct binding to p65 and c-Rel but not p506. However, we found that IκBβ could be immunoprecipitated only with p65 and c-Rel, but not p50 (Fig. 3a). Both immunoprecipitations with anti-p65 and anti-c-Rel antibodies pull down IκBβ, IκBα and p50. Thus, there are p65:p50 and inducible c-Rel:p50 complexes that are associated with IκBα or other IκBs, but not IκBβ. Reciprocal immunoprecipitation of p65 with c-Rel and both p65 and c-Rel with IκBβ suggests a p65:c-Rel heterodimer associated with IκBβ (Fig. 3b). To demonstrate the association of IκBβ with p65:c-Rel, we performed sequential immunoprecipitations by first immunoprecipitating IκBβ and then immunprecipitating the eluted IκBβ complexes with anti-c-Rel antibody. The presence of p65 in the anti-c-Rel immunoprecipitate confirms the presence of IκBβ:p65:c-Rel complex (Fig. 3c). The IκBβ:p65:c-Rel complex was found in nuclear extracts suggesting that this could be a transcriptionally active complex. We had previously reported10 that IκBβ exists in two phosphorylation states: a hyperphosphorylated state in quiescent, unstimulated cells, and a hypophosphorylated newly synthesized state in LPS stimulated cells (Fig. 3c and Supplementary Fig. 5a). In the co-immunoprecipitation experiments shown here we found that both forms of IκBβ can bind p65 and c-Rel, although the hypophosphorylated form predominates in the IκBβ:p65:cRel complex following LPS stimulation.
Figure 3. IκBβ is recruited to the promoter of TNFα together with P65 and c-Rel.
a,b, Raw264.7 were stimulated with LPS and immunoprecipitated (IP) with anti-IκBβ (a), anti-p65 (b) or anti-c-Rel (b) antibodies and immunoblotted (IB) as indicated. c, LPS-stimulated Raw264.7 lysates were immunoprecipitated with anti-IκBβ; eluted with IκBβ peptide; immunoprecipitated with anti-c-Rel antibody; and immunoblotted as indicated. d, Raw264.7 lysates were subjected to ChIP as indicated and analyzed by qPCR targeting TNFα and IL-6 promoter κB sites; error bars indicate ±s.d (n=3). e, ChIP was performed as in (d) on WT and IκBβ−/− BMDM treated with LPS for 2 hours; error bars indicate ±s.d (n=3). f, BMDM treated as in (e) were immunoprecipitated with anti-p65 antibody. g, RAW264.7 were treated with LPS and nuclear extracts were subjected to EMSA TNFα κB3 or κB2 probes. Super shifts were performed using cells stimulated for 1hr. h, BMDM were treated with LPS and EMSA and supershifts with the κB2 probe were performed as in (g).
There are four κB sites upstream of TNFα coding region, three of which are crucial for NF-κB dependent TNFα expression25. Therefore, we performed chromatin immunoprecipitation (ChIP) with anti-p65, anti-c-Rel and anti-IκBβ antibodies in RAW264.7 cells and monitored the region encompassing these three κB sites. Following LPS stimulation, TNFα promoter region DNA is enriched by p65, c-Rel and IκBβ antibodies by 56, 70 and 7 fold respectively (Fig. 3d). In contrast, IκBβ is not recruited to the IL-6 promoter following LPS stimulation while p65 and c-Rel are recruited as expected (Fig 3d). Recruitment of p65, c-Rel and IκBβ to the TNFα promoter was also confirmed in WT bone marrow derived macrophages (BMDM; Fig 3e). In the IκBβ−/− BMDM, both p65 and c-Rel are recruited normally to the TNFα promoter. However, when we performed immunoprecipitation with anti-p65, c-Rel and IκBβ are pulled down in WT but not IκBβ−/− BMDM (Fig. 3f). Therefore, p65 and c-Rel fail to form a stable complex in IκBβ−/− cells. Thus, the p65 and c-Rel recruited to the TNFα promoter in IκBβ−/− cells is not a p65:c-Rel complex. These data suggest that optimal TNFα transcription requires a ternary complex of IκBβ:p65:c-Rel binding to the TNFα promoter.
In order to identify the κB site for p65:c-Rel binding we performed EMSAs using the three κB sites from the TNFα promoter as probes (κB2, κB2a and κB3, Supplementary Fig. 5b). We identified two distinct gel-shift patterns. κB3 and κB2a show two major bands (only κB3 is shown in Fig. 3g) while κB2 shows three major inducible shift bands. The components of the bands were identified by super-shift assay (Fig. 3g, right panel). The top band in the κB2 gel-shift is mostly p65:c-Rel. Interestingly, the κB2 site possesses features predicted to favor p65:c-Rel binding (Supplementary Fig. 5c). Similar κB binding sites in the CD40 and CXCL1 promoters also demonstrated coordinate recruitment of IκBβ, p65, and c-Rel (Supplementary Fig. 5d). Furthermore, deletion of the κB2 site from a TNFα promoter reporter abrogated IκBβ-dependent reporter gene expression (Supplementary Fig. 6). In IκBβ−/− BMDM, the p65:c-Rel complex binding to theκB2 in EMSA assays is missing (Fig. 3h), in agreement with the immunoprecipitation result. Therefore optimal TNFα transcription requires a p65:c-Rel complex, stabilized by hypophosphorylated IκBβ, binding to the κB2 site in the TNFα promoter.
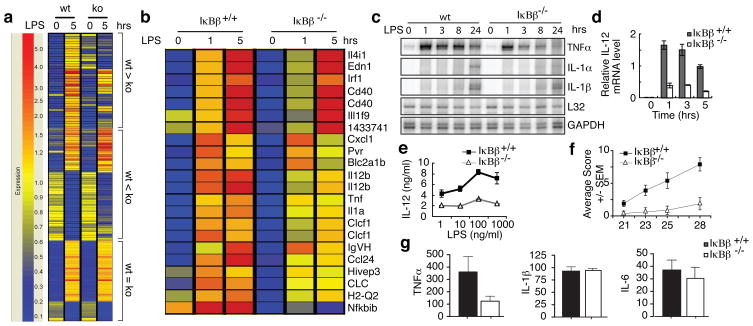
To identify other genes affected by IκBβ deficiency, we examined gene expression profiles in WT and IκBβ−/− BMDM. As expected, TNFα and IκBβ are among the genes whose expression is affected by IκBβ deficiency while IL-6 and IL-1β are not affected (Fig. 4a). Of the genes whose expression is reduced in the IκBβ−/− cells we identified 14 with expression patterns resembling TNFα (Fig. 4b). The expression of these genes was also reduced in p65, c-Rel or p65/c-Rel knock-out fetal liver macrophages suggesting that LPS-induced expression of these genes might depend on a mechanism similar to TNFα (data not shown). The expression of TNFα, IL-1α, IL-6 and IL-1β in response to LPS was further examined by RNase protection (Fig. 4c) and qRT-PCR assays (Supplementary Fig. 7) demonstrating that the reduction in persistent expression of TNFα in IκBβ−/− cells is unique. Reduced IL12b mRNA and protein secretion in the knockout TEPM was confirmed by qRT-PCR (Fig. 4d) and ELISA (Fig. 4e). Notably, transcription of IL12b, which has a κB site similar to κB2 of TNFα (Supplementary Fig. 5c), has previously been shown to require c-Rel and be partially dependent on p6526. Thus, only a select group of NF-κB dependent genes are diminished similarly to TNFα upon IκBβ deletion. As TNFα plays a key role in inflammation, we wanted to test whether IκBβ−/− deletion would affect the course of inflammatory diseases.
Figure 4. IκBβ knockout selectively affects only certain LPS responsive genes and attenuates collagen induced arthritis.
a, LPS responsive genes whose expression is either down-regulated, up-regulated or unchanged in IκBβ−/− BMDM. b, Host-pathogen interaction genes that are IκBβ dependent and LPS responsive genes whose expression pattern resembles TNFα. c, RNase protection assay using WT and IκBβ−/− BMDM stimulated with LPS. d, IL-12b relative mRNA level determined by qRT-PCR in samples prepared as in (c); error bars indicate ±s.d. (n=3). e, ELISA for IL-12p40 secreted from WT and IκBβ−/− TEMP stimulated with LPS for 20 hours; error bars indicate ±s.d. f, Arthritis clinical scoring in WT (n=10) or IκBβ−/− (n=8) DBA mice; error bars indicate ±SEM. g, Serum TNF-α, IL-1β, and IL-6 levels in WT or IκBβ−/− DBA mice in (f); error bars indicate ±SEM.
Rheumatoid arthritis (RA) is a common inflammatory disease with morbidity resulting from ongoing release of pro-inflammatory cytokines, including TNFα, and consequent destruction of joint tissue27. Previous studies have shown that NF-κB plays a key role in mouse models of arthritis and blocking NF-κB has a dramatic effect in preventing disease28,29. RA can also be effectively treated by anti-TNFα therapies, although there are significant side-effects30. The ability to block only persistent TNFα expression would be therapeutic without blocking beneficial TNFα responses including the expression of innate immune response genes. We therefore tested whether the lack of IκBβ altered the course of collagen-induced arthritis (CIA), a well-characterized mouse model of RA.
To induce CIA we immunized DBA/1J mice with bovine type II collagen. IκBβ−/− mice displayed delayed onset, lower incidence and decreased severity of CIA (Fig. 4f and Supplementary Fig. 8). Inflammation in the WT mice extended from the paws and digits to the ankle joints and distally through the limb (data not shown). In contrast, IκBβ−/− mice showed minimal visual signs of paw and joint swelling (Supplementary Fig. 8). Serum TNFα was markedly decreased in IκBβ−/− mice while other pro-inflammatory cytokines were not significantly affected (Fig. 4g and Supplementary Fig. 9). Therefore the absence of IκBβ limits the progression and severity of arthritis by reducing the chronic production of TNFα.
The results presented above demonstrate a dual role for IκBβ: during the early stages of LPS stimulation, NF-κB complexes released by IκBβ degradation contribute to the initial expression of TNFα (Supplementary Fig. 1). Then, newly synthesized hypophosphorylated IκBβ facilitates the formation of IκBβ:p65:c-Rel complexes which selectively bind to the κB2 site in the TNFα promoter augmenting transcription. As shown in the gene chip and RNAse protection assays, this is a relatively selective function and IκBβ−/− mice are, therefore, otherwise normal. Hence targeting IκBβ might be a promising new strategy to treat chronic inflammatory diseases such as arthritis.
Methods summary
Mice
IκBβ deficient mice were generated by standard homologous recombination in the CJ7 ES cell line using a targeting construct that replaced exon 2 through exon 5 with a G418-resistance gene. Screened ES cell clones were injected into blastocysts derived from C57BL/6 mice gave rise to IκBβ−/+/IκBβ+/+ chimeras. Germline transmission of the disrupted allele was obtained and verified by Southern blotting and PCR, and mice were backcrossed at least 10 generations onto the B57BL/6 background. Mice were backcrossed at least 8 generations onto the DBA background for CIA experiments. Mice were maintained in pathogen-free animal facilities at Yale Medical School.
Cells
WT and IκBβ knockout MEFs were generated from E12.5 embryos following timed breeding of IκBβ+/− animals. TEMPs were obtained from 6- to 8-week-old littermate mice three days after intraperitoneal injection with thioglycollate. BMDM were harvested by standard protocols and differentiated with 30% L929 supernatant-conditioned media.
Biochemistry
Cell fractionation, western blotting, EMSA, and immunoprecipitations were performed as previously described unless otherwise indicated6.
LPS-induced shock
LPS-induced shock was tested by intraperitoneal injection of 50 ug/g body weight LPS and monitoring for survival. In a separate identical experiment, the mice were bled at 1 hr and 2 hr after LPS treatment and the concentration of TNF-α, IL-6 and IL-1β in the serum was measured by ELISA.
Intracellular cytokine analysis
Pro-TNFα levels were analyzed in LPS stimulated TEMPs cells following LPS stimulation and brefeldin-A treatment. TNFα was detected following cell permeabilization using standard intracellular cytokine staining and flow cytometry.
qRT-PCR
RNA expression was quantified by quantitative two-step SYBR real-time RT-PCR, and relative mRNA levels were obtained by normalizing the readout for each specific gene by that of β-actin.
Microarray Analysis
Microarrays for gene expression analyses were performed on BMDMs stimulated with LPS and Affymetrix Mouse genome 430A 2.0 arrays as per the manufacturers protocol.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
We thank Dr. Aiping Lin at the Yale W.M. Keck Biostatistics Resource for analysis of microarray data. Supported by grants from the National Institutes of Health to SG.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author contributions PR characterized the mice and performed the majority of the experiments, MSH performed the immunoprecipitation experiments and helped in writing the paper, ML performed CIA experiments, DZ and APW performed generation of BMDM cells, AO performed some experiments, MLS and DB generated the knockout mice, CL and AH performed the RNAse protection assays, and SG conceived of the study and wrote the paper.
References
- 1.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132 (3):344. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Malek S, Chen Y, Huxford T, Ghosh G. IkappaBbeta, but not IkappaBalpha, functions as a classical cytoplasmic inhibitor of NF-kappaB dimers by masking both NF-kappaB nuclear localization sequences in resting cells. J Biol Chem. 2001;276 (48):45225. doi: 10.1074/jbc.M105865200. [DOI] [PubMed] [Google Scholar]
- 3.Tergaonkar V, Correa RG, Ikawa M, Verma IM. Distinct roles of IkappaB proteins in regulating constitutive NF-kappaB activity. Nat Cell Biol. 2005;7 (9):921. doi: 10.1038/ncb1296. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298 (5596):1241. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 5.Cheng JD, et al. Functional redundancy of the nuclear factor kappa B inhibitors I kappa B alpha and I kappa B beta. J Exp Med. 1998;188 (6):1055. doi: 10.1084/jem.188.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson JE, et al. I kappa B-beta regulates the persistent response in a biphasic activation of NF-kappa B. Cell. 1995;80 (4):573. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 7.Weil R, Laurent-Winter C, Israel A. Regulation of IkappaBbeta degradation. Similarities to and differences from IkappaBalpha. J Biol Chem. 1997;272 (15):9942. doi: 10.1074/jbc.272.15.9942. [DOI] [PubMed] [Google Scholar]
- 8.Kerr LD, et al. The rel-associated pp40 protein prevents DNA binding of Rel and NF-kappa B: relationship with I kappa B beta and regulation by phosphorylation. Genes Dev. 1991;5 (8):1464. doi: 10.1101/gad.5.8.1464. [DOI] [PubMed] [Google Scholar]
- 9.Tran K, Merika M, Thanos D. Distinct functional properties of IkappaB alpha and IkappaB beta. Mol Cell Biol. 1997;17 (9):5386. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suyang H, Phillips R, Douglas I, Ghosh S. Role of unphosphorylated, newly synthesized I kappa B beta in persistent activation of NF-kappa B. Mol Cell Biol. 1996;16 (10):5444. doi: 10.1128/mcb.16.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips RJ, Ghosh S. Regulation of IkappaB beta in WEHI 231 mature B cells. Mol Cell Biol. 1997;17 (8):4390. doi: 10.1128/mcb.17.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malek S, et al. X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex. J Biol Chem. 2003;278 (25):23094. doi: 10.1074/jbc.M301022200. [DOI] [PubMed] [Google Scholar]
- 13.Ernst MK, Dunn LL, Rice NR. The PEST-like sequence of I kappa B alpha is responsible for inhibition of DNA binding but not for cytoplasmic retention of c-Rel or RelA homodimers. Mol Cell Biol. 1995;15 (2):872. doi: 10.1128/mcb.15.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memet S, et al. IkappaBepsilon-deficient mice: reduction of one T cell precursor subspecies and enhanced Ig isotype switching and cytokine synthesis. J Immunol. 1999;163 (11):5994. [PubMed] [Google Scholar]
- 15.Hertlein E, et al. RelA/p65 regulation of IkappaBbeta. Mol Cell Biol. 2005;25 (12):4956. doi: 10.1128/MCB.25.12.4956-4968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klement JF, et al. IkappaBalpha deficiency results in a sustained NF-kappaB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16 (5):2341. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 1995;9 (22):2736. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 18.Goudeau B, et al. IkappaBalpha/IkappaBepsilon deficiency reveals that a critical NF-kappaB dosage is required for lymphocyte survival. Proc Natl Acad Sci U S A. 2003;100 (26):15800. doi: 10.1073/pnas.2535880100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25 (51):6758. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 20.Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 2008;8 (10):776. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans GF, Snyder YM, Butler LD, Zuckerman SH. Differential expression of interleukin-1 and tumor necrosis factor in murine septic shock models. Circ Shock. 1989;29 (4):279. [PubMed] [Google Scholar]
- 22.Kontoyiannis D, et al. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10 (3):387. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J Exp Med. 1990;171 (2):465. doi: 10.1084/jem.171.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu ZL, et al. Basal phosphorylation of the PEST domain in the I(kappa)B(beta) regulates its functional interaction with the c-rel proto-oncogene product. Mol Cell Biol. 1996;16 (11):5974. doi: 10.1128/mcb.16.11.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuprash DV, et al. Similarities and differences between human and murine TNF promoters in their response to lipopolysaccharide. J Immunol. 1999;162 (7):4045. [PubMed] [Google Scholar]
- 26.Sanjabi S, et al. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc Natl Acad Sci U S A. 2000;97 (23):12705. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennan FM, McInnes IB. Evidence that cytokines play a role in rheumatoid arthritis. J Clin Invest. 2008;118 (11):3537. doi: 10.1172/JCI36389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miagkov AV, et al. NF-kappaB activation provides the potential link between inflammation and hyperplasia in the arthritic joint. Proc Natl Acad Sci U S A. 1998;95 (23):13859. doi: 10.1073/pnas.95.23.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimi E, et al. Selective inhibition of NF-kappa B blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo. Nat Med. 2004;10 (6):617. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- 30.Feldmann M. Development of anti-TNF therapy for rheumatoid arthritis. Nat Rev Immunol. 2002;2 (5):364. doi: 10.1038/nri802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.