Summary
Background:
The inflammatory cytokine interleukin-6 (IL-6) is a main regulator of fibrinogen synthesis, though its interaction with fibrinogen genes (FGA, FGB, FGG) in relation to CVD risk is not well-studied in humans.
Methods and Results:
We investigated joint associations of common fibrinogen and IL6 tagSNPs with fibrinogen level, carotid intima-media thickness (IMT) and risk of myocardial infarction (MI) or ischemic stroke in 3900 European-American participants of the Cardiovascular Health Study. To identify combinations of genetic main effects and interactions associated with each outcome, we used logic regression. We also evaluated whether the relationship between fibrinogen SNPs and fibrinogen level varied by IL-6 level using linear regression models with multiplicative interaction terms. Combinations of fibrinogen and IL6 SNPs were associated with fibrinogen level (p<0.005), but not with IMT (p>0.30), MI (p=0.73) or stroke (p=0.21). Fibrinogen levels were higher in higher in individuals having FGB1437 (rs1800790) minor alleles and lacking FGA6534 (rs6050) minor alleles; these SNPs interacted with IL6 rs1800796 to influence fibrinogen level. Marginally significant (p=0.03) interactions between IL-6 level and SNPs located in promoter regions of FGA and FGG associated with fibrinogen levels were detected.
Conclusion:
We identified potential gene-gene interactions influencing fibrinogen levels. Although IL-6 responsive binding sites are present in fibrinogen gene promoter regions, we did not find strong evidence of interaction between fibrinogen SNPs and IL6 SNPs or levels influencing CVD risk.
Background
Fibrinogen is a key component of the inflammation and clotting pathways and an established risk factor for cardiovascular disease (CVD).(Danesh et al., 2005) Fibrinogen production is upregulated in response to cytokines released during inflammation, infection, neoplasia or tissue damage.(Ernst, 1993)
Three genes (FGA, FGB and FGG) on human chromosome 4 code for the two sets of α, β, and γ–chains that constitute fibrinogen. Although each gene is separately transcribed and translated, transcription is well coordinated so that when expression of one gene is elevated, the other genes are also upregulated.(Duan & Simpson-Haidaris, 2003, Fuller & Zhang, 2001) Interleukin-6 (IL-6) is the key regulator, though other mediators, such as glucocorticoids, IL-1β and TNF-α also modulate acute phase fibrinogen synthesis.(Fuller & Zhang, 2001) Acting via an intracellular pathway, IL-6 binds to its receptor and activates STAT proteins. In particular, STAT3, also known as acute phase response factor, is the major transcription factor mediating regulation of IL-6-responsive genes.(Duan & Simpson-Haidaris, 2006) STAT3 relays signals from the IL-6 receptor to the nucleus where it binds to response elements in the promoter regions of the fibrinogen genes. Human fibrinogen genes are highly responsive to the IL-6 cytokine; several IL-6 responsive binding sites in the promoter regions of the fibrinogen genes have been identified.(Duan & Simpson-Haidaris, 2003, Heinrich et al., 1990)
Though IL-6 is a key regulator of fibrinogen synthesis, few studies have investigated whether having specific combinations of variants in the IL6 and fibrinogen genes predisposes to CVD. In one such study, no interaction between an IL6 SNP and three fibrinogen SNPs influencing risk of MI was detected, though the study was small and included few variants.(Mannila et al., 2006)
Given the role of IL-6 in fibrinogen regulation, we hypothesized that genetic variation in the fibrinogen locus could affect binding to IL-6 response elements and potentially the regulation of fibrinogen levels in humans. Furthermore, the potential synergism between these genes on the fibrinogen phenotype may have implications for subclinical and clinical CVD development. In this study, we assessed the potential interaction of both common IL6 SNPs and the gene product, IL-6 cytokine levels, with fibrinogen SNPs in influencing fibrinogen levels. Fibrinogen and IL6 gene-gene interactions associated with markers of subclinical CVD, internal and common carotid artery intima-medial thickness (IMT), and risk of future ischemic stroke or myocardial infarction (MI) were also evaluated.
Methods
Study Population
The Cardiovascular Health Study is a population-based cohort of mainly white (European-descent) adults aged 65 years and older recruited from 4 field centers in North Carolina, California, Maryland and Pennsylvania.(Fried et al., 1991) Demographic, lifestyle and medical history information and fasting blood samples were collected on CHS participants at baseline. Blood collection procedures and laboratory methods have been reported previously.(Cushman et al., 1995) Hypertension was defined as systolic BP>=140mmHG or diastolic BP >=90mmHG or having a history of hypertension with concurrent use of anti-hypertensive medication. Diabetes status was defined as ‘established/new’ for participants using insulin or oral hypoglycemics and for participants with fasting glucose ≥ 126mg/dL, or as ‘normal.’ We restricted the current study to individuals of European descent, ‘whites,’ for issues of power; our genetic analyses precluded missing data and thus pooling of racial groups with different genotyped tagSNPs was not feasible without loss of gene coverage. Additionally, adequate power for interaction analyses (of moderate effect size) is a problem in the smaller minority cohort. Of the 4925 individuals whose self-identified race was ‘white,’ we excluded individuals who did not provide informed consent for participation in DNA studies, or for whom DNA was not available (n=211). Additionally, participants with a history of MI or stroke at baseline (n=610), those for whom baseline measures of fibrinogen were unavailable (n=44) and those with incomplete genotype data (n=160) were also excluded for a final sample of 3900. An additional 299 individuals without baseline measurements were dropped from analyses involving IL-6 levels for a total of 3621 participants.
Using baseline samples, plasma fibrinogen levels were measured using the Clauss assay which measures the functional clotting activity of fibrinogen. Both IL-6 and C-reactive protein (CRP) were measured in stored frozen serum samples using a commercial assay (Quantikine HS Human IL-6 Immunoassay, R&D Systems, Minneapolis, MN, USA) and a validated in-house high-sensitivity enzyme-linked-immunosorbent-assay (ELISA), respectively.(Macy et al., 1997) Analytic coefficients of variation were 9.7%, 6.3% and 5.1% for the assays, respectively. Maximum carotid artery intima-media thickness (IMT) in mm was determined at the baseline examination by high-resolution B-mode ultrasonography.(O'Leary et al., 1991) IMT measures were calculated for the common and internal carotid arteries by averaging maximum wall thicknesses obtained from scans of the near and far walls on the left and right sides.
Participants were followed through June 2003 (maximal follow-up of 14 years) with median follow-up of 13 years. Censoring date was defined by death, loss to follow-up, study drop out or event date. Incident, non-procedure-related (not occurring during surgery or re-vascularization) MI or ischemic stroke events were analyzed. MI was defined using standard CHS criteria: history of chest pain, cardiac enzyme levels, and characteristic changes on serial electrocardiograms. Stroke was validated based on criteria that included onset of symptoms, duration of deficits, and findings on computed tomography or magnetic resonance imaging. Participants with multiple incident events were censored on the date of the first event. All events were adjudicated by CHS committee as reported previously.(Ives et al., 1995)
SNP Selection Strategy and Genotyping
TagSNPs in the fibrinogen (n=16) and IL6 (n=10) genes were identified by the SNP discovery resource, SeattleSNPs, among European descent individuals (whites) using the LD select algorithm.(Carlson et al., 2004) Of these, 12 fibrinogen and 6 IL6 tagSNPs were common (minor allele frequency (MAF) ≥ 5%) in CHS whites. Genotyping was performed at the Laboratory for Clinical Biochemistry Research (University of Vermont) with the ABI TaqMan platform using an ABI 7900 real time thermal cycler under standard conditions (Applied Biosystems, Foster City, CA). Deviations from Hardy Weinberg equilibrium were tested with the Chi2-test; no severe deviations were apparent, but it was determined that a few of the tagSNPs were highly correlated (r2 ≥ 0.60). We excluded the following correlated SNPs with slightly lower MAF; specifically, FGA2224 (rs2070011), FGA9205 (rs2070022) and FGG10034 (rs2066865) and one IL6 tagSNP (rs1800795), leaving 9 fibrinogen and 5 IL6 tagSNPs for analysis. [Supplemental table and Figure]
Statistical Analysis
To investigate complex interaction between IL6 SNPs and fibrinogen SNPs, and simple interaction between IL-6 concentration and fibrinogen SNPs associated with CVD outcomes, we applied two separate statistical methods. Combinations of SNPs in fibrinogen and IL6 associated with fibrinogen level, carotid IMT or CVD events were evaluated by a data-driven approach to model building using logic regression.(Ruczinski et al., 2003) Briefly, logic regression builds potential models by searching for a variable (called a leaf) and/or Boolean expression (called a tree) that best predicts outcome (i.e. minimizes the scoring function); this process is repeated until additional variables do not improve model scores. It is adaptable to different forms of regression-based methods including linear regression and Cox proportional hazards methods. To simplify its interpretation and for discovery of combinations of predictors affecting relatively larger groups of individuals, we limited final model size a priori, by setting the maximum tree size to 2 and the number of leaves (i.e. variables) to 8, as suggested by Kooperberg et al.(Kooperberg et al., 2001) We used linear regression to analyze interactions between fibrinogen SNPs and IL6 SNPs associated with the continuous outcomes of baseline fibrinogen level and carotid IMT. MI and ischemic stroke events were analyzed using Cox proportional hazards models. Since logic regression is adaptive, appropriate model selection is necessary to prevent ‘over-fitting’ of the data. The logic regression software includes model selection tools such as cross-validation and conditional permutation tests which are used for interpretation of results. These tests account for multiple testing and thus reduce the chance of type I error. Once potential models have been created, a null model permutation test is run (n=200). Pseudo p-values derived from this test represent the proportion of times the ‘best fit model’ scores better than the ‘null model,’ suggesting evidence against the null model. Provided this null test indicated that the predictors had some discriminatory power, we chose a threshold of p<0.20, we pursued additional model selection tests using cross-validation (10-fold) and conditional permutation (n=200) tests conditioning on successive models of increasing size; a successive model was only chosen if it fit data better (had a smaller score) than the prior smaller model. Gene-gene interaction models were minimally adjusted for age, sex and recruitment site since acquired factors associated with fibrinogen level or risk of CVD events are unlikely to confound the genotypes. Both dominant and recessive genetic models were tested using this approach. For logic regression analyses, we used R software and the logic regression package (v.1.0.4) on the Linux platform; STATA/SE software v.8.2 (StataCorp, College Station, TX) was used for all other analyses.
To evaluate potential interactions between IL-6 levels and genetic variants in the fibrinogen genes with fibrinogen level, the significance of interaction terms on a multiplicative scale (fibrinogen SNPs in an additive genetic model multiplied by IL-6 concentration) was assessed using Wald's test. Only two-way interactions were tested. In sensitivity analyses, genotype was modeled using categorical variables for each genotype (common homozygote as reference) and the interaction effects were tested using a likelihood ratio test statistic, with two degrees of freedom. Results differed little from additive models, therefore for brevity, only results from additive genetic models results are presented. For the IL-6 level and fibrinogen SNP interactions, confounding of the IL-6 interaction term was an important consideration. For all models involving IL-6 level in the interaction term, we adjusted for age, sex and recruitment site a priori and investigated the following baseline variables for potential confounding of the IL-6 term: body mass index (BMI), current smoking, systolic blood pressure, diastolic blood pressure, medication use (anti-hypertensive or lipid-lowering agents and aspirin), diabetes or hypertension status and serum lipid levels (LDL-cholesterol, HDL-cholesterol, triglycerides and total cholesterol).
Results
Descriptives
In this subset of the CHS population without a prior history of MI or stroke, the majority of participants were female (60%).[Table 1] Diagnosis of hypertension was common in this older population (mean age at study entry= 73 years), though the prevalence of other CVD risk factors such as current smoking or diabetes was relatively low.
Table 1.
Baseline Characteristics of the Study Population
| Characteristic1 | N=3900 |
|---|---|
| Female | 2322 (59.5) |
| Age in years | 72.6 ± 5.5 |
| BMI in kg/m2 | 26.3 ± 4.5 |
| Current Smoker | 428 (11.0) |
| Diabetes (established/new) | 508 (13.0) |
| Hypertension | 2129 (54.8) |
| Hypertension Medication Use | 1584 (40.7) |
| Lipid-Lowering Medication Use | 189 (4.9) |
| Aspirin Use (>2 days in prior 2 weeks) | 1210 (31.1) |
| Total Cholesterol in mg/dL | 212.3 ± 39.0 |
| LDL Cholesterol in mg/dL | 130.1 ± 35.7 |
| HDL Cholesterol in mg/dL | 54.6 ± 15.8 |
| Triglycerides in mg/dL | 141.4 ± 74.8 |
| ln(CRP) in mg/L | 0.6 ± 1.0 |
Data are presented as number (%), or mean ± standard deviation.
Mean fibrinogen level was 317± 63 mg/dL and was approximately normally distributed, but due to the highly skewed distribution of IL-6 (median=1.6 pg/ml, IQR: 1.1-2.5), it was ln-transformed for analyses, mean ln(IL-6)= 0.6 ± 0.6. While several fibrinogen SNPs are strongly associated with fibrinogen levels in this population, IL6 SNPs did not appear to be strongly associated with plasma fibrinogen in univariate analyses. (data not shown)
Mean common carotid IMT was 0.99 ± 0.20 mm whereas internal carotid IMT was slightly skewed, median=1.32mm (IQR: 0.90-1.88), and was ln-tranformed for analysis. A total of 486 incident non-procedure related MIs and 419 ischemic strokes were observed; median follow-up times were approximately 13.1 years for both events.
Evaluation of gene-gene interactions in relation to fibrinogen levels, carotid IMT and CVD events
Using logic regression and modeling fibrinogen level as the outcome in linear regression models, the p-value for the null randomization test was significant, p<0.005.[Table 2] This finding suggests evidence of an association between fibrinogen level and at least one SNP or combination of SNPs so we pursued model selection to identify the minimal set of variables that best modeled the fibrinogen level outcome.
Table 2.
Logic Regression Results for the Within Gene and Gene-Gene Interactions between Fibrinogen SNPs and IL6 SNPs in Relation to Tested Outcomes
| Continuous Outcome | Signal Test p-value |
|---|---|
| Fibrinogen level in mg/dL | <0.005 |
| Ln(internal carotid IMT in mm) | 0.31 |
| Common carotid IMT in mm | 0.61 |
| Incident MI | 0.73 |
| Incident ischemic stroke | 0.21 |
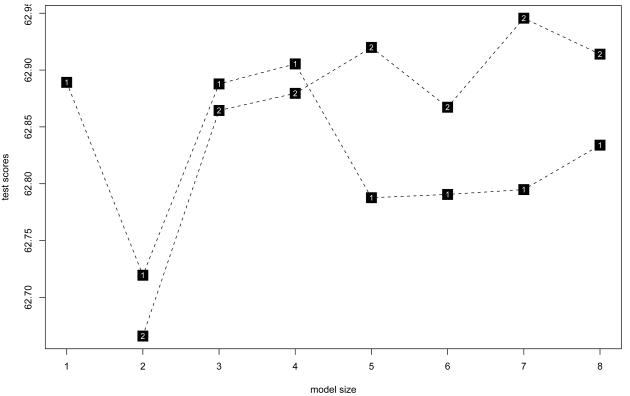
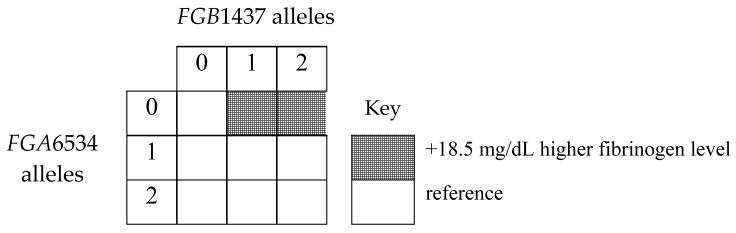
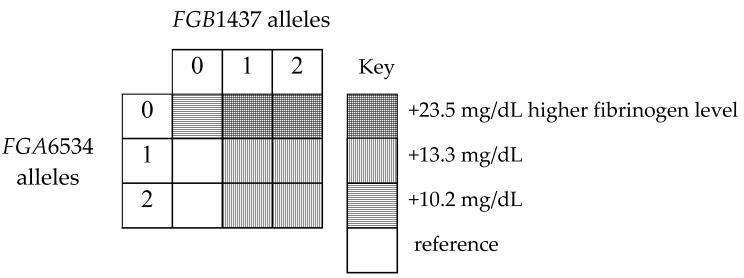
Of the top five parsimonious models evaluated using conditional permutation [Table 3] and/or cross-validation tests [Figure 1], the two best models included combinations of functional variants in FGA6534 (rs6050) and FGB1437 (rs1800790), both with dominant effects. Specifically, cross validation identified two models for the best fit: 1) 1 tree with 2 leaves and 2) 2 trees and 2 leaves.[Figure 2] The p-values estimated from conditional randomization testing of these models were p=0.055 and 0.180, respectively. The first model with ‘1 tree and 2 leaves’ includes one gene-gene interaction term and no main effect terms. It may be interpreted as individuals with no FGA6534 (rs6050) minor alleles and at least 1 minor allele of FGB1437 (rs1800790) have 18.5 mg/dL higher fibrinogen level on average than individuals who do not have that combination of genotypes, holding site, age and sex constant. The second model (2 trees and 2 leaves) includes main effect terms for the same FGA and FGB SNPs, but no interaction term. It suggests that after adjustment for site, age, sex and FGA6534 genotype, individuals with at least one FGB1437 (rs1800790) minor allele have 13.3 mg/dL higher levels of fibrinogen than individuals with no minor alleles. In addition, after adjustment for site, age, sex and FGB1437 (rs1800790) genotype, individuals with no FGA6534 (rs6050) minor alleles have 10.2 mg/dL higher fibrinogen level on average than individuals who have at least one minor allele. The pairwise correlation for these FGA and FGB SNPs was low, r2=0.03 though minor allele frequencies were similar, 25% and 22% respectively. These same SNPs appeared in the remaining top five models, along with IL6-1111 (rs1800796) (dominant genetic model) and FGA251 (rs2070006) (recessive genetic model). Specifically, permutation tests identified a model supporting interaction between FGA6534 (rs6050) and IL6-1111 (rs1800796), but because this model also included an additional term involving FGB1437 (rs1800790), it was not the most parsimonious compared with the two models listed above. We did not find strong evidence of interaction between fibrinogen SNPs and IL6 SNPs in relation to carotid IMT, MI or ischemic stroke; all global test p-values > 0.20.
Table 3.
Top Five Models from Conditional Permutation Tests
| Size | Model | p-value |
|---|---|---|
| 1 tree, 1 leaf | 14.8 * (≥1 FGB1437 minor allele) | 0.010 |
| 1 tree, 2 leaves | 18.5 * ((no FGA6534 minor alleles) & ≥1 FGB1437minor allele) | 0.055 |
| 1 tree, 3 leaves | −18.6*((no FGB1437 minor alleles) or ((no IL6_1111 minor alleles) & ≥1 FGA6534 minor allele)) |
0.105 |
| 1 tree, 4 leaves | −19.6 * ((≥1 FGA6534 minor allele & (no IL6_1111 minor alleles)) or (2 FGA251 minor alleles or (no FGB1437 minor alleles))) |
0.150 |
| 2 trees, 2 leaves | −13.3 * (no FGB1437 minor alleles) + 10.2 * (no FGA6534 minor alleles) |
0.180 |
Figure 1. Logic regression cross-validation testing results for models of fibrinogen and IL6 SNPs associated with fibrinogen level.
Model size (x-axis scale) indicates the number of leaves; shaded boxes indicate the number of trees in the models. The y-axis scale, ‘test score’ reflects the residual variance scores of each model. Lower test scores are indicative of better fitting models. In this figure, both 1 tree and 2 tree models with 2 leaves have the best scores (< 62.75) in 10-fold cross-validation testing.
Figure 2. Interaction between fibrinogen and IL6 SNPs in relation to fibrinogen level: best models identified by logic regression.
A. 1 tree with 2 leaves: score is 62.12
E(fibrinogen) = 243.35 + 18.50 ((no FGA6534 minor alleles) and ≥1 minor allele at FGB1437) + 1.22(site A) +17.67(site B) + 0.82(site C) + 0.91(age) − 4.25(male)
As shown below, this equation includes one gene-gene interaction term and no main effects. It suggests that holding site, age and sex constant, individuals with no FGA6534 minor alleles and at least 1 minor allele of FGB1437 have 18.5 mg/dL higher fibrinogen level on average than individuals who do not have that combination of genotypes.
B. 2 trees with 2 leaves: score is 62.03
E(fibrinogen) = 252.72 − 13.29(no FGB1437 minor alleles) + 10.21(no FGA6534 minor alleles) + 1.16(site A) +17.31(site B) + 0.47(site C) + 0.88(age) − 4.07(male)
As shown below, this equation includes a main effect for the same FGA and FGB SNPs, but no interaction term. It suggests that after adjustment for site, age, sex and FGA6534 genotype, individuals with no FGB1437 minor alleles have 13.3 mg/dL lower levels of fibrinogen than individuals with at least one FGB1437 minor allele. After adjustment for site, age, sex and FGB1437 genotype, individuals with no FGA6534 minor alleles have 10.2 mg/dL higher fibrinogen level on average than individuals who have at least one minor allele.
Effect of interaction between fibrinogen SNPs and IL-6 levels on fibrinogen levels
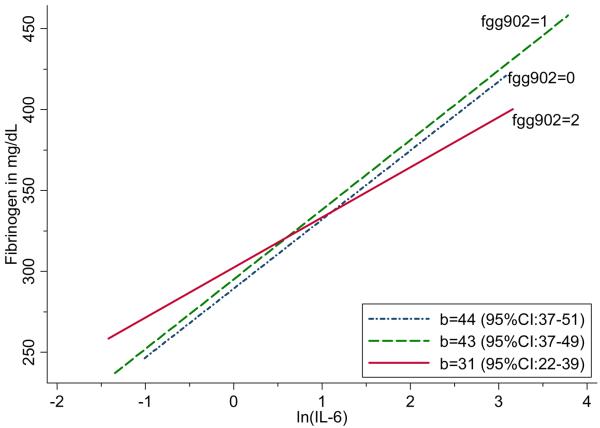
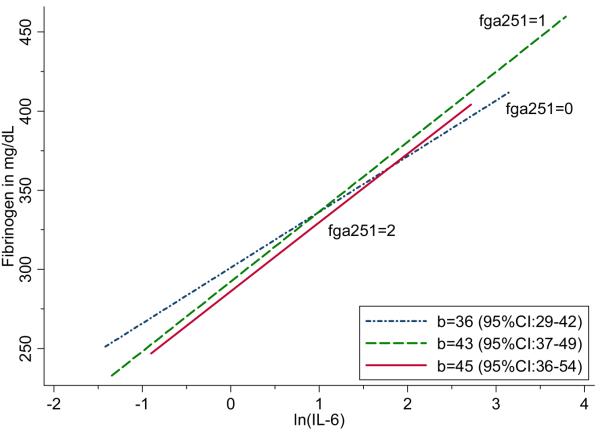
Baseline measures of plasma fibrinogen and IL-6 were strongly associated, for each 1 natural log unit increase in IL-6, mean fibrinogen was 41 mg/dL (95%CI: 37-45; p<0.0001) higher. Marginally significant interactions were found between IL-6 levels and two fibrinogen SNPs FGA251 (rs2070006), p=0.029, and FGG902 (rs1800792), p=0.033, associated with fibrinogen level.[Table 4] For interpretation of these findings, we plotted the relationship between fibrinogen level and ln(IL-6), by fibrinogen genotype. The slope describing the relationship between ln(IL-6) and fibrinogen is not as steep among carriers of two FGG902 alleles compared with other genotypes.[Figure 3] Conversely, the slope describing the association between ln(IL- 6) and fibrinogen is lower among individuals homozygous for the rare FGA251 allele (rs2070006).[Figure 4]
Table 4.
Interactions between Fibrinogen SNPs and IL-6 Plasma Concentration on Fibrinogen Plasma Concentration
| Interaction Term |
β (95%CI)1 p-value |
β (95%CI)2 p-value |
|---|---|---|
| FGA251 × lnIL6 | 5.8 (0.6, 10.9) 0.029 |
5.0 (−0.1, 10.2) 0.057 |
| FGA3807 × lnIL6 | −5.4 (−12.3, 1.4) 0.119 |
−5.2 (−12.0, 1.6) 0.137 |
| FGA5498 × lnIL6 | 6.5 (−0.6, 13.6) 0.074 |
5.9 (−1.2, 12.9) 0.103 |
| FGA6534 × lnIL6 | 3.4 (−2.7, 9.5) 0.270 | 2.9 (−3.2, 8.9) 0.349 |
| FGB1038 × lnIL6 | −1.0 (−8.2, 6.3) 0.797 |
−0.9 (−8.1, 6.3) 0.801 |
| FGB1437 × lnIL6 | 0.3 (−6.2, 6.9) 0.920 |
0.5 (−6.1, 7.0) 0.884 |
| FGB9952 × lnIL6 | −3.6 (−9.2, 2.2) 0.222 |
−3.3 (−9.0, 2.4) 0.255 |
| FGG902 × lnIL6 | −5.6 (−10.8, −0.5) 0.033 |
−5.0 (−10.2, 0.2) 0.058 |
| FGG9340 × lnIL6 | 4.6 (−1.2, 10.5) 0.121 |
4.4 (−1.5, 10.3) 0.140 |
Model adjusted for age, sex, site and ln(IL6).
Model adjusted for age, sex, site, ln(IL6), BMI, current smoking and diabetes.
Figure 3. Association between fibrinogen level and ln(IL-6), by FGG902 (rs1800792) genotype.
Figure 4. Association between fibrinogen level and ln(IL-6), by FGA251 (rs2070006) genotype.
Interaction terms were slightly attenuated by adjustment for potential confounders of IL-6 including BMI, current smoking and diabetes status[Table 4], but given their imprecision, we are cautious in our interpretation of adjusted vs. unadjusted models.
Sensitivity Analyses
The minor alleles of IL6 SNPs rs1554606 and rs1800795 have been previously associated with elevated IL-6 plasma levels in CHS Caucasians.(Walston et al., 2007) We found that these tagSNPs were highly correlated (r2=0.87) and chose to exclude rs1800795, which has a slightly lower MAF of 0.41 than rs1554606, MAF= 0.44. When we included rs1800795 instead of rs1554606 in logic regression models, results did not change. Similarly, the fibrinogen SNPs FGA6534 (rs6050) and FGG10034 (rs2066865) which also tag a common regional haplotype, were highly correlated in this population (r2=0.86). We excluded FGG10034, but when substituted for FGA6534 in the logic regression models with fibrinogen level as an outcome, it performed similarly to FGA6534 in that it was present in the three top models along with FGB1437, and had a comparable impact on fibrinogen levels; however the model supporting its interaction with IL61111 was ranked fourth with a non-significant p-value of 0.250. Interaction by genotype-sex was tested and not detected in any of the logic regression models.
Discussion
Elevated levels of fibrinogen(Tzoulaki et al., 2007, Kannel, 2005, Danesh et al., 2005) and IL- 6(Zakai et al., 2007, Tzoulaki et al., 2007) have been moderately associated with increased risks of CVD but whether they interact to influence risk has not been extensively studied in humans. In this study, we used two methods to assess interaction between these biologically related proteins associated with risk of CVD. Using logic regression to investigate combinations of variants in the fibrinogen and IL6 genes associated with CVD outcomes, our exploratory results support potential interaction between two fibrinogen SNPs, FGB1437 (rs1800790) and FGA6534 (rs6050) in determining fibrinogen levels, but no associations with subclinical or clinical CVD. These SNPs appeared in one model which included their main effects only and a second model including only their interaction term. It is unclear which model is best, since their scores were of similar magnitude and as demonstrated in Fig.2, their implications are somewhat similar. The main effect findings for these SNPs which tag different common haplotypes have been previously reported in CHS,(Carty et al., 2008) as has the haplotype describing the particular combination of SNPs identified in the current study. However, the current analysis is novel in that it includes additional predictors and genetic models, an assessment of complex interaction in the absence of main effects and conditional permutation tests which account for multiple tests performed. Though not statistically significant after correction for multiple testing, a model supporting interaction between FGA6534 and IL6-1111 (rs1800796) associated with fibrinogen level may be worthy of additional investigation.
In our standard analysis of interaction on a multiplicative scale between fibrinogen SNPs and baseline IL-6 level, we found marginally significant interactions in minimally adjusted models (unadjusted for multiple testing) between IL-6 level and fibrinogen SNPs located in the promoter regions of FGA and FGG so that in individuals having the homozygous rare FGG902 (rs1800792) genotype or lacking the rare FGA251 (rs2070006) allele, the slope describing the association between fibrinogen level and ln(IL-6) level was not as steep as with the other genotypes. These results are consistent with a recessive effect for FGG902 whereas the similar slopes for individuals with either 1 or 2 minor alleles are consistent with a dominant gene effect for FGA251, though the overlap of the confidence intervals should be noted. As reported previously(Carty et al., 2008), in single SNP analyses without interaction terms, each FGG902 allele was significantly associated with higher fibrinogen and each FGA251 minor allele was associated with significantly lower fibrinogen levels. Consideration of these findings in context with the smaller slope describing the relationship between IL-6 and fibrinogen levels among FGG902 rare homozygotes, may suggest that FGG902, located in the promoter region of the FGG gene, potentially interferes with IL-6 mediated binding. At a cutoff of r2>0.64, it is not in LD with any validated FGG SNPs, though it is in LD with FGA and FGB SNPs. However, these interaction results should be viewed as exploratory as they would not be significant following correction for multiple testing using the Bonferroni method.
Our fibrinogen level findings are generally consistent with results from laboratory studies identifying IL-6 response elements in the promoter regions of human FGA(Liu & Fuller, 1995), FGG(Zhang et al., 1995, Duan & Simpson-Haidaris, 2003, Mizuguchi et al., 1995) and to a lesser extent, FGB.(Huber et al., 1990) While the promoter SNPs studied are not located in the putative IL-6 response element sites, it is possible that they could indirectly modulate the IL-6 response by interacting with factors bound to the response element. Specifically, the FGG902 variant (rs1800792) is located at approximately −251 base pairs (bp), close to an IL-6 response element reported at −306 to −301 bp in the 5′ flanking region of human FGG. (Mizuguchi et al., 1995) Similarly the FGA251 variant, located at −201 bp, is within a 5′ flanking FGA region (from −217 to +1bp) reported to have significant promoter activity and be inducible by IL-6, (Hu et al., 1995) whereas the two FGB promoter SNPs rs1800791 and rs1800790, located at approximately −301 bp and −485 bp are not especially close to the putative IL-6 response region reported at −142 to −137 bp.(Dalmon et al., 1993) For FGB, we also did not detect evidence supporting in vitro findings indicating that the A allele of FGB1437 (rs1800790) is associated with higher IL-6- induced expression of fibrinogen than the G allele(van't Hooft et al., 1999, Brown & Fuller, 1998) or that a variant at FGB -148 (which is in complete LD with FGB1437) is associated with lower IL-6-induced levels of the fibrinogen-β protein.(Verschuur et al., 2005) The contrasts between our findings and in vitro studies may be related to issues of power or biological complexity of the in vivo regulation of fibrinogen synthesis as detailed below.
The heritability of fibrinogen level is reported as 20-50%,(de Lange et al., 2001, de Maat et al., 2004, Yang et al., 2003, Reed et al., 1994, Bladbjerg et al., 2006) yet several recent population-based association studies report that common variants in three fibrinogen genes explain less than 5% of the variance in fibrinogen level.(Reiner et al., 2006, Carty et al., 2008) One explanation for this discrepancy is that fibrinogen levels are affected by combinations of other inherited genes such as those involved in its regulation and break-down. Even in healthy individuals, fibrinogen level is heterogeneous due to variable rates of formation (or transcription) and removal (processing, stability and degradation) of the fibrinogen protein.(de Maat & Verschuur, 2005) Furthermore, the various molecular forms of fibrinogen resulting from splice variants, post-translational modification and proteolytic degradation have functional consequences with respect to fibrinolysis, cellular interactions, clotting time, and coagulation characteristics.(de Maat & Verschuur, 2005) Other environmental factors not accounted for in these analyses could also affect fibrinogen levels, such as glucocorticoids (GCs), which act synergistically with IL-6 to up-regulate fibrinogen gene expression.(Otto et al., 1987, Simpson Haidaris, 1997)
Similarly, IL6 SNPs alone explain little of the variance in IL-6 levels which presumably are reflective of other genetic and environmental factors. Laboratory studies suggest that an IL6 polymorphism (rs1800795) is associated with inter-individual differences in the degree of IL-6 response to stressful stimuli.(Fishman et al., 1998) This SNP was previously found to be significantly associated with higher fibrinogen levels (Jenny et al., 2002), IL-6 and CRP levels in CHS.(Walston et al., 2007) However, a previous study tested, but was unable to detect interaction between rs1800795 and three fibrinogen SNPs associated with risk of MI, though the study was small and included few variants.(Mannila et al., 2006) Rs1800795, excluded from our main analyses due to its high correlation with rs1554606 (r2 = 0.87), was also associated with higher IL-6 and CRP levels.(Walston et al., 2007) We did not identify any interactions involving rs1554606 or rs1800795 (in sensitivity analyses), but rather identified a potential interaction between another IL6promoter SNP, rs1800796 and an FGA SNP. This IL6SNP also has been associated with higher fibrinogen and CRP levels in a Chinese population.(Wong et al., 2007) Although this SNP is not in strong LD with either of the aforementioned IL6SNPs, our finding is consistent with the IL6promoter region being potentially important for the downstream expression of acute phase reactants.
In our gene-gene interaction analyses, we chose to use logic regression since it is particularly useful for binary variables such as those in genetic analyses.(Kooperberg et al., 2007) In addition, it can be used to detect within and between gene effects and easily incorporates testing of both dominant and recessive genetic models. It also can identify interactions among three or more variables and provides tools for use during model selection that account for multiple testing. A limitation of the logic regression approach is that it requires that continuous predictors such as IL-6 level be categorized into binary (or a series of binary) variables for modeling. This categorization may be more susceptible to misclassification error or a loss of power due to the increased number of degrees of freedom. For this reason, we analyzed interactions involving IL-6 levels using a different approach.
Because elevated fibrinogen levels are associated with increased risks of CVD and stroke, elucidation of the molecular mechanisms that regulate fibrinogen expression may be relevant for control of diseases associated with high levels of fibrinogen.(Humphries et al., 1999) Thus, in addition to furthering our understanding of how fibrinogen is up-regulated in response to inflammatory stimuli, the mechanisms that down-regulate fibrinogen gene expression could be important for normalization of elevated plasma fibrinogen levels in individuals at increased risk for cardiovascular disease.(Duan & Simpson-Haidaris, 2003) We hypothesized that IL-6 variants could potentially influence fibrinogen levels and CVD through their effects on IL-6 levels or IL-6 structure/binding and subsequent regulation of IL-6 induced genes. For our gene-gene interactions, we estimated that we had 98% power to estimate fibrinogen betas of approximately 11mg/dL for the interaction term; however, power dropped as low as approximately 58% for combinations of SNPs with the lowest minor allele frequencies. Similarly, we were well powered for fibrinogen SNP-IL6 level interactions, with at least 80% power to detect interaction terms accounting for 0.2% of the variance in fibrinogen levels, though power decreased for low frequency SNPs. In spite of the in vitro evidence of a biological interaction, we did not find strong evidence of interaction in risk of subclinical or clinical disease in this older population. Previous studies of IL6 and fibrinogen SNPs also did not reveal strong main effect associations between variants and complex disease in CHS,(Walston et al., 2007, Carty et al., 2008) though significant findings have been reported in other populations.(Mannila et al., 2006) It is likely that the interactions of many genetic variants, some of which may not encode proteins and thus can be difficult to identify, contribute to development of complex diseases such as CVD.(Freimer & Sabatti, 2007) Lacking blood samples drawn during an acute inflammatory event, when both IL-6 and fibrinogen would be upregulated, we relied on single baseline measures of these proteins. Future research may be better directed at larger populations for better power to detect the likely small effects of combinations of genes on risk of clinical CVD or on individuals having combinations of certain genotypes that make them more responsive to acute stimuli.(Humphries et al., 1999) Given the exploratory nature of our findings, validation of our results in an independent data set and using other statistical methods for determining interaction is necessary.
Supplementary Material
Acknowledgements
The authors wish to express sincere appreciation to the investigators and participants of the Cardiovascular Health Study. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. The research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, and grant number U01 HL080295 from the National Heart, Lung, and Blood Institute, (NHLBI) with additional contribution from the National Institute of Neurological Disorders and Stroke. CLC received support from the NHLBI training grant, T32 HL007902. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or the National Institutes of Health.
References
- Bladbjerg EM, De Maat MP, Christensen K, Bathum L, Jespersen J, Hjelmborg J. Genetic influence on thrombotic risk markers in the elderly--a Danish twin study. J Thromb Haemost. 2006;4:599–607. doi: 10.1111/j.1538-7836.2005.01778.x. [DOI] [PubMed] [Google Scholar]
- Brown ET, Fuller GM. Detection of a complex that associates with the Bbeta fibrinogen G-455-A polymorphism. Blood. 1998;92:3286–93. [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty CL, Cushman M, Jones D, Lange LA, Hindorff LA, Rice K, Jenny NS, Durda JP, Walston J, Carlson CS, Nickerson D, Tracy RP, Reiner AP. Associations between common fibrinogen gene polymorphisms and cardiovascular disease in older adults - The Cardiovascular Health Study. Thromb Haemost. 2008;99:388–95. doi: 10.1160/TH07-08-0523. [DOI] [PubMed] [Google Scholar]
- Cushman M, Cornell ES, Howard PR, Bovill EG, Tracy RP. Laboratory methods and quality assurance in the Cardiovascular Health Study. Clin Chem. 1995;41:264–270. [PubMed] [Google Scholar]
- Dalmon J, Laurent M, Courtois G. The human beta fibrinogen promoter contains a hepatocyte nuclear factor 1-dependent interleukin-6-responsive element. Mol. Cell. Biol. 1993;13:1183–93. doi: 10.1128/mcb.13.2.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh J, Lewington S, Thompson SG, Lowe GD, Collins R, Kostis JB, Wilson AC, Folsom AR, Wu K, Benderly M, Goldbourt U, Willeit J, Kiechl S, Yarnell JW, Sweetnam PM, Elwood PC, Cushman M, Psaty BM, Tracy RP, Tybjaerg-Hansen A, Haverkate F, De Maat MP, Fowkes FG, Lee AJ, Smith FB, Salomaa V, Harald K, Rasi R, Vahtera E, Jousilahti P, Pekkanen J, D'agostino R, Kannel WB, Wilson PW, Tofler G, Arocha-Pinango CL, Rodriguez-Larralde A, Nagy E, Mijares M, Espinosa R, Rodriquez-Roa E, Ryder E, Diez-Ewald MP, Campos G, Fernandez V, Torres E, Marchioli R, Valagussa F, Rosengren A, Wilhelmsen L, Lappas G, Eriksson H, Cremer P, Nagel D, Curb JD, Rodriguez B, Yano K, Salonen JT, Nyyssonen K, Tuomainen TP, Hedblad B, Lind P, Loewel H, Koenig W, Meade TW, Cooper JA, De Stavola B, Knottenbelt C, Miller GJ, Cooper JA, Bauer KA, Rosenberg RD, Sato S, Kitamura A, Naito Y, Palosuo T, Ducimetiere P, Amouyel P, Arveiler D, Evans AE, Ferrieres J, Juhan-Vague I, Bingham A, Schulte H, Assmann G, Cantin B, Lamarche B, Despres JP, Dagenais GR, Tunstall-Pedoe H, Woodward M, Ben-Shlomo Y, Davey Smith G, Palmieri V, Yeh JL, Rudnicka A, Ridker P, Rodeghiero F, Tosetto A, Shepherd J, et al. Plasma fibrinogen level and the risk of major cardiovascular diseases and nonvascular mortality: an individual participant meta-analysis. JAMA. 2005;294:1799–809. doi: 10.1001/jama.294.14.1799. [DOI] [PubMed] [Google Scholar]
- De Lange M, Snieder H, Ariens RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001;357:101–5. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- De Maat MP, Bladbjerg EM, Hjelmborg JB, Bathum L, Jespersen J, Christensen K. Genetic influence on inflammation variables in the elderly. Arterioscler Thromb Vasc Biol. 2004;24:2168–73. doi: 10.1161/01.ATV.0000143856.01669.e7. [DOI] [PubMed] [Google Scholar]
- De Maat MP, Verschuur M. Fibrinogen heterogeneity: inherited and noninherited. Curr Opin Hematol. 2005;12:377–83. doi: 10.1097/01.moh.0000169287.51594.3b. [DOI] [PubMed] [Google Scholar]
- Duan HO, Simpson-Haidaris PJ. Functional analysis of interleukin 6 response elements (IL-6REs) on the human gamma-fibrinogen promoter. Journal of Biological Chemistry. 2003;278:41270–81. doi: 10.1074/jbc.M304210200. [DOI] [PubMed] [Google Scholar]
- Duan HO, Simpson-Haidaris PJ. Cell type-specific differential induction of the human gamma-fibrinogen promoter by interleukin-6. J Biol Chem. 2006;281:12451–7. doi: 10.1074/jbc.M600294200. [DOI] [PubMed] [Google Scholar]
- Ernst E. The role of fibrinogen as a cardiovascular risk factor. Atherosclerosis. 1993;100:1–12. doi: 10.1016/0021-9150(93)90062-y. [DOI] [PubMed] [Google Scholar]
- Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–76. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimer NB, Sabatti C. Human genetics: variants in common diseases. Nature. 2007;445:828–830. doi: 10.1038/nature05568. [DOI] [PubMed] [Google Scholar]
- Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TM, Mittelmark MB, Newman A, O'leary DH, Psaty B, Rautaharju P, Tracy RT, Weiler PG. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- Fuller GM, Zhang Z. Transcriptional control mechanism of fibrinogen gene expression. Ann N Y Acad Sci. 2001;936:469–79. doi: 10.1111/j.1749-6632.2001.tb03534.x. [DOI] [PubMed] [Google Scholar]
- Genome Variation Server. Supported by SeattleSNPs Program for Genomic Applications (PGA) grants HL66682 and HL66642 from the National Heart, Lung, and Blood Institute; Jan 11, 2008. GVS version 3.11. [Google Scholar]
- Heinrich PC, Castell JV, Andus T. Interleukin-6 and the acute phase response. Biochem J. 1990;265:621–36. doi: 10.1042/bj2650621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CH, Harris JE, Davie EW, Chung DW. Characterization of the 5′-flanking region of the gene for the alpha chain of human fibrinogen. J Biol Chem. 1995;270:28342–9. doi: 10.1074/jbc.270.47.28342. [DOI] [PubMed] [Google Scholar]
- Huber P, Laurent M, Dalmon J. Human 1 fibrinogen gene expression: upstream sequences involved in its tissue-specific expression and its dexamethasone and interleukin-6 stimulation. J. Biol. Chem. 1990;265:5695–5701. [PubMed] [Google Scholar]
- Humphries SE, Luong LA, Montgomery HE, Day IN, Mohamed-Ali V, Yudkin JS. Gene-environment interaction in the determination of levels of plasma fibrinogen. Thromb Haemost. 1999;82:818–25. [PubMed] [Google Scholar]
- Ives DG, Fitzpatrick AL, Bild DE, Psaty BM, Kuller LH, Crowley PM, Cruise RG, Theroux S. Surveillance and ascertainment of cardiovascular events. The Cardiovascular Health Study. Ann Epidemiol. 1995;5:278–85. doi: 10.1016/1047-2797(94)00093-9. [DOI] [PubMed] [Google Scholar]
- Jenny NS, Tracy RP, Ogg MS, Luong Le A, Kuller LH, Arnold AM, Sharrett AR, Humphries SE. In the elderly, interleukin-6 plasma levels and the −174G>C polymorphism are associated with the development of cardiovascular disease. Arterioscler Thromb Vasc Biol. 2002;22:2066–71. doi: 10.1161/01.atv.0000040224.49362.60. [DOI] [PubMed] [Google Scholar]
- Kannel WB. Overview of hemostatic factors involved in atherosclerotic cardiovascular disease. Lipids. 2005;40:1215–20. doi: 10.1007/s11745-005-1488-8. [DOI] [PubMed] [Google Scholar]
- Kooperberg C, Bis JC, Marciante KD, Heckbert SR, Lumley T, Psaty BM. Logic regression for analysis of the association between genetic variation in the renin-angiotensin system and myocardial infarction or stroke. Am J Epidemiol. 2007;165:334–43. doi: 10.1093/aje/kwk006. [DOI] [PubMed] [Google Scholar]
- Kooperberg C, Ruczinski I, Leblanc ML, Hsu L. Sequence analysis using logic regression. Genetic Epidemiology. 2001;21(Suppl1):S626–31. doi: 10.1002/gepi.2001.21.s1.s626. [DOI] [PubMed] [Google Scholar]
- Liu Z, Fuller GM. Detection of a Novel Transcription Factor for the Aalpha Fibrinogen Gene in Response to Interleukin-6. J. Biol. Chem. 1995;270:7580–7586. doi: 10.1074/jbc.270.13.7580. [DOI] [PubMed] [Google Scholar]
- Macy E, Hayes T, Tracy R. Variability in the measurement of C-reactive protein in healthy subjects: implication for reference interval and epidemiological applications. Clin Chem. 1997;43:52–8. [PubMed] [Google Scholar]
- Mannila MN, Eriksson P, Ericsson CG, Hamsten A, Silveira A. Epistatic and pleiotropic effects of polymorphisms in the fibrinogen and coagulation factor XIII genes on plasma fibrinogen concentration, fibrin gel structure and risk of myocardial infarction. Thromb Haemost. 2006;95:420–7. doi: 10.1160/TH05-11-0777. [DOI] [PubMed] [Google Scholar]
- Mizuguchi J, Hu CH, Cao Z, Loeb KR, Chung DW, Davie EW. Characterization of the 5'-flanking region of the gene for the gamma chain of human fibrinogen. J Biol Chem. 1995;270:28350–6. doi: 10.1074/jbc.270.47.28350. [DOI] [PubMed] [Google Scholar]
- O'leary DH, Polak JF, Wolfson SK, Jr., Bond MG, Bommer W, Sheth S, Psaty BM, Sharrett AR, Manolio TA. Use of sonography to evaluate carotid atherosclerosis in the elderly. The Cardiovascular Health Study. CHS Collaborative Research Group. Stroke. 1991;22:1155–63. doi: 10.1161/01.str.22.9.1155. [DOI] [PubMed] [Google Scholar]
- Otto JM, Grenett HE, Fuller GM. The coordinated regulation of fibrinogen gene transcription by hepatocyte-stimulating factor and dexamethasone. J Cell Biol. 1987;105:1067–72. doi: 10.1083/jcb.105.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed T, Tracy RP, Fabsitz RR. Minimal genetic influences on plasma fibrinogen level in adult males in the NHLBI twin study. Clinical Genetics. 1994;45:71–77. doi: 10.1111/j.1399-0004.1994.tb03997.x. [DOI] [PubMed] [Google Scholar]
- Reiner AP, Carty CL, Carlson CS, Wan JY, Rieder MJ, Smith JD, Rice K, Fornage M, Jaquish CE, Williams OD, Tracy RP, Lewis CE, Siscovick DS, Boerwinkle E, Nickerson DA. Association between patterns of nucleotide variation across the three fibrinogen genes and plasma fibrinogen levels: the Coronary Artery Risk Development in Young Adults (CARDIA) study. J Thromb Haemost. 2006;4:1279–1287. doi: 10.1111/j.1538-7836.2006.01907.x. [DOI] [PubMed] [Google Scholar]
- Ruczinski I, Kooperberg C, Leblanc ML. Logic Regression. Journal of Computational and Graphical Statistics. 2003;12:475–511. [Google Scholar]
- Simpson Haidaris PJ. Induction of Fibrinogen Biosynthesis and Secretion From Cultured Pulmonary Epithelial Cells. Blood. 1997;89:873–882. [PubMed] [Google Scholar]
- Tzoulaki I, Murray GD, Lee AJ, Rumley A, Lowe GD, Fowkes FG. Relative value of inflammatory, hemostatic, and rheological factors for incident myocardial infarction and stroke: the Edinburgh Artery Study. Circulation. 2007;115:2119–27. doi: 10.1161/CIRCULATIONAHA.106.635029. [DOI] [PubMed] [Google Scholar]
- Van't Hooft F, Von Bahr SJF, Silveira A, Iliadou A, Eriksson P, Hamsten A. Two common, functional polymorphisms in the promoter region of the B-fibrinogen gene contribute to regulation of plasma fibrinogen concentration. Arterioscler Thromb Vasc Biol. 1999;19:3063–78. doi: 10.1161/01.atv.19.12.3063. [DOI] [PubMed] [Google Scholar]
- erschuur M, De Jong M, Felida L, De Maat MP, Vos HL. A hepatocyte nuclear factor-3 site in the fibrinogen beta promoter is important for interleukin 6-induced expression, and its activity is influenced by the adjacent −148C/T polymorphism. J Biol Chem. 2005;280:16763–71. doi: 10.1074/jbc.M501973200. [DOI] [PubMed] [Google Scholar]
- Walston JD, Fallin MD, Cushman M, Lange L, Psaty B, Jenny N, Browner W, Tracy R, Durda P, Reiner A. IL-6 gene variation is associated with IL-6 and C-reactive protein levels but not cardiovascular outcomes in the Cardiovascular Health Study. Hum Genet. 2007;122:485–94. doi: 10.1007/s00439-007-0428-x. [DOI] [PubMed] [Google Scholar]
- Wong LYF, Leung RYH, Ong KL, Cheung BMY. Plasma levels of fibrinogen and C-reactive protein are related to interleukin-6 gene −572C>G polymorphism in subjects with and without hypertension. J Hum Hypertens. 2007;21:875–882. doi: 10.1038/sj.jhh.1002233. [DOI] [PubMed] [Google Scholar]
- Yang Q, Tofler GH, Cupples LA, Larson MG, Feng D, Lindpaintner K, Levy D, D'agostino RB, O'donnell CJ. A genome-wide search for genes affecting circulating fibrinogen levels in the Framingham Heart Study. Thromb Res. 2003;110:57–64. doi: 10.1016/s0049-3848(03)00288-3. [DOI] [PubMed] [Google Scholar]
- Zakai NA, Katz R, Jenny NS, Psaty BM, Reiner AP, Schwartz SM, Cushman M. Inflammation and hemostasis biomarkers and cardiovascular risk in the elderly: the Cardiovascular Health Study. J Thromb Haemost. 2007;5:1128–35. doi: 10.1111/j.1538-7836.2007.02528.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Fuentes NL, Fuller GM. Characterization of the IL-6 Responsive Elements in the Gamma Fibrinogen Gene Promoter. J. Biol. Chem. 1995;270:24287–24291. doi: 10.1074/jbc.270.41.24287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.