Abstract
Generally, F-box proteins are the substrate recognition subunits of SCF (Skp1-Cul1-F-box protein) ubiquitin ligase complexes, which mediate the timely proteolysis of important eukaryotic regulatory proteins1,2. Mammalian genomes encode roughly 70 F-box proteins, but only a handful have established functions3,4. The F-box protein family obtained its name from Cyclin F (also called Fbxo1), in which the F-box motif (the ~40 amino acid domain required for binding to Skp1) was first described5. Cyclin F, which is encoded by an essential gene, also contains a cyclin box domain, but in contrast to most cyclins, it does not bind or activate any cyclin-dependent kinases (CDKs)5–7. However, like other cyclins, Cyclin F oscillates during the cell cycle, with protein levels peaking in G2. Despite its essential nature and status as the founding member of the F-box protein family, Cyclin F remains an orphan protein, whose functions are unknown. Starting from an unbiased screen, we identified CP110, a protein essential for centrosome duplication, as an interactor and substrate of Cyclin F. Utilizing a mode of substrate binding distinct from other F-box protein-substrate pairs, CP110 and Cyclin F physically associate on the centrioles during the G2 phase of the cell cycle, and CP110 is ubiquitylated via the SCFCyclin F ubiquitin ligase complex, leading to its degradation. siRNA-mediated depletion of Cyclin F in G2 induces centrosomal and mitotic abnormalities, such as multipolar spindles and asymmetric, bipolar spindles with lagging chromosomes. These phenotypes were reverted by co-silencing CP110 and were recapitulated by expressing a stable mutant of CP110 that is unable to bind Cyclin F. Finally, expression of a stable CP110 mutant in cultured cells also promotes the formation of micronuclei, a hallmark of chromosome instability. We propose that SCFCyclin F–mediated degradation of CP110 is required for the fidelity of mitosis and genome integrity.
To identify substrates of the SCFCyclin F ubiquitin ligase, FLAG-HA-tagged Cyclin F was transiently expressed in either HeLa or HEK-293T cells and immunopurified for analysis by Multidimensional Protein Identification Technology (MudPIT)8. In both cases, MudPIT revealed the presence of peptides corresponding to Skp1 and Cul1, whereas, in agreement with previous reports6,9, no peptides corresponding to CDKs were identified (see also Supplementary Fig. 1a). Instead, MudPIT revealed the presence of peptides derived from CP110 (Supplementary Table 1). Combining both analyses, 21 total spectra, corresponding to 12 unique CP110 peptides, were identified. In two additional experiments, we immunopurified a Cyclin F mutant lacking the cyclin box [Cyclin F(1–270)], and although Skp1 and Cul1 still co-immunoprecipitated with Cyclin F(1–270), CP110 was not present (Supplementary Table 1).
CP110 localizes to the distal ends of the centrioles, and its depletion interferes with centrosome re-duplication generated either by arresting cells in S phase for prolonged time periods or by overexpressing Plk4 (refs. 10,11), indicating that CP110 has a pivotal role in new centriole formation. Similarly, the fly ortholog of CP110 is necessary for both centriole duplication and centrosome maturation12. Finally, CP110 has additional roles in the regulation of centriole length and cilium formation13,14.
To investigate whether the binding between CP110 and Cyclin F is specific, we expressed in HEK-293T fourteen F-box proteins that were then immunoprecipitated to evaluate their interaction with CP110. We found that the only F-box protein able to co-immunoprecipitate endogenous CP110 was Cyclin F (Supplementary Fig. 1b). Using synchronized HeLa cells, the interaction between endogenous Cyclin F and endogenous CP110 was observed exclusively in G2 and M, as monitored by immunoblotting for cell cycle markers and flow cytometry (Fig. 1a and data not shown). Subsequently, we mapped the Cyclin F binding motif of CP110. A series of binding experiments, using multiple CP110 deletion mutants, narrowed the binding motif to a region of human CP110 located between amino acids 565–620 (Supplementary Fig. 2). This region contains one putative RxL motif, an established cyclin binding domain. A mutant in this motif [CP110(RxL/AxA)] failed to co-immunoprecipitate endogenous Cyclin F (Supplementary Fig. 2a, last lane), indicating that the RxL motif, located at residues 588–590, mediates binding to Cyclin F. Cyclins recruit RxL-containing proteins through a hydrophobic patch present in the cyclin box domain15. We observed that Cyclin F requires its cyclin box to bind endogenous CP110 (Supplementary Fig. 3a–b and Supplementary Table 1). Moreover, Cyclin F displays a conserved hydrophobic patch and a Cyclin F mutated in this domain [Cyclin F(M/A;L/A)] lost the ability to bind CP110 (Supplementary Fig. 3c–d).
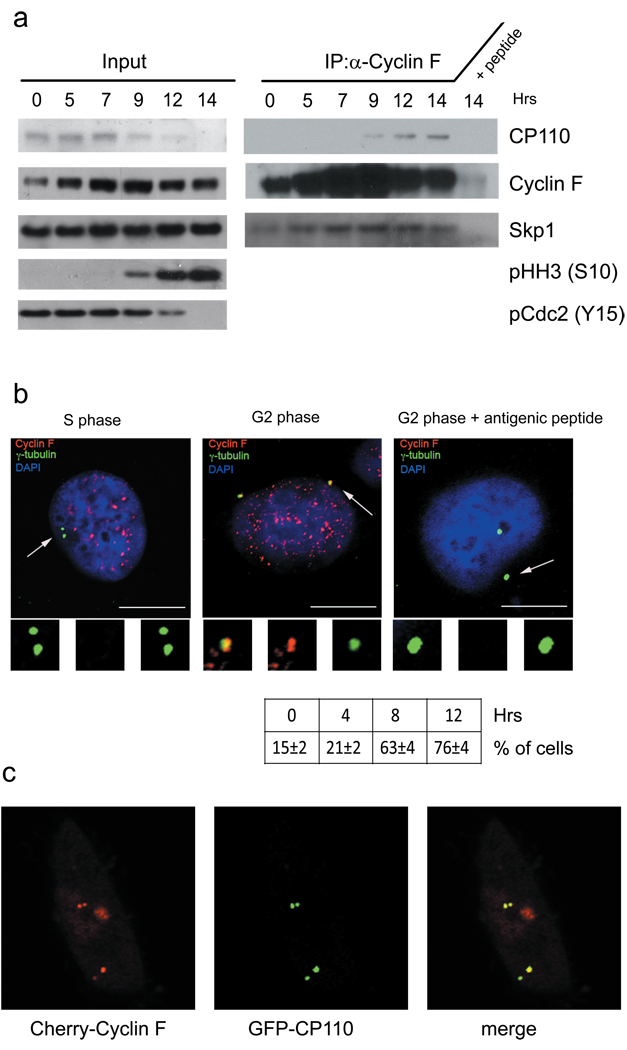
Figure 1. Cyclin F and CP110 interact and colocalize to the centrosomes.
a, HeLa cells were synchronized at G1/S using a double-thymidine block before release into fresh medium. Cells were collected at the indicated times, lysed, immunoprecipitated with anti-Cyclin F antibody, and immunoblotted as indicated. Last lane shows immunoprecipitation pre-incubated with the antigenic peptide. Left panel show 10% of the material used for immunoprecipitations (input).
b, U-2OS cells were synchronized as in (a), fixed, and incubated with an anti-Cyclin F antibody (red) and anti-γ-tubulin antibody (green). The panel shows staining after pre-incubation with the antigenic peptide. DNA was stained with DAPI. Insets show magnified views of the centrosomes indicated by arrows. Scale bar = 10 µM. The table shows the percentage of cells with centrosomal Cyclin F (where 100% was the total cells staining positive for nuclear Cyclin F) at different times after the release from the double-thymidine block (n = ~50 per time point).
c, Cyclin F and CP110 colocalize to the centrioles. U-2OS cells were transfected with GFP-CP110 and Cherry-Cyclin F. In merged images, yellow shows colocalization of CP110 and Cyclin F.
Because of the centrosomal localization of CP110, we investigated the subcellular localization of Cyclin F. Synchronized U-2OS cells were co-stained with antibodies to Cyclin F and γ-tubulin (a centrosome marker). As expected, both S and G2 phase cells displayed two γ-tubulin dots, which were distinct (but adjacent) in S phase and separated by at least 2 µM in G2. We found that, similar to CP110 (ref.10,11), endogenous Cyclin F partially colocalized with γ-tubulin (Fig. 1b); however, in contrast to CP110, which is exclusively centrosomal (ref.10,11, Fig. 1c, and Supplementary Fig. 4), Cyclin F also localized to the nucleus. Moreover, Cyclin F staining on the centrosomes was rare in S phase cells and increased in G2 cells, whereas the intensity of CP110 staining was predominant in S and decreased in G2 (Fig. 1b and Supplementary Fig. 4), correlating with the total CP110 levels detected by immunoblotting. We also observed that Cherry-Cyclin F colocalized with GFP-CP110 and was present on both the mother and daughter centrioles (Fig. 1b–c). Finally, we found that the centrosomal localization of Cyclin F does not require its binding to CP110 and is present at the N-terminus since: (i) Cyclin F(1–270) and Cyclin F(M/A;L/A) (which both do not interact with CP110) still localized to the centrioles and (ii) Cyclin F(271–786) (which contains the CP110-binding domain) completely lost its centrosomal localization and binding to CP110 (Supplementary Fig. 3a and data not shown).
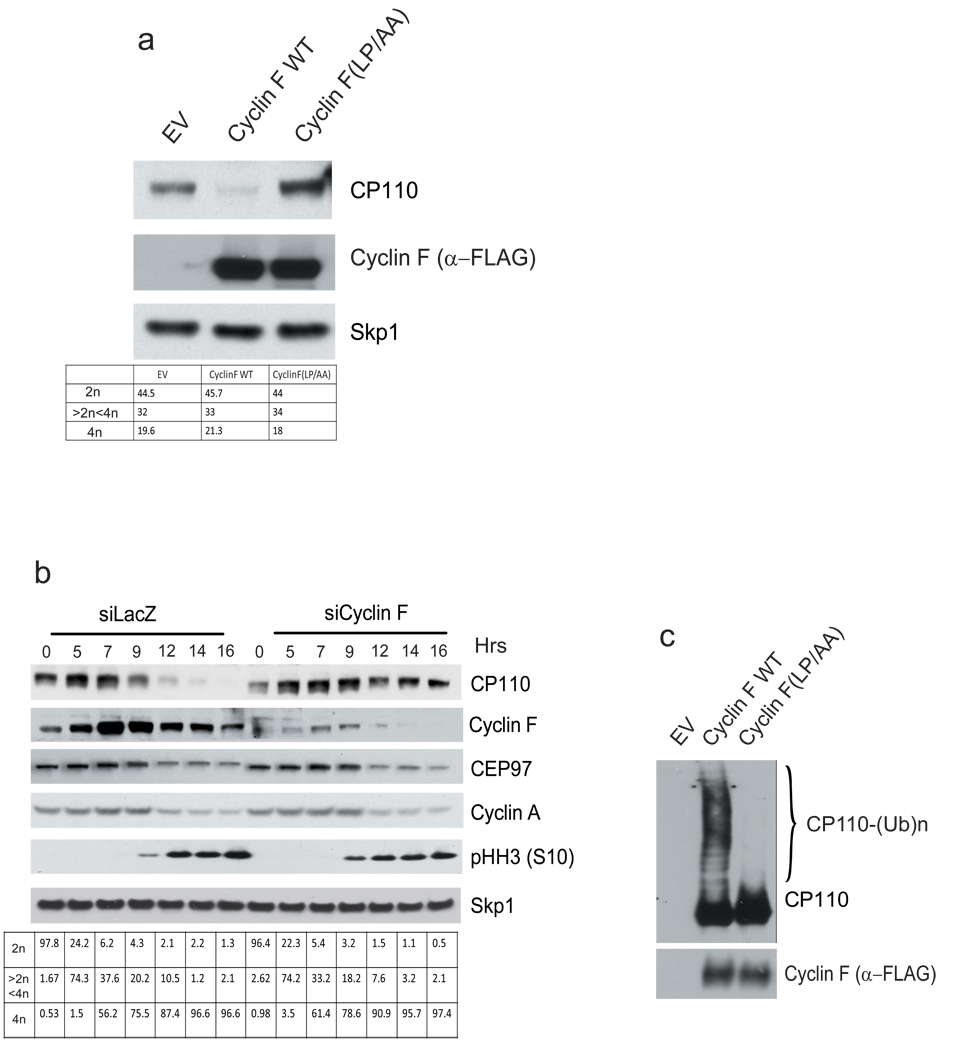
As part of our investigation of CP110 binding to Cyclin F, we noticed that, compared to wild type Cyclin F, the Cyclin F(LP/AA) mutant (in which the first two amino acids of the F-box domain were mutated to alanine) bound less Skp1 and Cul1 (as expected) but more CP110 (Supplementary Fig. 3b). This result suggested that CP110 is targeted for proteolysis by Cyclin F, because Cyclin F(LP/AA) is unable to form an active ubiquitin ligase: it can sequester CP110 in a more stable complex that is easier to detect. In agreement with this interpretation, expression of wild type Cyclin F resulted in a dramatic reduction of endogenous CP110 levels in four different cell lines. Moreover, expression of either Cyclin F(LP/AA) or Cyclin F(M/A;L/A) had no effect on CP110 levels (Fig. 2a and Supplementary Fig. 5).
Figure 2. CP110 is targeted for ubiquitylation and degradation by SCFCyclin F during the G2 phase of the cell cycle.
a, U-2OS cells were transfected with an empty vector (EV), FLAG-tagged Cyclin F, or FLAG-tagged Cyclin F(LP/AA). Twenty-four hours after transfection, cells were collected, lysed, and immunoblotted as indicated. DNA content was monitored by flow cytometry.
b, HeLa cells were transfected with short interfering RNAs (siRNAs) to either LacZ or Cyclin F (oligo #2). Cells synchronized as in (1a) were then collected at the indicated times, lysed, and immunoblotted as indicated. DNA content was monitored by flow cytometry.
c, HEK-293T cells were transfected as in (2a), lysed, immunoprecipitated with anti-FLAG resin, and used in a ubiquitylation assay.
To further test whether Cyclin F might regulate the degradation of CP110, we used three different siRNA oligos to reduce the expression of Cyclin F in synchronized HeLa cells. We also silenced Cyclin F expression in synchronized U-2OS and RPE1-hTERT cells using the most effective of the three oligos. In all cases, depletion of Cyclin F inhibited the G2-specific degradation of CP110 (Fig. 2b and Supplementary Fig. 6). Finally, immunopurified wild type Cyclin F, but not Cyclin F(LP/AA), promoted the in vitro ubiquitylation of CP110 (Fig. 2c).
Together, the results in Figs. 1–2 and Supplementary Figs. 1–6 demonstrate that Cyclin F mediates the degradation of CP110 in G2 by forming an active SCF ubiquitin ligase complex.
To study the biological significance of Cyclin F-mediated degradation of CP110, we investigated whether overexpression of Cyclin F would affect centrosome reduplication in cells in which DNA synthesis is inhibited by hydroxyurea16. Therefore, we forced the expression of Cyclin F in U-2OS cells that were subsequently treated for 48 hours with hydroxyurea, inducing a block in S phase [when endogenous protein levels are low (Supplementary Fig. 7) and Cyclin F is rarely localized to the centrosome (Fig. 1b)]. Overexpression of Cyclin F induced its temporal mislocalization to the centrosome (not shown). Cells were analyzed for centrosome number by dual-color, indirect immunofluorescence using an antibody to γ-tubulin and an antibody to Centrin 2 (a centriole marker). Control S-phase-arrested U-2OS cells underwent centrosome overduplication as determined by the presence of more than two γ-tubulin foci and more than four Centrin 2 foci. In contrast, cells expressing high levels of Cyclin F did not display centrosome overduplication. Importantly, co-expression of a CP110 mutant unable to bind Cyclin F reverted the Cyclin F-mediated inhibition of centrosome overduplication (Supplementary Fig. 7), suggesting that Cyclin F acts as an inhibitor of centrosome reduplication via its ability to restrain the expression of CP110.
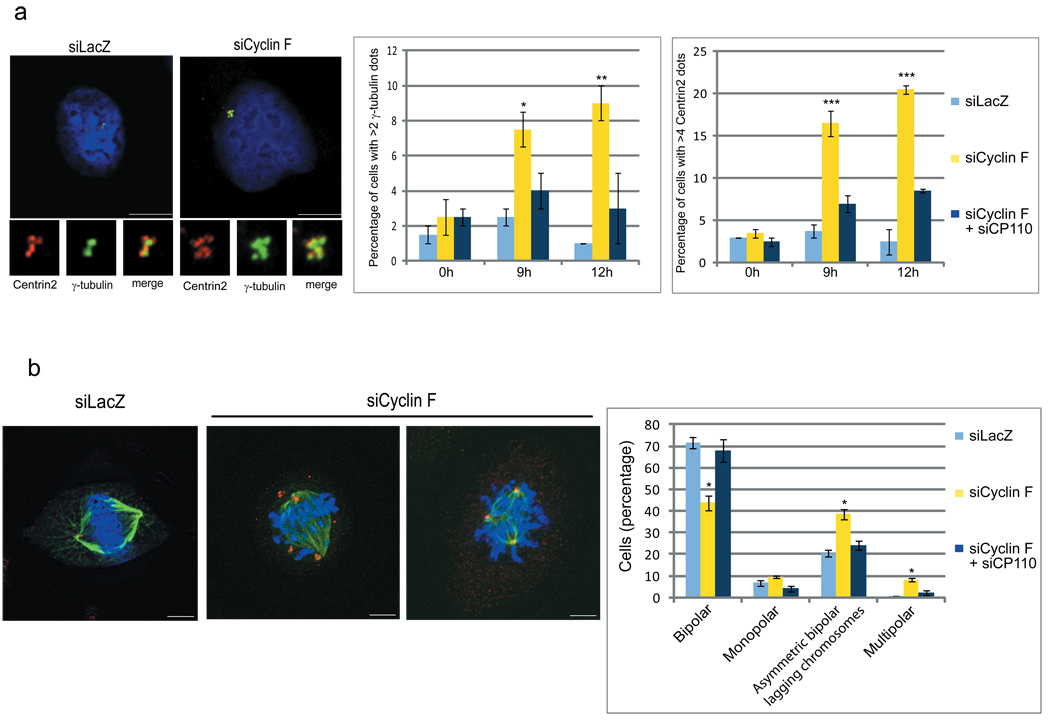
In a complementary approach, synchronized and siRNA-treated U-2OS cells were analyzed for centrosome defects. At both 9 and 12 hours after release from a G1/S block (when most cells were in G2 and early M, respectively), treatment with siRNAs targeting Cyclin F produced a significant increase in the percentage of cells showing more than two γ-tubulin foci and more than four foci for both Centrin 2 and CP110 (Fig. 3a and Supplementary Fig. 8). Significantly, expression of a wild type, but siRNA-insensitive, Cyclin F rescued the induction of excess CP110 dots. However, expression of a siRNA-resistant Cyclin F(M/A;L/A) failed to rescue the phenotype (Supplementary Fig. 8). Finally, in agreement with the siRNA results, we observed that Cyclin F−/− mouse embryonic fibroblasts (MEFs) displayed CP110 accumulation and increased number of γ-tubulin and Centrin 2 foci compared to Cyclin FFlox/− MEFs (Supplementary Fig. 9).
Figure 3. Cyclin F silencing induces centrosome and mitotic aberrations.
a, U-2OS cells were transfected with siRNAs to a LacZ, Cyclin F, or both Cyclin F and CP110, synchronized as in (1a), fixed at the indicated times after release from the block, and incubated with anti-Centrin 2 antibody (red) and anti-γ-tubulin antibody (green). DNA was stained with DAPI. Insets show magnified views of centrosomes. Scale bar = 10 µM. The graphs on the right show quantification of three experiments. Error bars indicate +/−SD. *= p=0.003; **= p=0.001; ***= p<0.001.
b, Experiments were performed as in (a), except that cells were collected 14 hours after release from the block and stained with anti-Centrin 2 antibody (red) and anti-α-tubulin antibody (green). Scale bar = 5 µM. The graphs on the right show the percentages of cells with various abnormal mitotic phenotypes. Error bars indicate +/−SD. *= p<0.001.
During mitosis, the assembly of a bipolar spindle is mediated by the centrosomes (from which microtubules nucleate) and ensures the proper segregation of the genetic material. Cells react to centrosome overduplication by centrosome clustering, to prevent the formation of multipolar spindles17–19. However, before centrosome clustering, cells with extra centrosomes pass through transient, multipolar intermediates that promote merotelic and syntelic attachments, with consequent lagging chromosomes20. Therefore, we analyzed mitotic figures using an antibody to α-tubulin (to stain mitotic spindles), an antibody to Centrin 2, and DAPI. Mitotic cells in which Cyclin F levels were reduced showed an increase in both multipolar spindles and asymmetric, bipolar spindles with lagging chromosomes (Fig. 3b and Supplementary Fig. 10). Significantly, when CP110 was silenced together with Cyclin F, the number of Centrin 2/γ-tubulin foci and mitotic aberrations reverted to levels approximating controls (Fig. 3a–b and Supplementary Figs. 10–11), strongly indicating that the G2 accumulation of CP110 in Cyclin F-depleted cells is responsible for the observed phenotypes.
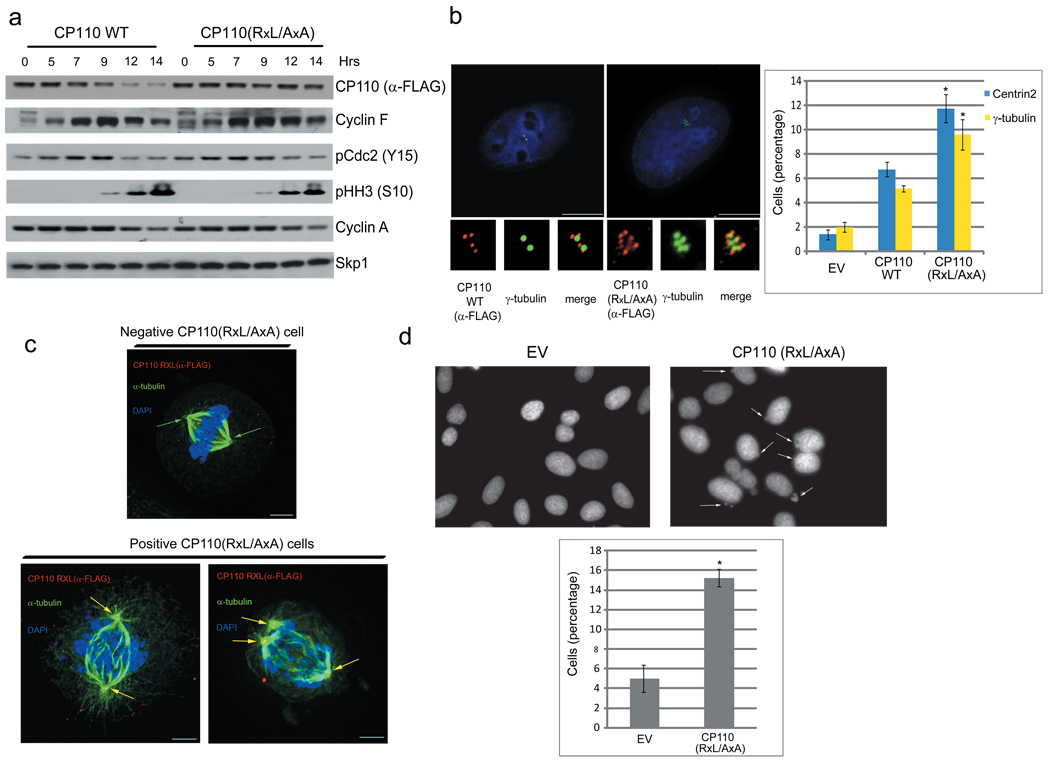
To further investigate the effects produced by the failure to degrade CP110 in G2, we analyzed synchronized U-2OS cells expressing either FLAG-tagged wild-type CP110 or FLAG-tagged CP110(RxL/AxA). Wild-type CP110 was degraded in G2 and M, while CP110(RxL/AxA), in agreement with its inability to bind Cyclin F (Supplementary Fig. 2), was stable (Fig. 4a). Importantly, expression of the stable CP110 mutant recapitulated the centrosome phenotypes observed upon Cyclin F silencing, namely an increased number of foci positive for Centrin 2, CP110, and γ–tubulin (Fig. 4b). Moreover, 100% of the mitotic cells that stained positive for CP110(RxL/AxA) showed abnormal spindles and lagging chromosomes (n= 29/29), whereas only 25% of mitotic cells that did not express CP110(RxL/AxA) showed mitotic aberrations (n=7/28) (Fig. 4c). Finally, expression of CP110(RxL/AxA) promoted the formation of micronuclei, a hallmark of chromosomal instability (Fig. 4d).
Figure 4. The failure to degrade CP110 causes centrosome and mitotic defects.
a, HeLa cells were transfected with FLAG-CP110 or FLAG-CP110(RxL/AxA), synchronized at G1/S as in (1a), collected at the indicated times, lysed, and immunoblotted as indicated.
b, U-2OS cells were transfected with FLAG-CP110 or FLAG-CP110(RxL/AxA), collected 48 hours after transfection, and stained with anti-FLAG antibody to visualize CP110 (red) and anti-γ-tubulin (green) antibody. Insets show magnified views of centrosomes. Scale bar = 10 µM. The graphs on the right show quantification of cells with excess Centrin 2 and γ-tubulin dots. Error bars indicate +/−SD. *= p<0.001 (n=3).
c, Experiments were performed as in (b), except that cells were transfected with only FLAG-CP110(RxL/AxA), collected 14 hours after release from the block, and stained with anti-α-tubulin antibody (green) and anti-FLAG antibody to visualize CP110-positive cells (red). The yellow in merged images shows colocalization of CP110 and α-tubulin (yellow arrows). Green arrows show spindles negative for CP110. CP110 negative and CP110 positive cells from the same coverslip are shown. Scale bar = 5 µM.
d, U-2OS cells were transfected with an empty vector (EV) or FLAG-CP110(RxL/AxA), fixed, and stained with DAPI to visualize the DNA. Micronuclei are highlighted by arrows. The graph on the bottom shows quantification of cells containing micronuclei. Error bars indicate +/−SD. *= p<0.0001.
Chromosome instability can result from centrosome overduplication18–20. Here, we demonstrate that a defect in the SCFCyclin F-mediated degradation of CP110 results in centrosome and chromosome aberrations. Thus, we propose that Cyclin F plays a role in limiting centrosome duplication to once per cell cycle to maintain chromosome stability.
It is known that the Cyclin E is necessary for centrosome duplication (via an unknown mechanism)21,22. The work presented here indicates that Cyclin F limits centrosome duplication by targeting CP110 for proteolysis. Thus, Cyclin E and Cyclin F have opposing roles, explaining why the former is expressed in S (when centrosome duplication occurs) and the latter in G2 (when centrosome duplication is restricted).
In summary, our study shows that Cyclin F forms an active SCF ubiquitin ligase complex (via its F-box motif), and it binds CP110 using a mode of substrate recognition identical to that used by the canonical cyclins involved in protein phosphorylation (i.e. via the hydrophobic patch present in the cyclin box domain and the RxL motif present in the substrate), thus unifying the functions of the two Cyclin F homology domains. Importantly, we show both temporal and spatial regulation of the SCFCyclin F-mediated proteolysis of CP110 and demonstrate a physiological role for this event in controlling genome integrity.
Methods Summary
Biochemical Methods
Extract preparation, immunoprecipitation, and immunoblotting have previously been described23,24
Plasmids
CP110 cDNA was from OpenBioSystems (clone MHS1010-7295919). Cyclin F cDNA was amplified by PCR using a cDNA library from HEK-293T cells. CP110 and Cyclin F mutants were generated either using the QuikChange Site-directed Mutagenesis kit (Stratagene) or by standard PCR methods. pCBF GFP-CP110 WT and CP110 truncation mutants were previously described10,13.
Immunofluorescence microscopy
For indirect immunofluorescence staining, cells were grown on glass coverslips and then fixed with methanol for 10 minutes. The cells were permeabilized with PBS/1% Triton X-100 for 10 minutes and blocked for one hour in PBS/0.1% Triton X-100 containing 3% BSA prior to incubation with primary antibodies. Alexa Fluor 568 conjugated goat anti-rabbit and Alexa Fluor 488-conjugated goat anti-mouse IgGs (Invitrogen) were used as secondary antibodies. DAPI was used to counterstain DNA. Slides were mounted with Prolong-Gold (Invitrogen). Image acquisition was performed using a Zeiss Axiovert 200 M microscope (63× objective lens, N.A. 1.4, 1.6× Optovar), equipped with a cooled Retiga 2000R CCD (QImaging). Mitotic images were deconvoluted using Metamorph (Molecular Devices). The images represent the maximum projection of 12 deconvoluted planes. Confocal microscopy (used in Fig. 2 and Supplementary Fig. 4) was performed using a Zeiss LSM 510, equipped with Zeiss LSM 510 software.
Supplementary Material
Acknowledgements
The authors thank S. Elledge for Cyclin FFlox/− and Cyclin F−/− MEFs and J.R. Skaar for reading the manuscript. MP is grateful to T.M. Thor for continuous support. This work was funded by fellowships from the American Italian Cancer Foundation to VD’A and VD, a grant from the March of Dimes (#1-FY08-372) to BD and grants from the National Institutes of Health (R01-GM057587, R37-CA076584, and R21-AG032560) to MP. AS, LF, and MPW are supported by the Stowers Institute for Medical Research. VD’A is a Leukemia & Lymphoma Society Scholar. MP is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Author Contributions. VD’A and VD performed and planned all experiments and helped to write the manuscript. MP coordinated the study, oversaw the results, and wrote the manuscript. SV and BD provided reagents, advice and assistance with the analysis of γ-tubulin and Centrin 2 foci. AS, LF, and MPW performed the mass spectrometry analysis of the Cyclin F complex purified by VD’A. All authors discussed the results and commented on the manuscript.
Author Information. The authors declare no competing financial interests.
References
- 1.Cardozo T, Pagano M. The SCF Ubiquitin Ligase: Insights into a Molecular Machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 2.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 3.Jin J, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skaar JR, D'Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009;137:1358–1358. doi: 10.1016/j.cell.2009.05.040. e1351. [DOI] [PubMed] [Google Scholar]
- 5.Bai C, et al. Skp1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 6.Fung TK, Siu WY, Yam CH, Lau A, Poon RY. Cyclin F is degraded during G2-M by mechanisms fundamentally different from other cyclins. J Biol Chem. 2002;277:35140–35149. doi: 10.1074/jbc.M205503200. [DOI] [PubMed] [Google Scholar]
- 7.Tetzlaff MT, et al. Cyclin F disruption compromises placental development and affects normal cell cycle execution. Mol Cell Biol. 2004;24:2487–2498. doi: 10.1128/MCB.24.6.2487-2498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 9.Bai C, Richman R, Elledge SJ. Human cyclin F. Embo J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 11.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Dobbelaere J, et al. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Kohlmaier G, et al. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci U S A. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balczon R, et al. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 18.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 19.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 20.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 22.Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cells. 2009;27:135–142. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guardavaccaro D, et al. SCFβTrcp-mediated degradation of REST supports chromosomal stability by inducing the mitotic checkpoint. Nature. 2008;452:365–369. [Google Scholar]
- 24.Bassermann F, et al. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.